Yaagyanika Gehlot1, Bhairvi Sharma1, P. Muthu1, Hariharan Muthusamy1 and S. Latha2
1Department of Biomedical Engineering SRM Institute of Science and Technology, Kattankulathur, Chennai, Tamil Nadu, India.
2Department of Electronics and Communication Engineering SRM Institute of Science and Technology, Kattankulathur, Chennai, Tamil Nadu, India.
Corresponding Author E-mail: muthu.p@ktr.srmuniv.ac.in
DOI : https://dx.doi.org/10.13005/bpj/1526
Abstract
Silver nitrous aqueous solution is used to biosynthesize Silver nanoparticles (Ag-NPs) through a green and easy way using tuber powder extracts of Curcuma Longa (C. longa). The aim is to model an Artificial Neural Network (ANN) using seven existing algorithms in MATLAB for forecasting the size of the silver nanoparticle with volume of both C. longa extraction and AgNO3, time of stirring and temperature of reaction as input functions. Several techniques including Quasi-Newton, Conjugate Gradient and Levenberg-Maquardt are employed for training the designed ANN model, a feed-forward backpropagation network with different combinations of architecture and transfer functions. Each algorithm is fashioned to obtain the best performance by calculating the Regression (R), Mean Square Error (MSE), Mean Absolute Error (MAE) and Error Sum of Squares (SSE), thereby comparing the results and propounding the optimum algorithm technique for the discussed application in nanoengineering. Finally, based on the findings, the optimum network is proposed through the simulation results.
Keywords
Artificial Neural Network; Feed-forward Back Propagationl Learning Algorithms; Nanoengineering, Silver Nanoparticles
Download this article as:| Copy the following to cite this article: Gehlot Y, Sharma B, Muthu P, Muthusamy H, Latha S. Biomedical Prediction of Radial Size of Powdered Element using Artificial Neural Network. Biomed Pharmacol J 2018;11(3). |
| Copy the following to cite this URL: Gehlot Y, Sharma B, Muthu P, Muthusamy H, Latha S. Biomedical Prediction of Radial Size of Powdered Element using Artificial Neural Network. Biomed Pharmacol J 2018;11(3). Available from: http://biomedpharmajournal.org/?p=22862 |
Introduction
The Nanoparticles (NPs) are a broad class of materials including particulate substances having a dimension less than 100 nm at least.1 Noble metals like Silver in metal Nanoparticles mainly have been used for experimental purposes because of their robust properties in optics. This creates large amount of applications in areas such as photography, dentistry, electronics, food industries, clothing etc.2 The shape and size of metal nanoparticle is measured typically using discrete techniques such as Scanning Electron Microscopy, Transmission Electron Microscopy etc.3
In the last decade an increasing use of artificial intelligence tools was observed in nanotechnology research. Artificial Intelligence can be used in classification of material properties of nanoscale, designing, simulation, nanocomputing etc. 4 Artificial neural network (ANN) is an efficient as well as dynamic simulation tool which allows one to classify, predict or estimate relationships among inputs and outputs.2 They can expertly solve difficult problems such as stock exchange prediction, image compression, face recognition etc. These tasks may be carried out without any prior information.
Artificial Neural Network
An ANN is a computational technique that uses the assistance of a learning paradigm along with processing nodes and attempts to present an affiliation between the input and output data.5 Majorly, there are two learning paradigms that an ANN can employ; supervised learning and unsupervised learning.6
A basic ANN network comprises of three primitive layers; input layer, hidden layer and output layer as illustrated in Fig. 1. These layers contain various mathematical functions, nodes which are also called artificial neurons, associated with weights or coefficients that builds the structure of the neural network.7 When an input and the corresponding target is provided to the ANN model (the training in this case is supervised), the error is calculated from the difference between the system output and the target response. This information of the error is fed back (Back propagation or BP learning) during the training phase and consequently the weights are adjusted accordingly, thereby improving the system parameters. Reiteration is done until the desired performance is achieved.8
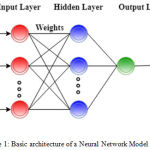 |
Figure 1: Basic architecture of a Neural Network Model.
|
Several types of ANNs have been designed with different configurations either with a single-layer or multiple layer neurons. A multilayer perceptron (MLP) is the best model for complex problems. By introducing more number of hidden layers a MLP outlives the drawback of the single-layer perceptron. In a conventional feed-forward MLP network, the input responses are multiplied with the weights and these multiplied signals from each input are then summed and guided to a transfer function which gives the output result for that particular neuron.7
Learning Algorithms
There are several types of training algorithms that can be adopted to train an ANN. MATLAB provides 9 different types of algorithms for an Engine Data Set problem, out of which top 7 algorithms are explored in this study.
