Manuscript accepted on :October 24, 2017
Published online on: --
Kristina Simanjuntak1, Jusman E. Simanjuntak2, Rosmalena3 and Vivitri D. Prasasty4
1Faculty of Medicine, Pembangunan Nasional “Veteran” University of Indonesia Jalan RS Fatmawati, 12450, Jakarta Selatan, Indonesia.
2Faculty of Health Science, Pembangunan Nasional “Veteran” University of Indonesia Jalan RS Fatmawati, 12450, Jakarta Selatan, Indonesia.
3Department of Medical Chemistry, Faculty of Medicine, University of Indonesia, Jalan Salemba Raya 6, Jakarta 10430, Indonesia.
4Faculty of Biotechnology, Atma Jaya Catholic University of Indonesia Jalan Jenderal Sudirman 51, Jakarta 12930, Indonesia.
Corresponding Author E-mail: vivitri.dewi@atmajaya.ac.id
DOI : https://dx.doi.org/10.13005/bpj/1318
Abstract
High mobility group box 1 (HMGB1) is known as non-histone nuclear protein which has many biological functions, and it plays a significant role in many diseases inhibition, such as inflammatory and cancer diseases. HMGB1 in cancer cells could induce cell proliferation, cell differentiation, carcinogenesis, and tumorigenesis. In addition, HMGB1 function and location highly depends on the redox states. Our work focuses on molecular interaction studies of quercetin and its derivatives with HGMB1 protein target. Early reports showed that quercetin could inhibit tumors and cancers in different experimental examinations. However, clinical studies of quercetin showed low performance in its bioavailability. Therefore, structural modification of quercetin is needed to enhance its pharmacological properties. Here, we found 9 molecules of quercetin derivatives (QD) which have favorable interaction with HGMB1 and improved pharmacological properties. These findings indicate that QD might be the potential candidates against HMGB1 as therapeutic target for anticancer.
Keywords
HMGB1; Quercetin Derivatives; Drug Design; Molecular Docking; Anticancer
Download this article as:| Copy the following to cite this article: Simanjuntak K, Simanjuntak J. E, Rosmalena R, Prasasty V. D. Structure–Based Drug Design of Quercetin and its Derivatives Against HMGB1. Biomed Pharmacol J 2017;10(4). |
| Copy the following to cite this URL: Simanjuntak K, Simanjuntak J. E, Rosmalena R, Prasasty V. D. Structure–Based Drug Design of Quercetin and its Derivatives Against HMGB1. Biomed Pharmacol J 2017;10(4). Available from: http://biomedpharmajournal.org/?p=17671 |
Introduction
High mobility group box 1 (HMGB1) is a non-histone chromosomal and highly conserved protein that serves various roles in intracellular and extracellular systems. This group of proteins was appointed as the “high mobility group,” due to their rapid mobility properties in polyacrylamide gel electrophoresis. HMGB1 is discovered firstly as achromatin-associated protein, extensive studies of this group have directed to the identification of many critical functions of HMGB1. In eukaryotic organisms, HMGB1 protein is mainly found in the nucleus, where its function is chaperoning DNA molecules alleviating replication, recombination, transcription, DNA repair, and nucleosome formation stabilization by forming the DNA helical structure and promoting the binding of regulatory complexes to DNA. HMGB1 takes part in regulating autophagy and maintaining balance between autophagy and apoptosis within the cytoplasm compartment1, 2.
HMGB1 is also implicated as an important mediator in cancer. HMGB1 expression is high in many cancer cells, especially in colon cancer, breast cancer, cervical cancer, lung cancer, prostate cancer, and pancreatic cancer 3-6. Previous studies exhibited that there is a correlation between HMGB1 and various malignant cancer mechanisms. The principle behind this connection may be due to the role of extracellular HMGB1 in angiogenesis, cell migration, and macrophages recruitment, which could contribute to tumor growth and metastasis. HMGB1 has an elaborated role in carcinogenes is which could suppress tumorigenesis by interacting with tumor suppressor genes such as p53, p73 and RB. Moreover, HMGB1 has mixed effects on the superiorities of cancer including unlimited replicative potential, ability to blood vessels development (angiogenesis), deflection of programmed cell death (apoptosis), self-sufficiency in growth signals, insensitivity to inhibitors of growth, tissue invasion and metastasis which cause inflammation 7, 8.
