Alpika Tripathi1* , Geetika Srivastava2
, Geetika Srivastava2 , K.K. Singh3
, K.K. Singh3 and P.K. Maurya4
and P.K. Maurya4
1Department of Computer Science and Engineering, ASET, Amity University, Lucknow - 226010, India.
2Department of Physics and Electronics, Dr. RML Avadh University, Faizabad - 224001, India.
3Department of E and CE, ASET, Amity University, Lucknow - 226010, India.
4Department of Neurology, RML Institute of Medical Sciences, Lucknow - 226010, India.
Corresponding Author E-mail: alpika2k@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1674
Abstract
The objective of this paper is to make a distinction between EEG data of normal and epileptic subjects. Methods: The dataset is taken from 20-30 years healthy male/female subjects from EEG lab of Dept. of Neurology, Dr. RML Institute of Medical Sciences, Lucknow (India). The feature extraction has been done using the Hilbert Huang Transform (HHT) method. The experimental EEG signals have been decomposed till 5th level of Intrinsic Mode Function (IMF) followed by calculation of high order statistical values of each IMF. Relief algorithm (RBAs) is used for feature selection and classification is performed using Linear Support Vector Machine (Linear SVM). This paper gives an independent approach of classifying Epileptic EEG data with reduced computational cost and high accuracy. Our classification result shows sensitivity, specificity, selectivity and accuracy of 96.4%, 79.16%, 84.3% and 88.5% respectively. The proposed method has been analyzed to be very effective in accurate classification of epileptic EEG data with high sensitivity.
Keywords
Epilepsy; EEG; Hilbert Huang Transform (HHT); Relief-Based Feature Selection Algorithms (Rbas); Linear Support Vector Machine (SVM)
Download this article as:| Copy the following to cite this article: Tripathi A, Srivastava G, Singh K. K, Maurya P. K. Epileptic Seizure Data Classification Using RBAs and Linear SVM. Biomed Pharmacol J 2019;12(2). |
| Copy the following to cite this URL: Tripathi A, Srivastava G, Singh K. K, Maurya P. K. Epileptic Seizure Data Classification Using RBAs and Linear SVM. Biomed Pharmacol J 2019;12(2). Available from: https://bit.ly/2IMPKZs |
Introduction
PILEPSY is a physical condition that occurs in the brain and affects the nervous system. According to the 2009 report by the World Health Organization around 70 million people worldwide have epilepsy.1-3 Around 90% of this population lives in developing countries, and about three fourths of them do not have access to the necessary treatment. Epilepsy is defined by two or more such unprovoked seizures.4 The seizures are commonly defined as abnormal electrical and chemical activities in the brain. Like many other neurological disorders, epilepsy can be assessed by the electroencephalogram (EEG). The EEG signal is highly non-linear and non-stationary in nature, and hence, it is difficult to characterize and interpret it using conventional frequency domain analysis.5-7
EEG is recording of the electrical activity of the brain from the scalp. The recorded waveforms reflect the cortical electrical activity and helps in identification of brain conditions. The most common method used for recording EEG is 10-20 system which is internationally recognized method that allows EEG electrode placement to be standardized.8
The system is based on the relationship between the location of an electrode and the underlying area of outer layer of the brain. The number ‘10’ & ‘20’ refer to the distance between adjacent electrode to be either 10% or 20% of the total front-back or right-left distance of the scalp . The electrode placement is shown in Figure 1.
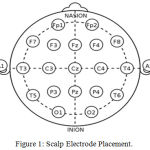 |
Figure 1: Scalp Electrode Placement.
|
In this paper authors have given a method for classification of epileptic and normal EEG data. The proposed method uses a combination of Hilbert Huang Transform (HHT) for features extraction, RBAs for feature selection and Linear SVM based classification of neural network modeling.
Hilbert-Huang Transform is a time frequency technique consisting of two parts, the Empirical Mode Decomposition (EMD), and the Hilbert Spectral Analysis (HSA).9,10 EMD decomposes an EEG signal into a finite set of band-limited signals termed intrinsic mode functions (IMFs), which are oscillatory components of input data. In the first step the mean frequency (MF) for each IMF has been computed using Fourier-Bessel expansion.11 The IMF oscillates in a narrow frequency band which is a reflection of quasi-periodicity and nonlinearity. The non-constant frequency means non-stationary. MF measure of the IMFs has been used as one of the features to differentiate between healthy and epileptic EEG signals. In the second part, the Hilbert transform is applied to the IMF, yielding a time-frequency representation (Hilbert spectrum) for each IMF.12,13
For feature selection authors have used Relief algorithm.14-17 Relief is an algorithm in which a filter-method approach is used for feature selection that is notably sensitive to feature interactions. It was originally designed for application to binary classification problems with discrete or numerical features. Relief was also described as generalizable to polynomial classification by decomposition into a number of binary problems.
Relief calculates a feature score for each feature which can then be applied to rank and select top scoring features for feature reduction. Alternatively, these scores may be applied as feature weights to guide downstream modeling. Relief feature scoring is based on the identification of feature value differences between nearest neighbor instance pairs. If a feature value difference is observed in a neighboring instance pair with the same class (a ‘hit’), the feature score decreases. Alternatively, if a feature value difference is observed in a neighboring instance pair with different class values (a ‘miss’), the feature score increases shown in Figure 2.
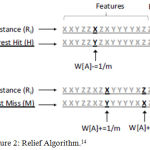 |
Figure 2: Relief Algorithm.14
|
The Support Vector Machine (SVM) is a popular classifier that can handle linear as well as non-linear class boundaries with the help of kernel functions.18 In this paper authors have used Linear SVM for data classification. The SVM tries to identify the maximum-margin hyper plane that separates the different classes. However, if the data cannot be linearly separated, non-linear kernel functions are used to transform the feature space, allowing a maximum-margin hyper plane to be established.19
HHT based feature extraction
The Hilbert–Huang transform (HHT) is an empirical data-analysis method. Its basis of expansion is adaptive, so it produces physically meaningful representations of data from nonlinear and non-stationary processes.20,21 Traditional data-analysis methods are all based on linear and stationary assumptions like Fourier transformation makes assumption of the signal period which creates spectral leakage. As is well known, the natural physical processes are mostly nonlinear and non-stationary like EEG signals from brain, yet the conventional data analysis methods provide very limited options for examining data from such processes. The available methods are either for linear but non-stationary, or nonlinear but stationary and statistically deterministic processes.