Conjugate Gradient
Conjugate Gradient (CG) starts by searching the negative of the descent in their first iteration. Before the next search is determined, a line search is implemented for acquiring the prime distance to travel forth the existing search direction, so that the two search directions are conjugate. The novel search direction is determined when the new steepest descent direction and the preceding search direction are combined.9 Several versions of CG are determined by computational means of the constant, βk.6
Powell/Beale Restarts (traincgb)
Powell/Beale (CGB) technique will restart if the current and previous gradient have very little orthogonality between them.9
Fletcher-Reeves Update (traincgf)
For the Fletcher-Reeves Update (CGF), the constant is calculated as the ratio of the norm squared of the present gradient to the norm squared of the previous gradient.7
Polak-Ribiére Update (traincgp)
Another practice of the CG algorithm is Polak-Ribiére Update (CGP). In CGP, the constant is calculated by the inner product of the previous gradient change with the current gradient divided by the norm squared of the previous gradient. CGP requires more storage than CGF.6
Scaled Conjugate Gradient (trainscg)
Scaled Conjugate Gradient (SCG) does not call for a line search at each and every iteration and employs the step size scaling mechanism which reduces the time consumption, making SCG the fastest among the second order algorithm. Although the number of iteration may increase for the algorithm to converge.9
Quasi-Newton
Newton’s technique provides improved optimization and converges faster than CG techniques but the Hessian matrix of the performance index at the present values of the biases as well as weights, which is the elementary step to the Newton’s method, takes more time hence making the method complex for feed forward ANN. Based on this a class of algorithms, quasi-Newton or secant method, does not require the computation of second derivatives. In each iteration of the algorithm the approximate Hessian Matrix is updated.6
Broyden–Fletcher–Goldfarb–Shanno (trainbfg)
In Broyden–Fletcher–Goldfarb–Shanno (BFGS), the approximate Hessian matrix is stored with an n x n dimension, where n represents the number of weights and biases in the ANN model. Although it converges in fewer iterations, it has more calculations and storage requirements than CG methods.7,9
One Step Secant Algorithm (trainoss)
The One Step Secant (OSS) technique adopts that at every iteration, the preceding Hessian matrix is the identity matrix thereby not storing the complete Hessian giving it an additional benefit of calculating the new search direction without calculating the matrix inverse.6
Levenberg-Marquardt (trainlm)
The Levenberg-Marquardt (LM) training algorithm is a numerical least-squares non-linear function minimization technique.10 LM method computes a Jacobian matrix that contains first derivatives of the network error with respect to the weights as well as biases. The calculation of Jacobian matrix by standard BP technique is less complicated than the Hessian matrix.6
LM algorithm first initializes the weights of the network following the computation of the outputs and errors for all the input responses. Subsequently, the Jacobian matrix is calculated and the new weights are obtained. A new error value is determined from these weights and a comparison between the new and the current error value is carried out. Accordingly, the regularization parameter, µ is reduced by a factor of β if error is smaller otherwise it is increased by β. It is re-iterated until the error is below the predefined value or a stopping condition is met.10
Other types of algorithms used are Variable Learning Rate (traingda, traingdx) and Resilient Backpropagation (trainrp).6
Network Design
Data Set
In this study, the sample data employed to train the ANN model is presented in [2, Table I]. The database is split into; training set, validating set and testing set. A training set is adopted for learning to fit the parameters and is specifically applied to alter the varying weights and errors of the network in each iteration.2,11 Validation set tunes the parameters. It is used to vary and enhance the structure of ANN like training function, transfer function, number of hidden layers and neurons etc.2,11 A test set is used only to assess the effectiveness and efficiency of the ANN.2 Table [2, I] presents the four parameters produced as a function to predict the size of the Ag-NPs along with the actual size of the nanoparticle obtained.
Table 1: Experimental Values For Prediction Of The Size Of Ag-Nps.