This exhibits that HMGB1 is a potential target for cancer treatment. Suppression of HMGB1 is impor tant and it becomes prospective strategy for treating various cancers. Recently, various synthetic and natural products have been used to inhibit HMGB1 expression. Quercetin is a secondary metabolite compound which has polyphenol groups, and it belongs to the flavonol class of flavonoids. The broad range of biochemical and pharmacological properties of quercetin and its metabolites are due to the relative substitution of various functional groups on the flavonol molecule 9.
Phytochemical investigations of various plant extracts have revealed that quercetin can exist in a free state as an aglycone, or as its derivative by conjugating with: carbohydrates as quercetin glycosides 10; alkyls as quercetin methyl or ethyl 11, 12; hydroxyl group as quercetin ethers 13-15; and a sulfate group as quercetin derived-sulfates 16. Our research aim is to investigate the interaction binding between quercetin derivatives (QD) and HMGB1 through molecular docking analysis. Furthermore, the best docking hit of QD were analysed the ADMET (absorption, distribution, metabolism, excretion and toxicity) prediction 17, 18 to evaluate its pharmacological properties of QD in different mechanisms in the body. Here, we report the highlight molecular nature of QD action mechanims with various pharmacological properties as HGMB1 inhibitors.
Method
Protein Preparation
The protein sequence of human High mobility group box 1 (UNIPROT ID: P09429) was retrieved from Protein Sequence Databank: Uniprot (http://www.uniprot.org/). The three-dimensional structure of human High mobility group box 1 was built by I-TASSER (Iterative Threading ASSEmbly Refinement) 19. The coordinates of hHGMB1 only consists of amino acid residues in .pdb format file.
Ligand Preparation
The structure of the ligands were obtained from PubChem 20, 21 in. sdf format files (Fig. 1). Ligands were converted into. pdb format using GUI Open Babel 22. Some of ligand structures were drawn and the 3D structures were optimized by using Chem Draw 23. Pharmacokinetic properties of all the ligands were analysed by using SwissADME 17.
Molecular Docking Analysis
Molecular docking was conducted by using Autodock Vina 24, 25. The grid box site parameters was set up on xyz center coordinates at 73x68x73, and grid box size at (40x40x40) Å. Before running docking, receptor and ligands were prepared using AutoDock Tools 26 with activated Polygon Stipples and visualized by PyMOL Viewer 27. All receptor and ligand files were saved as pdbqt. Binding energies were obtained from molecular docking.Furthermore, these values were calculated to have inhibitor constant (Ki) values by using equation below 28:
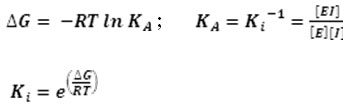
ADMET Prediction
Absorption, distribution, metabolism, excretion and toxicity (ADMET) prediction for the top docking of quercetin derivatives were predicted using ACD/I-Lab 18. This plat form predicts physicochemical descriptors as well as to predict ADMET parameters. ACD/I-Lab provides particular molecule’s properties with 95% of known drugs. Evaluation of ADMET properties, were based on absorption involved maximum passive absorption through intracellular and paracellular pathways, Volume distribution (Vd) was also involved in this work. Plasma Protein Binding percentage (%PPB) and P-gp specificity were used to evaluate metabolism and excretion profiles, respectively. Additionally, LD50 mouse and probability of health effects predictions of QD were calculated using ACD/I-Lab which is a web-based service that provides instant access to spectral and chemical databases, ADME predictions, and toxicity characteristics. A comparative analysis was performed for mouse LD50 (intraperitoneal, oral, intravenous, subcutaneous) and probability of health effect of blood, cardiovascular system, gastrointestinal system, kidney, liver and lung by QD‘s biological activities.
Results
The derivatives of Quercetin have been successfully built for further in silico analysis. The structures of Quercetin derivatives are based on the variation of their side group, as depicted in Fig. 1.