It is known that frequency of sinusoidal waveform is a well defined quantity. However, in practice, signals are not purely sinusoidal or stationary. Thus representing such non stationary signals as combination of different sinusoidal components will be a compromise with the accurate assessment of an event. For such signal the term frequency loses its effectiveness and a need for a parameter which accounts for the varying nature of the phenomena arises.
This gives rise to an idea of instantaneous frequency (IF) – which means the signal is either composed of a single frequency or a narrow band of frequencies. For each component instantaneous frequency can be defined.
HHT was motivated by the need to describe nonlinear distorted waves in detail, along with the variations of these signals that naturally occur in non-stationary processes.
The empirical mode decomposition method is necessary to deal with data from non-stationary and nonlinear processes.22,23 EMD decomposes signal X (𝑡) into a number of oscillatory component which is known as intrinsic mode function through a shifting process. Each IMF has its own distinct time scale. Furthermore EMD does not consider the stationary and the linearity of the signal.20 For an input signal X (𝑡) the process of calculating IMFs are given below:
Let’s set
X(𝑡) = X(𝑡)𝑜𝑙𝑑………………………… (1)
Find all the maxima and minima in
X(𝑡)𝑜𝑙d…………………………………(2)
Interpolate between minima and maxima using cubic spline interpolation which will generate local maxima envelope (𝑡) and local minima envelope
𝑒l(𝑡)…………..…………………………(3)
Calculate the mean of envelopes
𝑒𝑚𝑒𝑎𝑛 = ((𝑡)+𝑒𝑙(𝑡))/2………………….(4)
Now subtract 𝑒𝑚𝑒𝑎𝑛 from X(𝑡)𝑜𝑙𝑑 we will get X(𝑡)𝑛𝑒𝑤as,
X(𝑡)𝑛𝑒𝑤 = X(𝑡)𝑜𝑙𝑑 –𝑒𝑚𝑒𝑎𝑛……….,………(5)
Now set
X(𝑡)𝑜𝑙𝑑 = X(𝑡)𝑛𝑒𝑤 ……………..………..(6)
Repeat the process (2 to 4) until standard deviation
SD = (∑│X(t)new –X(t)old │2 / ∑ X(t)2old ) < α…..(7)
Where α is value that between 0.2-0.3
The first IMF is defined as IMF1 = X(𝑡)𝑛𝑒𝑤 which is the smallest temporal scale of X(𝑡). By subtracting IMF1 from X(𝑡) we will get residual signal R(𝑡) which can be expressed as ,
R(𝑡) = X(𝑡) – IMF1.
After acquiring R(𝑡) we put it in the same process above to get new IMF which means each IMF will have different frequencies against time. So the original signal can be rewritten as
![]()
Hilbert Huang Transform (HHT) is a very new and powerful tool for analyzing data from non-stationary and nonlinear processing realm and capable of filtering data based on empirical mode decomposition (EMD). The EMD is based on the sequential extraction of energy associated with various intrinsic time scales of the signal; therefore total sum of the intrinsic mode functions (IMFs) matches the signal very well and ensures completeness.
Studies show that we can discriminate between normal EEG data and abnormal EEG by statistically analyzing the IMF. Their statistical use is motivated by the fact that the distributions of samples in the data are characterized by their asymmetry, dispersion and concentration around the mean. After analyzing visually we can see IMF obtained from normal and pathological EEG are quite different from one another. These differences can easily extracted by statistical methods like Mean Function (MF), Standard Deviation (SD), Variance (VAR), Kurtosis (KUR), Skewness (SKW).24-26
Mean: Computes the average values of the signal at various frequency levels.
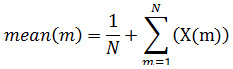
Standard Deviation: For calculating the variations of signal at various levels.
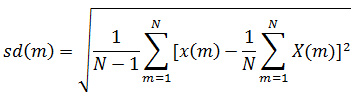
Variance: It is the square of the standard deviation.
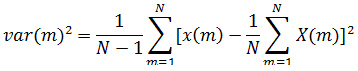
Kurtosis: Coefficients of EEG signal do not follow the normal distribution, and have a heavy tail characteristic is justified by the value of kurtosis parameters.
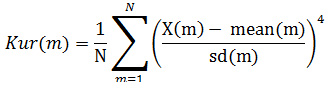
Skewness: It is a measure of the asymmetry. If the probability distribution of a real-valued random variable around its mean is not symmetrical, the data is said to be skewed.
The same processing steps are further applied on other 16 channels of EEG recording of Normal and Epileptic subjects.
Relief Based Feature Selection Algorithm
Take a data set with n instances of p features, belonging to two known classes. Within the data set, each feature should be scaled to the interval [0 1] (binary data should remain as 0 and 1). The algorithm will be repeated k times. Start with a p-long weight vector (G) of zeros.
At each iteration, take the feature vector (V) belonging to one random instance, and the feature vectors of the instance closest to V (by Euclidean distance) from each class. The closest same-class instance is called ‘near-hit’, and the closest different-class instance is called ‘near-miss’. Update the weight vector such that
Gi = Gi – (vi – nearHiti)2 + (vi – nearMissi)2
Thus the weight of any given feature decreases if it differs from that feature in nearby instances of the same class more than nearby instances of the other class, and increases in the reverse case.
After k iterations, divide each element of the weight vector by k. This becomes the relevance vector. Features are selected if their relevance is greater than a threshold T.
Kira and Rendell’s experiments showed a clear contrast between relevant and irrelevant features, allowing T to be determined by inspection.27 However, it can also be determined by Chebyshev’s inequality for a given confidence level (α) that a T of 1/sqrt(α*k) is good enough to make the probability of a Type I error less than α, although it is stated that T can be much smaller than that.
Classification using Linear SVM
Support Vector Machine (SVM) is a supervised machine learning algorithm which can be used for both classification and regression based problems. However, it is mostly used in classification problems. In this algorithm, we plot each data item as a point in n-dimensional space (where n is number of features you have) with the value of each feature being the value of a particular coordinate. Then, we perform classification by finding the hyper-plane that differentiate the two classes very well (look at the below snapshot) shown in Figure 3[A].
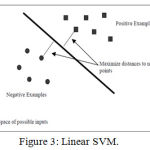 |
Figure 3: Linear SVM.
|
Support Vectors are simply the co-ordinates of individual observation. Support Vector Machine is a frontier which best segregates the two classes (hyper-plane/ line).24 In the linear case, the margin is defined by the distance of the hyper plane to the nearest of the positive and negative examples. The formula for the output of a linear SVM is
v = ѡ̄ * xˉ – b (1)
where w is the normal vector to the hyper plane and x is the input vector. The separating hyper plane is the plane v=0. The nearest points lie on the planes v = ±1. The margin m is thus
k = 1/║ѡ║2
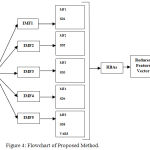 |
Figure 4: Flowchart of Proposed Method.
|
Result and Discussion
This paper gives the feature extraction results produced by applying decomposition of signal till fifth level of IMFs by applying HHT on EEG signals, and then RBA is used for feature selection, followed by Linear SVM for classification. The normal and abnormal input data is applied after removing artifacts.