| S.No. | Volume. C. Longa Extract (mL) | Temperature (°C) | Stirring Time (h) | Volume of AgNO3(mL) | Ag-NPs Particle Size (Actual)(nm) |
| Training Set | |||||
| 1. | 20 | 40 | 48 | 5 | 5.52 |
| 2. | 20 | 50 | 48 | 10 | 6.08 |
| 3. | 20 | 70 | 24 | 20 | 7.35 |
| 4. | 10 | 25 | 24 | 1 | 8.18 |
| 5. | 10 | 30 | 24 | 2 | 8.41 |
| 6. | 10 | 60 | 12 | 15 | 9.35 |
| 7. | 10 | 70 | 12 | 20 | 9.78 |
| 8. | 5 | 25 | 24 | 1 | 10.46 |
| 9. | 5 | 40 | 12 | 5 | 10.86 |
| 10. | 5 | 70 | 6 | 20 | 11.82 |
| 11. | 2 | 25 | 6 | 1 | 12.37 |
| 12. | 2 | 30 | 6 | 2 | 12.49 |
| 13. | 2 | 40 | 3 | 5 | 12.73 |
| 1. | 2 | 50 | 3 | 10 | 12.96 |
| 15. | 2 | 70 | 3 | 20 | 13.78 |
| 16. | 1 | 25 | 3 | 1 | 14.36 |
| 17. | 1 | 30 | 1 | 2 | 14.55 |
| 18. | 1 | 40 | 1 | 5 | 14.65 |
| 19. | 1 | 50 | 1 | 10 | 14.85 |
| 20. | 1 | 70 | 1 | 20 | 15.32 |
| Validating Set | |||||
| 21. | 20 | 30 | 48 | 2 | 5.18 |
| 22. | 10 | 50 | 24 | 10 | 9.11 |
| 23. | 5 | 60 | 6 | 15 | 11.69 |
| 24. | 2 | 60 | 3 | 15 | 13.47 |
| 25. | 1 | 60 | 1 | 15 | 14.93 |
| Testing Set | |||||
| 26. | 20 | 25 | 24 | 1 | 4.90 |
| 27. | 20 | 60 | 48 | 15 | 6.67 |
| 28. | 10 | 40 | 24 | 5 | 8.85 |
| 29. | 5 | 30 | 12 | 2 | 10.74 |
| 30. | 5 | 50 | 6 | 10 | 11.23 |
Methodology
An appropriate ANN model requires a learning algorithm, transfer function, suitable number of hidden layers and neurons. The framework to build and elect the appropriate ANN model for the chosen application is shown in Fig. 2. The most common learning in ANN is the BP technique which uses a supervised learning. A supervised learning paradigm compares the output response to the target response to calculate the learning error. This learning error is used to adjust the network parameters to enhance the performance of the network.5 In this paper, the designed network has four input parameters and one output parameter. Thus, the ANN is constructed with 4 neurons in the input layer and the output layer with 1 neuron. The number of neurons in the hidden layer and the transfer function is tested against to find the best suitable architecture for the application. The final evaluation of each network operation is done using Mean Square Error (MSE), Mean Absolute Error (MAE), Error Sum of Squares (SSE) and Regression (R).
The values of these indices can be calculated using the following equations,
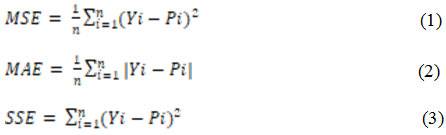
Where, n is the number of points, Yi is the value predicted from the ANN model and Pi is the actual value.2 R, the determination coefficient of linear regression, is a line between the predicted values from the ANN model and the target output. It fits better to the actual data when the R value tends to 1.12
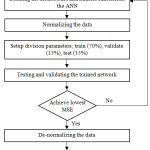 |
Figure 2: Flowchart of the methodology used.
|
Results and Discussions
All 7 algorithms used are coded in MATLAB with R2012b (8.0.0.783) version. The study is carried out by choosing one input, hidden and an output layer. The architecture of the ANN model is changed by altering the number of neurons in the hidden layer (10, 20, and 30) along with the transfer functions (purelin, logsig and tansig) in both hidden and output layer. Table II presents the values obtained by various architectures and transfer function arrangements of each algorithm. Normalization of all the input data in accordance with the transfer function is the first step of the calculation before using the neural networks. The last step is the de-normalization of the output data.2 For enhanced performance and selecting the optimum architecture for the application, the performance indicators ((1)-(3)) and R between the target response and the output obtained are analyzed.
Table 2: Results And Comparison Of Algorithms Using Different Architectures And Transfer Functions.