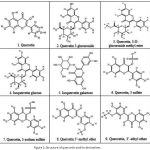 |
Figure 1: Structure of quercetin and its derivatives |
The pharmacophore properties showed no violation Lipinski‘s rule of five on Quercetin;Quercetin, 3-sulfate; Quercetin, 3-sodium sulfate; Quercetin, 3’-methyl ether; and Quercetin, 3’-ethyl ether. Meanwhile, Quercetin, 3-glucuronide; Quercetin, 3-O-glucuronide methyl esther;Isoquercetin glucose; and Isoquercetin galactose showed 3 violations following Lipinski‘s rule of five (Table 1).
Table 1: Pharmacophore properties following Lipinski rule of five with ACD/I-LAB
| No. | Compound Name | HBA | HBD | MW | Log P | Rotatable Bonds | TPSA | Violation of Ro5 |
| 1 | Quercetin | 7 | 5 | 302.24 | 2.07 | 1 | 127.45 | 0 |
| 2 | Quercetin, 3-glucuronide | 13 | 8 | 478.36 | 2.10 | 4 | 223.67 | 3 |
| 3 | Quercetin, 3-O-glucuronide methyl esther | 13 | 7 | 492.39 | 1.73 | 5 | 212.67 | 3 |
| 4 | Isoquercetin glucose | 12 | 8 | 464.38 | 1.75 | 4 | 206.6 | 3 |
| 5 | Isoquercetin galactose | 12 | 8 | 464.38 | 1.75 | 4 | 206.6 | 3 |
| 6 | Quercetin, 3-sulfate | 10 | 5 | 382.3 | 1.66 | 3 | 179.2 | 0 |
| 7 | Quercetin, 3-sodium sulfate | 10 | 1 | 382.3 | 1.66 | 7 | 135.2 | 0 |
| 8 | Quercetin, 3’-methyl ether | 7 | 4 | 316.26 | 1.76 | 2 | 116.45 | 0 |
| 9 | Quercetin, 3’-ethyl ether | 7 | 4 | 330.29 | 2.36 | 3 | 116.45 | 0 |
Table 2: Molecular Docking Analysis
| No. | Compound Name | Affinity Binding (kcal/mol) | Ki (M) | Hydrogen Bond | Residues involved in Hydrophobic interaction |
| 1 | Quercetin | -6.8 | 1.02×10-5 | 1 | Arg97, Ala148, Lys152, Lys180 |
| 2 | Quercetin, 3-glucuronide | -11.1 | 7.16×10-9 | 0 | Ala69, Glu72, Arg73, Lys76, Glu203, Glu204, Asp211 |
| 3 | Quercetin, 3-O-glucuronide methyl esther | -7.5 | 3.14×10-6 | 0 | Met63, Ala66, Lys65, Arg70, Glu210, Asp212, Asp213, Asp214, Glu215 |
| 4 | Isoquercetin glucose | -7.2 | 5.20×10-6 | 2 | Lys65, Ala66, Asp67, Glu210, Asp211, Asp212, Asp213, Asp213, Glu215 |
| 5 | Isoquercetin galactose | -6.6 | 1.43×10-5 | 2 | Lys68, Glu72, Met75, Lys76, Glu201, Asp211, Glu215 |
| 6 | Quercetin, 3-sulfate | -7.7 | 2.24×10-6 | 2 | Arg97, Ala148, Ala149, Lys152, Tyr155 |
| 7 | Quercetin, 3-sodium sulfate | -11.2 | 6.04×10-9 | 0 | Ala69, Glu72, Arg73, Lys76 |
| 8 | Quercetin, 3’-methyl ether | -9.8 | 6.44×10-8 | 0 | Lys96, Pro95, Arg97 |
| 9 | Quercetin, 3’-ethyl ether | -7.3 | 4.40×10-6 | 0 | Ala69, Glu72, Met75, Lys76, Glu215 |
Molecular docking analysis showed that Quercetin, 3 sodium sulfate showed the best interaction with HMGB1, base don its affinity energy (-11.2 kcal/mol) and Ki value (6.04×10-9 M). This interaction does not involve hydrogen bond, but it involves hydrophobic interaction with the residues in HMGB1 binding site, including: Ala69, Glu72, Arg73, and Lys76 (Table 2, Fig. 2).