Statistical values of 29 EEG signals of normal subjects upto 5th IMF has been shown in table 1 and values of 23 EEG signal of Epileptic subjects has been displayed in table 2. On the basis of these obtained values, the training and testing of Linear SVM classifier has been proposed. 70% of data set is used for training purpose and rest 30% were utilised for testing results.
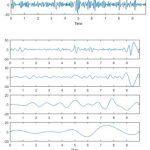 |
Figure 5: IMFs of Normal EEG Signal.
|
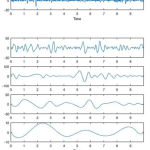 |
Figure 6: IMFs of Epileptic EEG Signal.
|
Once the calculation of IMFs is complete, the following important statistical parameters are evaluated. Table 1 & 2 are presenting the calculated statistical values of Healthy and Unhealthy subjects.
Table 1: High Order Statistical Values of 29 EEG signal of Normal Subjects upto 5th IMF.
| HOS Values of 29 EEG Signals | High Order Statistical Values | ||||
| IMF | MF | SD | VAR | KUR | SKW |
| Subjects | IMF 1 of Healthy Subjects | ||||
| NS1 | 0.794014 | 0.194748 | 0.0808656 | 0.0574944 | 0.0275939 |
| NS2 | 0.7134649 | 0.3835633 | 0.1539171 | 0.0777196 | 0.0382288 |
| NS3 | 0.7120425 | 0.392048 | 0.125765 | 0.0633654 | 0.0277442 |
| NS4 | 0.6561924 | 0.3911876 | 0.1600673 | 0.0619577 | 0.0186501 |
| NS5 | 0.7923439 | 0.39687 | 0.1419385 | 0.0608046 | 0.0248904 |
| NS6 | 0.69531 | 0.3801633 | 0.1490987 | 0.0560318 | 0.0249986 |
| NS7 | 0.6191468 | 0.3552116 | 0.1217889 | 0.0628417 | 0.0204335 |
| NS8 | 0.6190132 | 0.2576959 | 0.1470799 | 0.0867544 | 0.0133281 |
| NS9 | 0.7279635 | 0.3972242 | 0.1589645 | 0.0708214 | 0.0291047 |
| NS10 | 0.6938438 | 0.3249872 | 0.1626327 | 0.0611141 | 0.0319724 |
| NS11 | 0.6037114 | 0.2716271 | 0.1364642 | 0.0631772 | 0.0158963 |
| NS12 | 0.5959096 | 0.2387053 | 0.1123194 | 0.0494584 | 0.0204792 |
| NS13 | 0.7039527 | 0.3296545 | 0.1480881 | 0.0748475 | 0.0269281 |
| NS14 | 0.6844529 | 0.4312466 | 0.165582 | 0.0781406 | 0.0422987 |
| NS15 | 0.7843782 | 0.4633051 | 0.1519389 | 0.0567255 | 0.0207948 |
| NS16 | 0.7207146 | 0.356144 | 0.1542686 | 0.0732491 | 0.023715 |
| NS17 | 0.7776593 | 0.4194125 | 0.157747 | 0.0816223 | 0.0360981 |
| NS18 | 0.7018462 | 0.4333557 | 0.1727692 | 0.0736394 | 0.0344944 |
| NS19 | 0.7561062 | 0.3031047 | 0.1599127 | 0.0667163 | 0.0298385 |
| NS20 | 0.5615601 | 0.2440335 | 0.1393101 | 0.0625563 | 0.0232113 |
| NS21 | 1.0151271 | 0.5419102 | 0.2060411 | 0.1060046 | 0.0350871 |
| NS22 | 0.7082527 | 0.4058785 | 0.1250664 | 0.061215 | 0.0242943 |
| NS23 | 0.58942 | 0.2640501 | 0.1342691 | 0.0745797 | 0.0292519 |
| NS24 | 0.6863921 | 0.2591281 | 0.1569117 | 0.0679083 | 0.0330215 |
| NS25 | 0.6579692 | 0.3820743 | 0.1577376 | 0.0544039 | 0.0247068 |
| NS27 | 0.6185096 | 0.3248713 | 0.1347292 | 0.0557921 | 0.0208321 |
| NS28 | 0.7167416 | 0.2263721 | 0.1505363 | 0.0735477 | 0.0337342 |
| NS29 | 0.4230327 | 0.3507278 | 0.1417878 | 0.0668265 | 0.026326 |
| Subjects | IMF 2 of Healthy Subjects | ||||
| NS1 | 1.3671451 | 2.8716697 | 6.5527059 | 9.3498067 | 8.6381729 |
| NS2 | 1.1840138 | 1.3916471 | 2.8017808 | 2.4379485 | 2.2303501 |
| NS3 | 0.843453 | 0.9167091 | 2.4823977 | 3.5295424 | 3.3642967 |
| NS4 | 0.8421751 | 0.9637028 | 2.2007558 | 2.8008546 | 5.2072605 |
| NS5 | 0.5406368 | 0.9089775 | 2.0956593 | 4.9641066 | 2.7387957 |
| NS6 | 0.6736103 | 0.8451676 | 2.1182751 | 3.4507008 | 3.1589237 |
| NS7 | 0.9872215 | 1.0550772 | 2.5285452 | 2.9370106 | 5.9665401 |
| NS8 | 4.5190525 | 6.5279076 | 9.970391 | 5.4465119 | 20.088837 |
| NS9 | 0.5998085 | 0.8185307 | 1.6512623 | 2.3169359 | 3.1302619 |
| NS10 | 0.8223158 | 1.0699668 | 1.8031497 | 2.367291 | 2.5566929 |