| Algorithm | Training
Function |
H | Transfer Function | Best Validation at Epoch | Epoch | R on
Training |
R on Validation | R on
Testing |
|
| Hidden | Output | ||||||||
| Conjugate Gradient | traincgb | 10 | logsig | purelin | 0.0033 at epoch 14 | 20 | 0.9864 | 0.9877 | 0.9711 |
| logsig | logsig | 0.0582 at epoch 16 | 22 | 0.8845 | 0.9633 | 0.9969 | |||
| logsig | tansig | 0.0099 at epoch 12 | 18 | 0.9815 | 0.9697 | 0.9649 | |||
| tansig | purelin | 0.0245 at epoch 7 | 13 | 0.9740 | 0.8949 | 0.9554 | |||
| tansig | logsig | 0.0027 at epoch 7 | 13 | 0.9753 | 0.9807 | 0.9802 | |||
| tansig | tansig | 0.0021 at epoch 10 | 16 | 0.9791 | 0.9977 | 0.8021 | |||
| 20 | logsig | purelin | 0.0022 at epoch 31 | 37 | 0.9942 | 0.9915 | 0.9942 | ||
| logsig | logsig | 0.054 at epoch 21 | 27 | 0.8904 | 0.9408 | 0.9457 | |||
| logsig | tansig | 0.0226 at epoch 3 | 9 | 0.9263 | 0.9901 | 0.9967 | |||
| tansig | purelin | 0.0195 at epoch 12 | 18 | 0.9925 | 0.8846 | 0.9703 | |||
| tansig | logsig | 0.0026 at epoch 26 | 32 | 0.9687 | 0.9941 | 0.9043 | |||
| tansig | tansig | 0.0031 at epoch 7 | 13 | 0.9855 | 0.9975 | 0.9556 | |||
| 30 | logsig | purelin | 0.0029 at epoch 11 | 17 | 0.9800 | 0.9892 | 0.9901 | ||
| logsig | logsig | 0.0053 at epoch 10 | 25 | 0.8769 | 0.9189 | 0.9645 | |||
| logsig | tansig | 0.0439 at epoch 4 | 10 | 0.9556 | 0.9353 | 0.9667 | |||
| tansig | purelin | 0.0024 at epoch 34 | 40 | 0.9969 | 0.9886 | 0.9398 | |||
| tansig | logsig | 0.0233 at epoch 15 | 21 | 0.9521 | 0.8570 | 0.9058 | |||
| tansig | tansig | 0.0620 at epoch 5 | 11 | 0.9674 | 0.7895 | 0.9405 | |||
| traincgf | 10 | logsig | purelin | 0.0014 at epoch 15 | 21 | 0.9880 | 0.9981 | 0.9784 | |
| logsig | logsig | 0.0022 at epoch 28 | 34 | 0.9077 | 0.9805 | 0.8597 | |||
| logsig | tansig | 0.0376 at epoch 12 | 18 | 0.9278 | 0.9710 | 0.9496 | |||
| tansig | purelin | 0.0030 at epoch 22 | 28 | 0.9905 | 0.9869 | 0.9605 | |||
| tansig | logsig | 0.0122 at epoch 7 | 13 | 0.8446 | 0.9763 | 0.9576 | |||
| tansig | tansig | 0.0056 at epoch 17 | 23 | 0.9921 | 0.9963 | 0.9871 | |||
| 20 | logsig | purelin | 0.0021 at epoch 11 | 17 | 0.9748 | 0.9805 | 0.9729 | ||
| logsig | logsig | 0.0563 at epoch 11 | 17 | 0.8751 | 0.9625 | 0.8621 | |||
| logsig | tansig | 0.0419 at epoch 17 | 23 | 0.9844 | 0.9325 | 0.9494 | |||
| tansig | purelin | 0.0067 at epoch 17 | 23 | 0.9694 | 0.9557 | 0.9883 | |||
| tansig | logsig | 0.0075 at epoch 5 | 11 | 0.8501 | 0.9856 | 0.9855 | |||
| tansig | tansig | 0.0456 at epoch 2 | 8 | 0.9355 | 0.9439 | 0.9823 | |||
| 30 | logsig | purelin | 0.0057 at epoch 6 | 12 | 0.9776 | 0.9891 | 0.9697 | ||
| logsig | logsig | 0.0109 at epoch 15 | 21 | 0.9228 | 0.8781 | 0.8880 | |||
| logsig | tansig | 0.3116 at epoch 10 | 16 | 0.9444 | 0.9129 | 0.8875 | |||
| tansig | purelin | 0.0179 at epoch 43 | 49 | 0.9943 | 0.9211 | 0.9739 | |||
| tansig | logsig | 0.05417 at epoch 9 | 15 | 0.9145 | 0.9727 | 0.9466 | |||
| tansig | tansig | 0.0152 at epoch 6 | 12 | 0.9748 | 0.9746 | 0.9537 | |||
| traincgp | 10 | logsig | purelin | 0.0020 at epoch 9 | 15 | 0.9679 | 0.9922 | 0.9927 | |
| logsig | logsig | 0.0700 at epoch 2 | 3 | 0.90502 | 0.8122 | 0.8055 | |||
| logsig | tansig | 0.0206 at epoch 25 | 31 | 0.9848 | 0.9788 | 0.9905 | |||
| tansig | purelin | 0.0142 at epoch 6 | 12 | 0.9437 | 0.9612 | 0.9716 | |||
| tansig | logsig | 0.0036 at epoch 16 | 22 | 0.9924 | 0.9824 | 0.9634 | |||