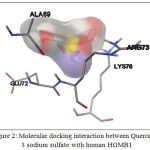 |
Figure 2: Molecular docking interaction between Quercetin, 3 sodium sulfate with human HGMB1 |
ADME and toxicity predictions of Quercetin derivatives showed various results (Table 3 and Table 4). These analyses were conducted by using ACD/I-LAB program.
Table 3: ADME Prediction with ACD/I-LAB
| No. | Compound Name | Absorptiona (%) | Distributionb(L/kg) | Metabolismc(%) | Excretion Probabilityd |
| 1 | Quercetin | 100 | 0.60 | 93.43 | 2.5 |
| 2 | Quercetin, 3-glucuronide | 0 | 0.25 | 88.35 | 5 |
| 3 | Quercetin, 3-O-glucuronide methyl esther | 12 | 0.62 | 85.36 | 0.5 |
| 4 | Isoquercetin glucose | 7 | 0.61 | 82.9 | 0.5 |
| 5 | Isoquercetin galactose | 7 | 0.61 | 82.9 | 0.5 |
| 6 | Quercetin, 3-sulfate | 1 | 0.25 | 99.82 | 5 |
| 7 | Quercetin, 3-sodium sulfate | 2 | 0.25 | 99.36 | 5 |
| 8 | Quercetin, 3’-methyl ether | 100 | 0.62 | 94.9 | 2.5 |
| 9 | Quercetin, 3’-ethyl ether | 100 | 0.62 | 94.9 | 2.5 |
aMaximum passive absorption b Volume of distribution (Vd) c%Plasma Protein Binding (%PPB) dP-gp specificity (AB/logP), value between 2.5-5 is possibly inhibitor
Table 4: Toxicity Prediction with ACD/I-LAB
| No. | Compound Name | LD50 (mg/Kg) | |||
| Intraperitoneal | Oral | Intravenous | Subcutaneous | ||
| 1 | Quercetin | 450 | 670 | 350 | 160 |
| 2 | Quercetin, 3-glucuronide | 470 | 3600 | 1800 | 1900 |
| 3 | Quercetin, 3-O-glucuronide methyl esther | 390 | 3300 | 1200 | 740 |
| 4 | Isoquercetin glucose | 730 | 1200 | 740 | 310 |
| 5 | Isoquercetin galactose | 730 | 1200 | 740 | 310 |
| 6 | Quercetin, 3-sulfate | 760 | 1500 | 270 | 730 |
| 7 | Quercetin, 3-sodium sulfate | 1100 | 1600 | 210 | 1500 |
| 8 | Quercetin, 3’-methyl ether | 570 | 550 | 290 | 120 |
| 9 | Quercetin, 3’-ethyl ether | 560 | 570 | 270 | 120 |
ADME and toxicity predictions of Quercetin derivatives showed various results (Table 3 andTable 4).These analyses were conducted by using ACD/ILAB program.
Discussion
Quercetin is categorized by a benzo-(g)pyrone skeletal structure with C6-C3-C6 carbon form, consisting of two benzene rings, A and B, linked by three carbons pyrone ring C as shown in Fig. 1. Quercetin is referred to pentahydroxy flavonol due to the presence of five hydroxyl groups on its flavonol skeletal framework at 3, 30, 40, 5, and 7 carbons 29-31.
The broad range of biochemical and pharmacological properties of quercetin and its derivatives are due to functional group substitutions of flavonol molecule 32. However, glycosylation of other hydroxyl groups has also been found from plants 33, 34. The sugar moieties could be in the form of monosaccharides, disaccharides, or polysaccharides. The most common monosaccharides substituents are glucose, galactose, rhamnose, and xylose. Isoquercetin glycosides also has been reported having radical scavenging properties 35, 36.