| NS11 | 1.0965751 | 1.460845 | 2.553014 | 2.7946504 | 5.9919763 |
| NS12 | 1.1829003 | 1.7480142 | 3.5795746 | 4.8171016 | 4.7400878 |
| NS13 | 1.0971638 | 1.4864917 | 2.6012838 | 2.1556349 | 2.5493749 |
| NS14 | 0.4312608 | 0.5038683 | 0.9079241 | 1.3058499 | 1.4082351 |
| NS15 | 1.5704348 | 1.487021 | 2.6870926 | 7.916428 | 14.940614 |
| NS16 | 0.8077475 | 1.0170576 | 2.5210509 | 2.3879847 | 2.7999426 |
| NS17 | 0.4905736 | 0.7537514 | 1.8861905 | 1.9574766 | 1.9970076 |
| NS18 | 0.7166195 | 1.0073588 | 2.0143632 | 2.1873461 | 2.7866116 |
| NS19 | 0.680081 | 1.1124491 | 1.9422344 | 2.3306878 | 3.7269487 |
| NS20 | 3.0825562 | 4.1088006 | 5.6119314 | 4.6420457 | 3.914372 |
| NS21 | 3.0527935 | 3.1810749 | 2.9614952 | 2.9901266 | 2.0240628 |
| NS22 | 0.6201656 | 0.6909153 | 2.2992595 | 3.5640906 | 3.9328668 |
| NS23 | 2.1344174 | 2.6372075 | 4.8936201 | 3.6146932 | 3.2462856 |
| NS24 | 0.5554465 | 1.3915533 | 2.3443719 | 2.6064464 | 1.9867625 |
| NS25 | 1.0111484 | 1.1234713 | 2.0467199 | 3.0696111 | 2.481803 |
| NS27 | 1.1047307 | 1.2129053 | 2.3732381 | 3.3731465 | 5.2515313 |
| NS28 | 0.6727835 | 1.6808005 | 3.1633969 | 3.2929552 | 2.621261 |
| NS29 | 0.5791149 | 0.5605072 | 1.4387479 | 1.9286429 | 3.2353766 |
| Subjects | IMF 3 of Healthy Subjects | ||||
| NS1 | 1.8690857 | 8.246487 | 42.937955 | 87.418885 | 74.618031 |
| NS2 | 1.4018887 | 1.9366815 | 7.8499754 | 5.9435931 | 4.9744615 |
| NS3 | 0.7114129 | 0.8403556 | 6.1622985 | 12.45767 | 11.318492 |
| NS4 | 0.7092588 | 0.928723 | 4.8433261 | 7.8447862 | 27.115562 |
| NS5 | 0.2922881 | 0.8262401 | 4.3917878 | 24.642354 | 7.5010021 |
| NS6 | 0.4537508 | 0.7143082 | 4.4870895 | 11.907336 | 9.978799 |
| NS7 | 0.9746063 | 1.1131878 | 6.3935406 | 8.6260312 | 35.5996 |
| NS8 | 20.421835 | 42.613577 | 99.408696 | 29.664492 | 403.56139 |
| NS9 | 0.3597703 | 0.6699926 | 2.7266671 | 5.3681922 | 9.7985398 |
| NS10 | 0.6762032 | 1.144829 | 3.251349 | 5.6040665 | 6.5366786 |
| NS11 | 1.2024769 | 2.1340681 | 6.5178805 | 7.8100709 | 35.90378 |
| NS12 | 1.399253 | 3.0555538 | 12.813354 | 23.204467 | 22.468433 |
| NS13 | 1.2037684 | 2.2096577 | 6.7666774 | 4.6467616 | 6.4993125 |
| NS14 | 0.1859859 | 0.2538833 | 0.8243261 | 1.705244 | 1.9831261 |
| NS15 | 2.4662655 | 2.2112313 | 7.2204666 | 62.669832 | 223.22196 |
| NS16 | 0.652456 | 1.0344061 | 6.3556978 | 5.7024708 | 7.8396784 |
| NS17 | 0.2406624 | 0.5681411 | 3.5577147 | 3.8317147 | 3.9880392 |
| NS18 | 0.5135434 | 1.0147717 | 4.0576589 | 4.7844829 | 7.7652043 |
| NS19 | 0.4625101 | 1.237543 | 3.7722746 | 5.4321057 | 13.890147 |
| NS20 | 9.5021525 | 16.882242 | 31.493774 | 21.548588 | 15.322308 |
| NS21 | 9.3195484 | 10.119238 | 8.770454 | 8.940857 | 4.0968302 |
| NS22 | 0.3846054 | 0.477364 | 5.2865944 | 12.702742 | 15.467441 |
| NS23 | 4.5557378 | 6.9548636 | 23.947518 | 13.066007 | 10.53837 |
| NS24 | 0.3085208 | 1.9364205 | 5.4960795 | 6.7935628 | 3.9472251 |
| NS25 | 1.0224211 | 1.2621877 | 4.1890623 | 9.4225121 | 6.1593459 |
| NS27 | 1.22043 | 1.4711394 | 5.6322589 | 11.378117 | 27.578581 |
| NS28 | 0.4526377 | 2.8250902 | 10.00708 | 10.843554 | 6.871009 |
| NS29 | 0.3353741 | 0.3141683 | 2.0699954 | 3.7196636 | 10.467662 |
| Subjects | IMF 4 of Healthy Subjects | ||||
| NS1 | 26.1211 | 173.58881 | 105.7402 | 86.502577 | 14.866722 |
| NS2 | 4.5384237 | 5.8235914 | 3.3288413 | 3.9677612 | 3.7352665 |
| NS3 | 4.4268523 | 6.0777767 | 6.4834451 | 4.3057737 | 4.129212 |
| NS4 | 13.246323 | 6.5958737 | 3.7837047 | 3.3066044 | 4.5568696 |
| NS5 | 5.8715379 | 9.3080921 | 4.438701 | 2.8511137 | 3.3463041 |
| NS6 | 4.7410993 | 5.602009 | 3.5239203 | 3.5818778 | 3.7913422 |