| tansig | tansig | 0.0190 at epoch 2 | 8 | 0.9204 | 0.9852 | 0.8885 | |||
| 20 | logsig | purelin | 0.0245 at epoch 6 | 12 | 0.96421 | 0.9669 | 0.954 | ||
| logsig | logsig | 0.0117 at epoch 5 | 11 | 0.90012 | 0.8776 | 0.8624 | |||
| logsig | tansig | 0.0383 at epoch 16 | 22 | 0.9648 | 0.9761 | 0.8404 | |||
| tansig | purelin | 0.0144 at epoch 13 | 19 | 0.9787 | 0.9465 | 0.9966 | |||
| tansig | logsig | 0.0090 at epoch 9 | 15 | 0.9696 | 0.9914 | 0.9392 | |||
| tansig | tansig | 0.0171 at epoch 9 | 15 | 0.9677 | 0.9962 | 0.9835 | |||
| 30 | logsig | purelin | 0.004 at epoch 8 | 14 | 0.9777 | 0.98295 | 0.86807 | ||
| logsig | logsig | 0.0516 at epoch 9 | 15 | 0.8800 | 0.98295 | 0.8680 | |||
| logsig | tansig | 0.1150 at epoch 5 | 11 | 0.9097 | 0.9547 | 0.9543 | |||
| tansig | purelin | 0.0112 at epoch 22 | 28 | 0.9936 | 0.9039 | 0.9850 | |||
| tansig | logsig | 0.0399 at epoch 45 | 51 | 0.9970 | 0.7896 | 0.928 | |||
| tansig | tansig | 0.01950 at epoch 9 | 15 | 0.9890 | 0.9885 | 0.9848 | |||
| trainscg | 10 | logsig | purelin | 0.0017 at epoch 18 | 24 | 0.9852 | 0.9917 | 0.9808 | |
| logsig | logsig | 0.0372 at epoch 13 | 19 | 0.9109 | 0.9056 | 0.8883 | |||
| logsig | tansig | 0.0112 at epoch 17 | 23 | 0.9831 | 0.9925 | 0.8805 | |||
| tansig | purelin | 0.0010 at epoch 12 | 18 | 0.9882 | 0.9885 | 0.9932 | |||
| tansig | logsig | 0.0176 at epoch 26 | 26 | 0.7811 | 0.9223 | 0.9363 | |||
| tansig | tansig | 0.0612 at epoch 17 | 23 | 0.9806 | 0.9985 | 0.9007 | |||
| 20 | logsig | purelin | 0.0025 at epoch 22 | 28 | 0.9843 | 0.9897 | 0.9264 | ||
| logsig | logsig | 0.0042 at epoch 12 | 18 | 0.8961 | 0.9898 | 0.9918 | |||
| logsig | tansig | 0.0385 at epoch 12 | 18 | 0.9618 | 0.9428 | 0.9344 | |||
| tansig | purelin | 0.0050 at epoch 47 | 53 | 0.999 | 0.9554 | 0.9797 | |||
| tansig | logsig | 0.0746 at epoch 16 | 21 | 0.8717 | 0.8212 | 0.8214 | |||
| tansig | tansig | 0.037 at epoch 10 | 16 | 0.9785 | 0.9782 | 0.9802 | |||
| 30 | logsig | purelin | 0.0031 at epoch 14 | 20 | 0.9844 | 0.9793 | 0.9679 | ||
| logsig | logsig | 0.0056 at epoch 12 | 18 | 0.9055 | 0.9891 | 0.9556 | |||
| logsig | tansig | 0.0614 at epoch 7 | 13 | 0.8326 | 0.9154 | 0.8425 | |||
| tansig | purelin | 0.0380 at epoch 7 | 13 | 0.9321 | 0.8106 | 0.9403 | |||
| tansig | logsig | 0.0055 at epoch 8 | 14 | 0.9033 | 0.8995 | 0.8428 | |||
| tansig | tansig | 0.1157 at epoch 12 | 18 | 0.9405 | 0.9755 | 0.9374 | |||
| quasi-Newton
|
trainbfg | 10 | logsig | purelin | 0.0016 at epoch 21 | 27 | 0.9920 | 0.9663 | 0.9917 |
| logsig | logsig | 0.0440 at epoch 7 | 13 | 0.8967 | 0.8775 | 0.9213 | |||
| logsig | tansig | 0.0282 at epoch 22 | 28 | 0.9757 | 0.977 | 0.9918 | |||
| tansig | purelin | 0.0069 at epoch29 | 35 | 0.9961 | 0.9538 | 0.9941 | |||
| tansig | logsig | 0.0367 at epoch 18 | 24 | 0.8885 | 0.9245 | 0.9792 | |||
| tansig | tansig | 0.0142 at epoch 26 | 32 | 0.9820 | 0.9817 | 0.9786 | |||
| 20 | logsig | purelin | 0.0063 at epoch 25 | 31 | 0.9896 | 0.9625 | 0.9624 | ||
| logsig | logsig | 0.0103 at epoch 9 | 15 | 0.9003 | 0.9195 | 0.9395 | |||
| logsig | tansig | 0.486 at epoch 17 | 23 | 0.9906 | 0.9308 | 0.8701 | |||
| tansig | purelin | 0.0195 at epoch 33 | 39 | 0.9969 | 0.9450 | 0.8070 | |||
| tansig | logsig | 0.0422 at epoch 21 | 27 | 0.8890 | 0.8934 | 0.9289 | |||
| tansig | tansig | 0.0019 at epoch 18 | 24 | 0.9846 | 0.9965 | 0.9578 | |||