Quercetin (3,31,41,5,7-pentahydroxy flavone) is natural flavonoid and its common derivative as flavone (2-phenylchromen-4-one). Quercetin contains five hydroxyl groups that are responsible for its biological activities and derivative diversification. Flavonoids generally consist of two benzene rings linked by pyran or pyrone rings30, 32.
In addition, the conformational analysis of quercetin showed the presence of 12 conformations of this molecule having Gibbs energies in the range of 0 to 5.33 kcal/ mol 37,38. Furthermore, quercetin exhibits strong intramolecular Hydrogen bond interactions, which displaying its biological multifunctional activities and renders its ability to form strong complex interactions, including with metals, affecting its bioavailability and transport system in the cells (39-41). Among these H-bonds, two bonds are with carbonyl groups and third one is between hydroxyl groups (42, 43). Moreover, the derivatives of quercetin glycosides may also contain acyl and sulfur substituents becomes sugar moieties. In the case of quercetin derivatives, the hydroxyl groups of quercetin are attached with alcohols via ether bonds. Although quercetin is lipophilic, the glycosylation of quercetin derivatives can increase the hydrophilicity and enable the molecules to transport through all parts of the plant (44, 45).
Pharmacophore properties of quercetin and its derivatives showed various properties according to Lipinski‘s rule of five (Ro5). Ro5 consist of HBA/ HBD value up to 10 and 5, respectively; MW less than 500, Log P value less than 5 and total polar surfacearea (TPSA) value less than 140 Å (46). From the results showed that there was no violation of Ro5 for quercetin; Quercetin, 3-sulfate; Quercetin, 3-sodium sulphate; Quercetin, 3’-methyl ether; and Quercetin, 3’-ethyl ether, while the 4 other derivatives showed 3 violations of Ro5 as shown on Table 1.
Analysis of molecular docking was performed by using Autodock Vina. The best docking is Quercetin, 3-sodium sulfate with Affinity binding -11.2 kcal/mol. This favorable binding was contributed by hydrophobic interactions from four amino acid residues from HGMB1 protein including Ala69, Glu72, Arg73, and Lys76 (Fig.2). The inhibition constant (Ki) of Quercetin, 3-sodium sulfate is 6.04×10-9 M. The result of docking interaction, Ki value, and HGMB1 residues involved in hydrophobic interaction is shown on Tabel 2.
Prediction of ADME properties of quercetin and its derivatives were done by using ACD/I-Lab platform as shown on Table 3. Maximum passive absorptions of Quercetin, Quercetin, 3’-methyl ether and Quercetin, 3’-methyl ether were found 100%. Meanwhile, other quercetin derivatives were found less than 15%.Volume distribution rates of quercetin and its derivatives we found within 0.25 – 0.65 L/kg. Plasma binding protein ability rate in metabolisms prediction from quercetin and its derivatives were within 85.36 – 99.82. Excretion probabilities by involving P-gp specificity (AB/logP) from quercetin and its derivatives had value between 2.5-5 were possibly inhibitor, meanwhile P-gp specificity value less than 2.5 indicated were not inhibitors as well.
Table 5: Health Effects Prediction with ACD/I-LAB
| Probability of Health Effects | |||||||
| No. | Compound Name | Blood | Cardiovasculr | Gastrointestil | Kidney | Lung | Liver |
| 1 | Quercetin | 0.69 | 0.27 | 0.45 | 0.54 | 0.38 | 0.09 |
| 2 | Quercetin, 3-glucuronide | 0.96 | 0.9 | 0.68 | 0.75 | 0.81 | 0.82 |
| 3 | Quercetin, 3-O-glucuronide methyl esther | 0.95 | 0.98 | 1 | 0.68 | 0.9 | 0.77 |
| 4 | Isoquercetin glucose | 0.96 | 0.93 | 0.99 | 0.62 | 0.8 | 0.84 |
| 5 | Isoquercetin galactose | 0.96 | 0.87 | 0.88 | 0.55 | 0.8 | 0.74 |
| 6 | Quercetin, 3-sulfate | 0.19 | 0.27 | 0.2 | 0.56 | 0.2 | 0.27 |
| 7 | Quercetin, 3-sodium sulfate | 0.79 | 0.48 | 0.99 | 0.51 | 0.23 | 0.38 |
| 8 | Quercetin, 3’-methyl ether | 0.6 | 0.64 | 0.44 | 0.5 | 0.37 | 0.27 |
| 9 | Quercetin, 3’-ethyl ether | 0.29 | 0.61 | 0.41 | 0.49 | 0.4 | 0.34 |
Moreover, toxicity analyses of quercetin and its derivatives were done by using ACD/I-Lab platform as shown on Table 4 and health effects prediction on Table 5. Quercetin, 3-sodium sulfate was found as acceptable ADME value, meanwhile toxicity of Quercetin, 3-sodium sulfate seems high on oral system with LD50 = 1600 mg/kg, whatsoever other derivatives gave toxicity prediction values in various deposition in the body tissues. To evaluate health effect in the body system responses, we also analysed the health effect prediction in blood, cardiovascular, gastrointestinal, kidney, lung, and liver. Quercetin, 3-sodium sulfate might give high effect to gastrointestinal which causes dysfunction of gastrointestinal.