| NS7 | 5.019083 | 4.3211797 | 7.0526305 | 3.8742989 | 6.3995483 |
| NS8 | 12.323221 | 26.213322 | 6.9748686 | 5.203992 | 14.349722 |
| NS9 | 5.2998872 | 8.1148395 | 4.0907434 | 3.5577341 | 2.7479037 |
| NS10 | 5.5523941 | 15.273256 | 3.2721178 | 4.860471 | 3.6195343 |
| NS11 | 5.1316229 | 7.6528964 | 3.4040442 | 3.4681068 | 11.761679 |
| NS12 | 24.611047 | 7.1365169 | 4.9725586 | 7.1197671 | 7.7796655 |
| NS13 | 4.9092302 | 7.0940397 | 3.7071417 | 4.1142901 | 4.2781562 |
| NS14 | 142.10583 | 27.677388 | 4.7715764 | 3.4914967 | 2.9643649 |
| NS15 | 8.9146693 | 7.9038536 | 10.159726 | 7.0833235 | 5.5552036 |
| NS16 | 7.5069103 | 10.522004 | 5.2477441 | 3.1655753 | 4.1332655 |
| NS17 | 6.2864369 | 3.7169318 | 3.4123277 | 3.6338549 | 3.3516207 |
| NS18 | 7.3879998 | 3.6279616 | 3.2416907 | 4.0381763 | 12.696512 |
| NS19 | 124.96265 | 7.2477353 | 3.3237369 | 3.8798556 | 5.3328726 |
| NS20 | 4.8953379 | 6.7082148 | 3.0854336 | 4.6323791 | 5.6055095 |
| NS21 | 19.483011 | 18.089693 | 4.9513198 | 4.9412625 | 2.9780993 |
| NS22 | 18.376056 | 6.0605848 | 5.1925368 | 7.9395411 | 9.4866847 |
| NS23 | 4.3802035 | 6.3862992 | 3.1073743 | 2.9512085 | 8.2121815 |
| NS24 | 5.8519008 | 7.5350298 | 3.1145678 | 3.5716336 | 4.0131338 |
| NS25 | 4.8353898 | 4.4480728 | 3.2057832 | 6.4106097 | 4.777637 |
| NS27 | 6.9280576 | 6.504447 | 5.4986645 | 7.9219861 | 13.547343 |
| NS28 | 81.381915 | 12.772316 | 3.7331439 | 3.2206681 | 3.218636 |
| NS29 | 859.94158 | 22.498134 | 4.7187115 | 4.8681671 | 4.070564 |
| Subjects | IMF 5 of Healthy Subjects | ||||
| NS1 | 0.9368617 | 2.2400095 | 1.8788759 | -4.6346204 | -0.4742972 |
| NS2 | -0.0641331 | 0.02556 | 0.0213764 | -0.1746376 | -0.1316439 |
| NS3 | -0.0329525 | -0.0372348 | -0.170347 | 0.03744 | -0.2469791 |
| NS4 | 0.0760672 | -0.0646503 | 0.078916 | -0.0567486 | 0.265975 |
| NS5 | 0.1335422 | -0.0651292 | -0.0394491 | -0.1116554 | -0.5056581 |
| NS6 | -0.0182776 | -0.1920655 | 0.0125293 | -0.0639116 | 0.2236741 |
| NS7 | 0.0518529 | 0.0462345 | 0.2674941 | 0.0297923 | -0.7641902 |
| NS8 | 0.0091883 | -0.2378462 | 0.0179487 | 0.0271938 | 1.0651003 |
| NS9 | -0.0182481 | -0.0174901 | 0.0300339 | 0.0872897 | -0.0599968 |
| NS10 | -0.0801845 | 0.6202513 | -0.0045994 | 0.2976808 | -0.0249067 |
| NS11 | -0.039921 | 0.3110126 | 0.086128 | -0.0891201 | -0.345381 |
| NS12 | 1.2827003 | -0.0110624 | -0.0726262 | 0.0017892 | -0.6870894 |
| NS13 | 0.0069424 | -0.0484375 | 0.0417263 | -0.1836737 | 0.3871868 |
| NS14 | -0.6105902 | 1.2346839 | 0.1682674 | 0.0953418 | -0.0021122 |
| NS15 | -0.1261543 | -0.4746706 | -0.3580666 | 0.2197198 | -0.0952142 |
| NS16 | 0.0352776 | 0.2623286 | 0.0164817 | 0.1063452 | -0.2706383 |
| NS17 | -0.0915483 | 0.0119703 | -0.0373728 | 0.0496959 | 0.2590188 |
| NS18 | 0.1835534 | 0.0202854 | -0.0084826 | 0.0108377 | 0.9936275 |
| NS19 | 4.2085615 | 0.0127075 | -0.059354 | -0.2144468 | -0.2042717 |
| NS20 | -0.0086416 | -0.0772461 | -0.0441348 | -0.0245628 | 0.4144225 |
| NS21 | 0.1078166 | 0.2286214 | 0.0188273 | 0.0214265 | 0.000411 |
| NS22 | -0.6105868 | -0.1642786 | 0.0458767 | 0.8285085 | -0.2906335 |
| NS23 | 0.0155542 | 0.1616218 | 0.0071227 | -0.0232878 | -1.1183538 |
| NS24 | -0.0677949 | -0.0092523 | 0.0438202 | -0.0172195 | -0.2904886 |
| NS25 | -0.0088769 | 0.0240307 | -0.0392672 | 0.3934843 | 0.5874376 |
| NS27 | -0.0855313 | -0.0039345 | -0.1527337 | 0.2219079 | -0.7106446 |
| NS28 | -0.3867619 | -0.1214849 | -0.0287683 | -0.0250265 | -0.1046807 |
| NS29 | 24.095343 | -0.4350748 | 0.0042242 | 0.0465327 | -0.1108155 |
Table 2: High Order Statistical Values of 23 EEG signal of Epileptic Subjects upto 5th IMF.