| 30 | logsig | purelin | 0.0028 at epoch 26 | 32 | 0.9955 | 0.9708 | 0.9906 | ||
| logsig | logsig | 0.0086 at epoch 15 | 21 | 0.9106 | 0.9682 | 0.8964 | |||
| logsig | tansig | 0.0070 at epoch 18 | 24 | 0.9572 | 0.9811 | 0.83 | |||
| tansig | purelin | 0.0022 at epoch 6 | 12 | 0.9664 | 0.9821 | 0.8168 | |||
| tansig | logsig | 0.0083 at epoch 7 | 13 | 0.8214 | 0.9959 | 0.9455 | |||
| tansig | tansig | 0.01 at epoch 4 | 10 | 0.9209 | 0.9851 | 0.9926 | |||
| trainoss | 10 | logsig | purelin | 0.0024 at epoch 12 | 18 | 0.97948 | 0.9947 | 0.97729 | |
| logsig | logsig | 0.01568 at epoch 7 | 13 | 0.9164 | 0.9802 | 0.97626 | |||
| logsig | tansig | 0.0186 at epoch 15 | 21 | 0.9835 | 0.9477 | 0.9428 | |||
| tansig | purelin | 0.0076 at epoch 7 | 13 | 0.9731 | 0.9663 | 0.9337 | |||
| tansig | logsig | 0.0109 at epoch 3 | 9 | 0.9009 | 0.9118 | 0.9434 | |||
| tansig | tansig | 0.0322 at epoch 4 | 10 | 0.9603 | 0.9877 | 0.9173 | |||
| 20 | logsig | purelin | 0.002 at epoch 34 | 40 | 0.9838 | 0.9881 | 0.9745 | ||
| logsig | logsig | 0.0574 at epoch 27 | 33 | 0.9009 | 0.8239 | 0.95261 | |||
| logsig | tansig | 0.0316 at epoch 13 | 19 | 0.9658 | 0.9579 | 0.9547 | |||
| tansig | purelin | 0.0045 at epoch 75 | 81 | 0.9931 | 0.9492 | 0.9996 | |||
| tansig | logsig | 0.0059 at epoch 4 | 10 | 0.8050 | 0.9429 | 0.9677 | |||
| tansig | tansig | 0.0803 at epoch 10 | 16 | 0.9554 | 0.8986 | 0.9422 | |||
| 30 | logsig | purelin | 0.0035 at epoch 17 | 23 | 0.97342 | 0.98861 | 0.98923 | ||
| logsig | logsig | 0.0993 at epoch 13 | 19 | 0.8720 | 0.96933 | 0.8588 | |||
| logsig | tansig | 0.0196 at epoch 3 | 9 | 0.9402 | 0.9858 | 0.9810 | |||
| tansig | purelin | 0.0157 at epoch 29 | 35 | 0.9933 | 0.8529 | 0.9927 | |||
| tansig | logsig | 0.048 at epoch 6 | 12 | 0.8102 | 0.9907 | 0.9165 | |||
| tansig | tansig | 0.0534 at epoch 35 | 41 | 0.9807 | 0.9444 | 0.9630 | |||
| Levenberg-Marquardt
|
trainlm | 10 | logsig | purelin | 0.0008 at epoch 15 | 21 | 0.9977 | 0.9968 | 0.9959 |
| logsig | logsig | 0.0020 at epoch 15 | 21 | 0.9060 | 0.9803 | 0.9163 | |||
| logsig | tansig | 0.0144 at epoch 11 | 17 | 0.9994 | 0.9913 | 0.9863 | |||
| tansig | purelin | 0.0033 at epoch 2 | 8 | 0.9871 | 0.9943 | 0.9764 | |||
| tansig | logsig | 0.005 at epoch 5 | 11 | 0.9132 | 0.9876 | 0.8244 | |||
| tansig | tansig | 0.0037 at epoch 10 | 16 | 0.9972 | 0.9975 | 0.9853 | |||
| 20 | logsig | purelin | 0.0016 at epoch 2 | 8 | 0.9986 | 0.9981 | 0.9800 | ||
| logsig | logsig | 0.0121 at epoch 2 | 8 | 0.9087 | 0.9234 | 0.9166 | |||
| logsig | tansig | 0.0410 at epoch 2 | 8 | 0.9409 | 0.9337 | 0.8826 | |||
| tansig | purelin | 0.0023 at epoch 2 | 8 | 0.9910 | 0.9957 | 0.9934 | |||
| tansig | logsig | 0.0091 at epoch 149 | 149 | 0.9405 | 0.8253 | 0.8044 | |||
| tansig | tansig | 0.0095 at epoch 11 | 17 | 0.9979 | 0.9364 | 0.8362 | |||
| 30 | logsig | purelin | 0.0036 at epoch 6 | 10 | 0.9998 | 0.9669 | 0.9375 | ||
| logsig | logsig | 0.0036 at epoch 5 | 11 | 0.9217 | 0.9675 | 0.9916 | |||
| logsig | tansig | 0.1298 at epoch 1 | 7 | 0.8216 | 0.945 | 0.9853 | |||
| tansig | purelin | 0.0040 at epoch 3 | 6 | 0.9991 | 0.9744 | 0.8632 | |||
| tansig | logsig | 0.012 at epoch 8 | 14 | 0.8805 | 0.9335 | 0.9389 | |||
| tansig | tansig | 0.0168 at epoch8 | 14 | 0.9990 | 0.9985 | 0.9953 | |||