| HOS Values of 23 EEG Signals | High Order Statistical Values | ||||
| IMF | MF | SD | VAR | KUR | SKW |
| Subjects | IMF 1 of Unhealthy Subjects | ||||
| ES1 | 0.8650899 | 0.4729091 | 0.0656301 | 0.0500698 | 0.0282743 |
| ES2 | 0.7204698 | 0.301023 | 0.1401935 | 0.0650751 | 0.0277056 |
| ES3 | 0.8391117 | 0.5123799 | 0.1675866 | 0.0580672 | 0.017728 |
| ES4 | 0.6846268 | 0.2982419 | 0.1178403 | 0.0341233 | 0.019428 |
| ES5 | 0.8246545 | 0.5175436 | 0.1532076 | 0.0706868 | 0.037721 |
| ES6 | 0.7866724 | 0.5715599 | 0.1436699 | 0.0708974 | 0.0364508 |
| ES7 | 0.7180737 | 0.5269248 | 0.1003643 | 0.0498809 | 0.0373853 |
| ES8 | 0.6780945 | 0.2309114 | 0.1450357 | 0.0151319 | 0.0131524 |
| ES9 | 0.6633649 | 0.2670126 | 0.1298747 | 0.0509755 | 0.029085 |
| ES10 | 0.6764995 | 0.3393412 | 0.1618375 | 0.0640588 | 0.0253861 |
| ES11 | 0.8552998 | 0.4695551 | 0.1592763 | 0.0396773 | 0.0178348 |
| ES12 | 0.9608706 | 0.3980764 | 0.1618239 | 0.0526249 | 0.0202914 |
| ES13 | 0.6468634 | 0.2672435 | 0.1478045 | 0.0621515 | 0.0111755 |
| ES14 | 0.7191737 | 0.4133107 | 0.1478768 | 0.0643535 | 0.0182317 |
| ES15 | 0.5935737 | 0.261775 | 0.1387919 | 0.0708448 | 0.0263439 |
| ES16 | 0.7620866 | 0.2757785 | 0.0989844 | 0.0334673 | 0.0162968 |
| ES17 | 0.6647294 | 0.3585806 | 0.1565834 | 0.0678156 | 0.0250318 |
| ES18 | 0.7578168 | 0.4157828 | 0.1355068 | 0.0425817 | 0.0257703 |
| ES19 | 0.8166415 | 0.4948268 | 0.09664 | 0.0278963 | 0.0233678 |
| ES20 | 0.6549819 | 0.2668567 | 0.1455575 | 0.0675608 | 0.0255792 |
| ES21 | 0.6143616 | 0.4561768 | 0.1217524 | 0.0705827 | 0.0390815 |
| ES22 | 0.8869436 | 0.0897994 | 0.089129 | 0.0387991 | 0.0219594 |
| ES23 | 0.9041045 | 0.5533729 | 0.1935902 | 0.0959522 | 0.0479147 |
| Subjects | IMF 2 of Unhealthy Subjects | ||||
| ES1 | 0.3303539 | 0.4068024 | 5.0004128 | 6.9876397 | 4.450579 |
| ES2 | 0.7759541 | 1.1785239 | 2.2471837 | 2.6743979 | 2.9395671 |
| ES3 | 0.9123532 | 1.0098045 | 1.2550563 | 2.205777 | 10.381173 |
| ES4 | 0.4983477 | 0.9938214 | 3.0678322 | 10.58149 | 16.008316 |
| ES5 | 0.3640596 | 0.4457771 | 1.0317171 | 1.9604556 | 2.8770458 |
| ES6 | 0.4965187 | 0.7479746 | 1.5959717 | 2.2057836 | 2.4487858 |
| ES7 | 0.4709187 | 0.4566298 | 1.689656 | 4.7747918 | 3.8819404 |
| ES8 | 2.2989287 | 2.7078515 | 4.1221566 | 13.077719 | 19.783741 |
| ES9 | 0.5902094 | 1.0359951 | 1.9711336 | 4.1579259 | 5.7722332 |
| ES10 | 1.1322866 | 1.3598762 | 2.584256 | 3.8131407 | 4.8031704 |
| ES11 | 0.7589179 | 0.8662426 | 1.49653 | 4.2199104 | 6.6306221 |
| ES12 | 0.4402255 | 0.4632958 | 0.8848784 | 2.8468135 | 5.2325725 |
| ES13 | 2.1736135 | 2.5147764 | 5.2851702 | 3.4996659 | 13.125115 |
| ES14 | 0.5805407 | 0.6980601 | 1.8219706 | 2.5253038 | 11.036471 |
| ES15 | 2.2502876 | 2.9015059 | 4.0772807 | 3.3953816 | 2.7857397 |
| ES16 | 0.3805414 | 0.7496236 | 3.5402665 | 12.479548 | 12.62304 |
| ES17 | 1.0604954 | 1.1549985 | 2.0075533 | 2.6702119 | 5.2474406 |
| ES18 | 0.5168601 | 0.6714232 | 1.6591487 | 6.2580204 | 5.0768957 |
| ES19 | 0.5728281 | 0.5966072 | 1.6938629 | 6.5959014 | 7.3144355 |
| ES20 | 1.3439738 | 2.0153848 | 3.1818819 | 2.2977474 | 2.7072257 |
| ES21 | 0.3307135 | 0.4728326 | 1.7315057 | 2.1334664 | 2.3625637 |
| ES22 | 0.3404253 | 2.8266346 | 2.919212 | 6.7144357 | 7.7731332 |
| ES23 | 1.0936969 | 1.0679932 | 1.7261892 | 1.9150612 | 1.8657758 |
| Subjects | IMF 3 of Unhealthy Subjects | ||||
| ES1 | 0.1091337 | 0.1654882 | 25.004129 | 48.827109 | 19.807653 |
| ES2 | 0.6021047 | 1.3889186 | 5.0498348 | 7.1524041 | 8.6410547 |
| ES3 | 0.8323884 | 1.0197052 | 1.5751663 | 4.8654523 | 107.76876 |
| ES4 | 0.2483504 | 0.987681 | 9.4115946 | 111.96793 | 256.26619 |
| ES5 | 0.1325394 | 0.1987172 | 1.0644402 | 3.8433861 | 8.2773925 |
| ES6 | 0.2465308 | 0.5594661 | 2.5471256 | 4.8654812 | 5.9965518 |
| ES7 | 0.2217644 | 0.2085108 | 2.8549373 | 22.798637 | 15.069461 |
| ES8 | 5.2850734 | 7.3324599 | 16.992175 | 171.02673 | 391.39639 |
| ES9 | 0.3483471 | 1.0732858 | 3.8853676 | 17.288348 | 33.318676 |
| ES10 | 1.2820729 | 1.8492632 | 6.6783789 | 14.540042 | 23.070446 |
| ES11 | 0.5759564 | 0.7503762 | 2.239602 | 17.807644 | 43.965149 |
| ES12 | 0.1937985 | 0.214643 | 0.7830098 | 8.104347 | 27.379815 |
| ES13 | 4.7245957 | 6.3241006 | 27.933024 | 12.247662 | 172.26864 |
| ES14 | 0.3370275 | 0.4872879 | 3.3195768 | 6.3771594 | 121.8037 |
| ES15 | 5.0637941 | 8.4187365 | 16.624218 | 11.528616 | 7.7603459 |
| ES16 | 0.1448118 | 0.5619355 | 12.533487 | 155.73913 | 159.34114 |
| ES17 | 1.1246505 | 1.3340216 | 4.0302702 | 7.1300316 | 27.535632 |
| ES18 | 0.2671444 | 0.4508091 | 2.7527745 | 39.162819 | 25.77487 |