Other values of the indices comprising MSE, MAE and SSE are recorded in Fig. 3, 4 and 5. The transfer function is applied to both hidden and output layer in the ANN model. Therefore for example, in Fig. 3. (2-1) explains the use of Logsig transfer function in the hidden layer and Purelin transfer function in the output layer. All the other combinations follow the same pattern.
The values of indices are computed using the MATLAB syntax in the code itself. As presented in Table II, the optimum network model for this application for traincgb is when the network has 10 neurons in the hidden layer and logsig; purelin as the activation function in the network. The MSE corresponding to this is 0.003. It can be seen that all the other readings for MSE are bigger than MSE reading for the optimum network found. The R values for this network are 0.9864, 0.9877 and 0.9711. For traincgf the optimum network is found to be 10, logsig; purelin with MSE value as 0.027 and R value as 0.9880, 0.9981 and 0.9784 whereas traincgp gives the optimal results when the network architecture and parameters are set to 10, tansig; logsig where MSE value is seen to be 0.0026 and R values as 0.9924, 0.9824 and 0.9634. The trainscg algorithm gives better results with 10, tansig; purelin as its architecture and activation function. The MSE value for the same is found to be 0.0028. 0.9882, 0.9885 and 0.9932 are the R values. However, it is seen that MSE values for trainbfg algorithm, 0.0018, is same for when the network is 10, logsig; purelin and 10, tansig; purelin. In this case, the optimal network is chosen by comparing the R values and the best validation giving the most favorable architecture in trainbfg as 10, logsig; purelin with best validation performance being 0.0016 at epoch 21 and 0.9920, 0.9663, 0.9917 being the R values. In trainoss the finest value of MSE is 0.0027 whereas R is 0.9838, 0.9881, 0.9745 with the network parameters as 20, logsig; purelin. Finally for trainlm, MSE value is recorded as 0.00007 with R values nearest to 1; 0.9977, 0.9968 and 0.9959 when the network had 10 number of neurons in the hidden layer and logsig; purelin as the activation function.
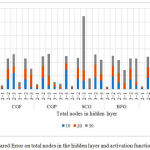 |
Figure 3: Effect of Mean Squared Error on total nodes in the hidden layer and activation function on each algorithm.
|
Effect of each of the seven algorithms on the output response by varying the architecture of the ANN model and the transfer function in hidden and output layer is shown in Fig. 3, 4 and 5. ANN models that are simulated using numerous training functions are altered in accordance with the number of neurons in their hidden layer. MSE of all the responses recorded is illustrated in Fig. 3. MSE is an important criterion for measuring the overall performance of a designed ANN model. Fig. 4 illustrates a graph between the MAE and total number of nodes in the hidden layer and activation function for all the 7 algorithms used to design the various ANN models. The absolute value of the difference between the target value provided to the ANN model to train and the actual value obtained is the absolute error. Fig. 5 illustrates a graph between error sum of squares, which computes the total deviation of the obtained values from the fitting line or the regression line, and total number of nodes in the hidden layer and activation function. Smaller the value of SSE, better will be the regression line. It is sometimes also referred to as residuals sum of square.