| ES19 | 0.328132 | 0.3559401 | 2.8691715 | 43.505915 | 53.500966 |
| ES20 | 1.8062657 | 4.0617758 | 10.124372 | 5.2796432 | 7.3290711 |
| ES21 | 0.1093714 | 0.2235707 | 2.998112 | 4.5516789 | 5.5817073 |
| ES22 | 0.1158894 | 7.9898633 | 8.5217987 | 45.083647 | 60.4216 |
| ES23 | 1.1961729 | 1.1406094 | 2.9797292 | 3.6674596 | 3.4811194 |
| Subjects | IMF 4 of Unhealthy Subjects | ||||
| ES1 | 4.5771176 | 7.0087635 | 18.72618 | 4.438525 | 3.6387834 |
| ES2 | 4.6721191 | 12.485297 | 3.7515448 | 2.8945809 | 3.2989253 |
| ES3 | 4.8363808 | 3.6414101 | 4.8090367 | 9.1757919 | 3.8358201 |
| ES4 | 25.708015 | 29.801328 | 6.3511609 | 12.355667 | 2.2715564 |
| ES5 | 8.4152284 | 3.0111605 | 5.1884963 | 11.195804 | 4.1664795 |
| ES6 | 8.058631 | 2.5071926 | 5.6127706 | 4.8551599 | 3.3116094 |
| ES7 | 294.40981 | 3.1362269 | 27.83137 | 5.3359899 | 2.957612 |
| ES8 | 4.9401601 | 11.098406 | 3.3255404 | 35.553052 | 18.566356 |
| ES9 | 14.952852 | 13.619801 | 5.7570903 | 8.0806325 | 12.965434 |
| ES10 | 4.5372324 | 6.8736788 | 3.6542007 | 6.0117279 | 12.628416 |
| ES11 | 3.7915257 | 3.8012139 | 5.0176857 | 10.534601 | 16.394638 |
| ES12 | 8.3659199 | 66.930757 | 9.4311181 | 14.648822 | 13.724967 |
| ES13 | 7.3759178 | 12.338818 | 2.9419575 | 21.578049 | 31.927595 |
| ES14 | 63.38271 | 4.2477574 | 3.713664 | 5.0928349 | 8.7693387 |
| ES15 | 6.158873 | 5.8291961 | 3.826798 | 2.9856206 | 4.9684164 |
| ES16 | 43.596893 | 24.808282 | 5.9428689 | 5.6386731 | 7.2970514 |
| ES17 | 5.4453413 | 6.0432796 | 3.93111 | 6.0310116 | 7.4071691 |
| ES18 | 17.473343 | 7.0504811 | 4.7559268 | 4.5921542 | 3.0711551 |
| ES19 | 223.20771 | 6.1305469 | 30.525167 | 43.536547 | 22.863897 |
| ES20 | 3.83648 | 6.6480955 | 3.4461044 | 3.1125352 | 3.3795001 |
| ES21 | 395.02359 | 7.7392785 | 15.905384 | 5.0630354 | 3.1100875 |
| ES22 | 6.965067 | 100.63262 | 22.440095 | 16.072997 | 8.0500811 |
| ES23 | 4.7254519 | 4.026748 | 6.3249347 | 10.183007 | 15.691209 |
| Subjects | IMF 5 of Unhealthy Subjects | ||||
| ES1 | 0.024654 | -0.4296459 | 0.3575977 | 0.0226011 | -0.0755703 |
| ES2 | 0.0061941 | 0.5499125 | -0.1601651 | 0.0755629 | 0.1800178 |
| ES3 | 0.0010872 | -0.0197224 | -0.0789725 | -0.140138 | 0.1755386 |
| ES4 | -0.7393064 | -0.4796606 | -0.0979807 | -0.8759199 | 0.090423 |
| ES5 | 0.2707581 | -0.001205 | -0.0570468 | 0.2302964 | 0.2013605 |
| ES6 | -0.0974631 | 0.0032945 | 0.0368402 | -0.1686396 | 0.1249878 |
| ES7 | 9.9175498 | 0.0201692 | -2.417516 | -0.1451854 | 0.0458859 |
| ES8 | 0.0410811 | -0.3725798 | 0.0468306 | -3.5959749 | 1.6056099 |
| ES9 | -0.7679569 | -0.2677045 | -0.2248263 | 0.1450071 | 0.2373168 |
| ES10 | 0.011263 | 0.2940156 | 0.0009562 | 0.3756074 | 0.2475115 |
| ES11 | -0.0123112 | 0.0664354 | -0.103722 | 1.323574 | 2.3883766 |
| ES12 | 0.3587948 | -1.3089528 | -0.3568748 | 0.9671277 | -1.8264115 |
| ES13 | 0.352495 | 0.236305 | -0.00324 | -2.3182493 | -4.0947717 |
| ES14 | -2.6589605 | 0.0485656 | 0.0581806 | -0.0709458 | -0.0135256 |
| ES15 | 0.1221658 | 0.0747326 | 0.0272607 | 0.0547411 | -0.2824818 |
| ES16 | -2.2468663 | 0.0203399 | 0.0164117 | 0.0324535 | -0.9939975 |
| ES17 | 0.076271 | -0.1215204 | 0.1040614 | -0.059463 | -0.3068512 |
| ES18 | 0.9846622 | -0.0624885 | 0.0260383 | 0.1778707 | -0.0116882 |
| ES19 | 6.9752414 | -0.1316119 | -0.0018436 | -3.9574945 | -1.6565017 |
| ES20 | -0.0276649 | -0.1214705 | -0.0007395 | 0.0384048 | -0.0818361 |
| ES21 | 14.24066 | 0.1333398 | -0.3670489 | 0.0921567 | -0.0049415 |
| ES22 | 0.2247009 | -3.5664924 | -0.315258 | 0.7120273 | -0.5549812 |
| ES23 | -0.0670528 | -0.0564401 | -0.0320062 | -0.5541185 | -1.4992487 |
For feature selection Relief algorithm calculates a feature score for each feature which can then be applied to rank and select top scoring features. The Hilbert–Huang transform (HHT) is a way to decompose a signal into intrinsic mode functions (IMF) along with a trend, and obtain instantaneous frequency data and used for feature extraction. For classifying the experimental EEG signals, the Linear SVM concept has been used. The computed statistical values are fed into Linear SVM classifier for training and testing. The performance of proposed methodology is evaluated in terms of parameters of confusion matrix & ROC Curve shown in Figure 7 & 8.

Where TP, TN, FP and FN stands for true positive, true negative, false positive and false negative respectively. Sensitivity is used to diagnose the correctly identified positive case, specificity is defined as the determination of negative cases accurately, selectivity is defined as the recognition of unidentified positive results and accuracy stands for the identification of correct classified instances.
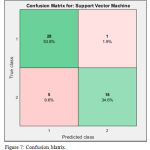 |
Figure 7: Confusion Matrix.
|
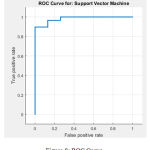 |
Figure 8: ROC Curve.
|
Table 3: Shows the proposed classifier output and compare with existing techniques
| Authors | Sensitivity | Selectivity | Accuracy |
| Syed Muhammad Usman et al., (2017) | 92.3% | – | – |
| Yildiz, Bergil & C.Oral (2017) | 88.06% | – | – |
| Bandarabadi, Mojtaba et al., (2015) | 75.8% | – | – |
| Our Findings | 96.5% | 84.8% | 88.5% |
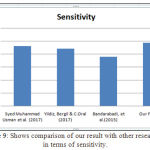 |
Figure 9: Shows comparison of our result with other researchers in terms of sensitivity.
|
Conclusion
The proposed method is suitable for separating normal and epileptic EEG data. The combined approach of HHT, RBAs with Linear SVM has found to be very effective in such classification with high sensitivity. The result of the classification process is based on using the statistical values obtained by HHT. This technique has been found to be suitable in the correct classification of epileptic and healthy EEG data. The data set is taken from Natus NeuroWorks EEG Recording Machine from RML Institute of Medical Sciences, Lucknow (U.P.), India. Our classification result shows sensitivity, selectivity and accuracy are 96.5%, 84.8% and 88.5% respectively.
Acknowledgements
The authors acknowledge Dr. A. K. Thacker, Head of Department, Department of Neurology, RMLIMS, Lucknow (U.P) for his valuable help in developing an understanding towards the Epilepsy. His contributions and work in the related field helped us to think a stage ahead and Ms. Nidhi, Technical Staff EEG Lab, RMLIMS, Lucknow (U.P) for providing EEG Recording of Healthy& Unhealthy Subjects.
Conflict of Interest
There is no conflict of interest.
References
- Fisher, W. van Emde Boas, W. Blume, C. Elger, P. Genton, P. Lee, J. Engel, “Epileptic seizures and epilepsy: definitions”, Proposed by the International League Against Epilepsy (ILAE) and the International Bureau for Epilepsy (IBE), Epilepsia, vol. 46 no. 4, pp. 470–472, 2005.
- A.S. Ruiz, R. Ranta, V. Louis-Dorr, “EEG montage analysis in the Blind Source Separation framework”, Bio signal Processing and Control vol. 6 no. 1, pp.77–84, 2010.
- http://www.who.int/mediacentre/factsheets/fs999/en/index.html (accessed April 2018), WHO Report
- Coyle, T.M. McGinnity, G. Prasad, “Improving the separability of multiple EEG features for a BCI by neural-time-series prediction-preprocessing”, Bio-signal Processing and Control vol.5 no.3, pp.196–204, 2010.
- Pardey, James, Stephen Roberts, and Lionel Tarassenko. “A review of parametric modelling techniques for EEG analysis.” Medical engineering & physics 18.1 (1996): 2-11.
- Lange, Nicholas, and Scott L. Zeger. “Non‐linear Fourier time series analysis for human brain mapping by functional magnetic resonance imaging.” Journal of the Royal Statistical Society: Series C (Applied Statistics) 46.1 (1997): 1-29.
- Bachmann, Maie, et al. “Non-linear analysis of the electroencephalogram for detecting effects of low-level electromagnetic fields.” Medical and Biological Engineering and Computing 43.1 (2005): 142-149.
- Homan, Richard W., John Herman, and Phillip Purdy. “Cerebral location of international 10–20 system electrode placement.” Electroencephalography and clinical neurophysiology 66.4 (1987): 376-382
- Huang, Norden Eh. Hilbert-Huang transform and its applications. Vol. 16. World Scientific, 2014.
- Huang, Norden E., and Zhaohua Wu. “A review on Hilbert‐Huang transform: Method and its applications to geophysical studies.” Reviews of geophysics 46.2 (2008).
- Pachori, Ram Bilas. “Discrimination between ictal and seizure-free EEG signals using empirical mode decomposition.” Research Letters in Signal Processing 2008 (2008): 14
- Huang, Z. Shen, and S. R. Long, “The empirical mode decompo-sition and the Hilbert spectrum for nonlinear and non-stationary time series analysis”, in Proc. Royal Society of London Ser., vol. 454, pp. 903–995, 1998.
- Li, Yi, et al. “Sleep stage classification based on EEG Hilbert-Huang transform.” Industrial Electronics and Applications, 2009. ICIEA 2009. 4th IEEE Conference on. IEEE, 2009.
- Urbanowicz, Ryan J., et al., “Relief-based feature selection: introduction and review”, arXiv preprint arXiv:1711.08421, 2017.
- Yu, Lei, and Huan Liu. “Feature selection for high-dimensional data: A fast correlation-based filter solution.” Proceedings of the 20th international conference on machine learning (ICML-03). 2003.
- Sun, Yijun., “Iterative RELIEF for feature weighting: algorithms, theories, and applications”, IEEE transactions on pattern analysis and machine intelligence 29.6 , 2007.
- Sun, Yijun, Sinisa Todorovic, and Steve Goodison. “Local-learning-based feature selection for high-dimensional data analysis”, IEEE transactions on pattern analysis and machine intelligence 32.9, pp. 1610-1626, 2010.
- Subasi, Abdulhamit, and M. Ismail Gursoy. “EEG signal classification using PCA, ICA, LDA and support vector machines.” Expert systems with applications 37.12 (2010): 8659-8666.
- Amari, Shun-ichi, and Si Wu. “Improving support vector machine classifiers by modifying kernel functions.” Neural Networks 12.6 (1999): 783-789.
- C. Pei and M. H. Yeh, “Discrete fractional Hilbert transform”, IEEE Trans. Circuits Syst. II, vol. 47, no. 11, pp. 1307–1311, Nov. 2000.
- Barnhart, B. L., “The Hilbert-Huang transform: theory, applications, development”, 2011.
- Flandrin, P., Rilling, G., & Goncalves, P, “Empirical mode decomposition as a filter bank”, Signal Processing Letters, IEEE, 11(2), 112-114, 2004.
- Huang, N. E., “Introduction to the Hilbert-Huang Transform and its related mathematical problems”, Hilbert-Huang transform and its applications, interdisciplinary mathematical sciences, 5, 1-24, 2005.
- Fu, Kai, et al. “Classification of seizure based on the time-frequency image of EEG signals using HHT and SVM.” Biomedical Signal Processing and Control 13 (2014): 15-22
- Jospin, Mathieu, et al. “Detrended fluctuation analysis of EEG as a measure of depth of anesthesia.” IEEE Transactions on Biomedical Engineering 54.5 (2007): 840-846.
- Persson, Isac, “Feature selection of EEG-signal data for cognitive load”, 2017.
- Kira, Kenji and Rendell, Larry , “A Practical Approach to Feature Selection”, Proceedings of the Ninth International Workshop on Machine Learning, p249-256, 1992