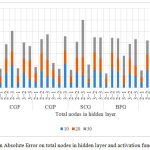 |
Figure 4: Effect of Mean Absolute Error on total nodes in hidden layer and activation function on each algorithm.
|
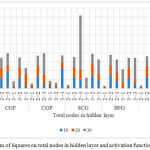 |
Figure 5: Effect of Error Sum of Squares on total nodes in hidden layer and activation function on each algorithm.
|
Conclusion
In this research, the size of the Ag-NPs is determined using ANN modeling from different combinations of architectures and transfer functions by means of a feed-forward neural network model which renders the effect of volume of C. longa extraction, stirring time, temperature, and volume of AgNO3 on the nanocomposites behavior. The ANN model is simulated, trained and tested with the learning algorithms like Quasi-Newton, Conjugate Gradient and Levenberg-Maquardt using the dataset. In the projected work it is evident that Levenberg-Maquardt is the best suited algorithm when considering engine data set type for the particular application. It converges in lesser epochs and indeed takes shorter time period than all the other training algorithms. Some suitable architectures gave worthy performances within the same algorithms as their R value is observed nearest to 1. The experiment shows that ANN is an effectual tool in pondering subjects related to nanoengineering as the size of the silver nanoparticle is predicted in the absence of the costly and time-consuming tests.
Acknowledgement
The authors would like to acknowledge & thank Dr. V. Ganapathy, Professor in SRM Institute of Science & Technology, Kattankulathur for his immense help in this work.
References
- Khan I., Saeed K., S khan I. Nano particles Properties applications and toxicities. Arabian Journal of Chemistry. 2017. Available: http://dx.doi.org/ 10.1016/j.arabjc. 2017.05.011.
- Shabanzadeh, Senu N., Shameli K., Ismail F. Application of Artificial Neural Network (ANN) for Prediction Diameter of Silver Nano particles Bio synthesized in Curcuma Long a Extract. Digest Journal of Nano materials and Bio structures. 2013;8(3):1133–1144.
- Steven J. O. Silver Nano particles Properties and Applications Sigma-Aldrich, Available: https://www. sigmaaldrich.com/technical-documents/articles/materials-science/nano materials/silver-nano particles .html. Accessed. 2018;16.
- Sacha G. M., Varona P. Artificial intelligence in nanotechnology. Nanotechnology. 2013;24:452002.
CrossRef - Hemmat M. E., Afranda M., Wongwisesb S., Naderic A., Asadid A., Rostamia S., Akbaria M. Applications of feed forward multi layer perceptron artificial neural networks and empirical correlation for prediction of thermal conductivity of Mg(OH)2–EG using experimental data. International Communications in Heat and Mass Transfer. 2015;67;46–50.
CrossRef - Demuth H., Beale M., Hagan M . Neural Network ToolboxTM 6 User’s Guide, COPYRIGHT. Available. 1992–2008. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.144.5444&rep=rep1&type=pdf.
- Sharma B., Venugopalan K. Comparison of Neural Network Training Functions for Hematoma Classification in Brain CT Images. IOSR Journal of Computer Engineering (IOSR-JCE). 2014;16(1):31-35.
CrossRef - Dongare D.,Kharde R. R., Amit D. K. Introduction to Artificial Neural Network. International Journal of Engineering and Innovative Technology (IJEIT). 2012;2(1):189-194.
- Richard J. S. An Introduction to the Conjugate Gradient Method Without the Agonizing Pain, edition 1 , Carnegie Mellon University, Pittsburgh, August 1994. Available: https://www.cs.cmu.edu/~quake-papers/painless-conjugate-gradient.pdf. Accessed. 2018;March 17.
- El-Nabarawy I., Ashraf M. A., Donald C. W. I. Levenberg-Marquardt and Conjugate Gradient Methods Applied to a High-Order Neural Network, The 2013 International Joint Conference on Neural Networks (IJCNN), Dallas TX. 2013;1-7.
- Brownlee J. What is the Difference Between Test and Validation Datasets – Machine Learning Mastery, [Online]. Available https://machine learning mastery.com/difference-test-validation-datasets/. Accessed. 2018; March 17.
- Shyam S. S., Oon-Doo B., Marcotte M. Neural networks for predicting thermal conductivity of bakery products. Journal of Food Engineering. 2002;52(3):299-304.
CrossRef