Conclusion
The molecular docking studies with quercetin and its derivatives into the binding cavity of human HGMB1 inducible showed the derivatives having more favorable interaction than quercetin with docking score involving hydrogen bond and ligandprotein interaction energy compared to quercetin. As earlier reported in literature, quercetin is known for having anticancer property and inhibiting the HGMB1 protein, the derivatives were docked at the binding cavity could have also possess some sort of anticancer property as it is 95% similar to quercetin retrieved form the NCBI PubChem database. The docked compounds used in the present study do not violate the Lipinski rule of five parameters. Also, from the ADME-Toxicity prediction using ACD/I-Lab revealed the docked compounds are in the acceptable range of various pharmacological parameters and they have similar behaviour of health effects and LD50 compared to quercetin.Therefore, we conclude that quercetin derivative compounds could be potential candidate molecules and could support for experimental testing against HGMB1 protein as anticancer agents.
Conflict of Interest
Authors declare that there is no conflict of interest.
Acknowledgement
Authors would like to thank the Scientific Affairs of Pembangunan Nasional “Veteran” University of Indonesia for their kind support in facilitating the valuable resources to accomplish this work.
References
- Pallier, C. et al. Association of chromatin proteins high mobility group box (HMGB) 1 and HMGB2 with mitotic chromosomes. Molecular biology of the cell 14, 3414-3426 (2003).
CrossRef - Taniguchi, N. et al. High mobility group box chromosomal protein 1 plays a role in the pathogenesis of rheumatoid arthritis as a novel cytokine. Arthritis and rheumatism 48, 971-981 (2003).
CrossRef - Ellerman, J.E. et al. Masquerader: high mobility group box-1 and cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 13, 2836-2848 (2007).
CrossRef - Dong Xda, E. et al. High mobility group box I (HMGB1) release from tumor cells after treatment: implications for development of targeted chemoimmunotherapy. J Immunother 30, 596-606 (2007).
CrossRef - Xia, Y., Papalopulu, N., Vogt, P.K. & Li, J. The oncogenic potential of the high mobility group box protein Sox3. Cancer research 60, 6303-6306 (2000).
- Pang, X., Zhang, Y. & Zhang, S. High-mobility group box 1 is overexpressed in cervical carcinoma and promotes cell invasion and migration in vitro. Oncology reports 37, 831-840 (2017).
CrossRef - Xu, M. et al. Inhibiting High-Mobility Group Box 1 (HMGB1) Attenuates Inflammatory Cytokine Expression and Neurological Deficit in Ischemic Brain Injury Following Cardiac Arrest in Rats. Inflammation 39, 1594-1602 (2016).
CrossRef - Xu, L. et al. Blockade of Extracellular High-Mobility Group Box 1 Attenuates Systemic Inflammation and Coagulation Abnormalities in Rats with Acute Traumatic Coagulopathy. Medical science monitor : international medical journal of experimental and clinical research 22, 2561-2570 (2016).
CrossRef - Enkhmaa, B. et al. Mulberry (Morus alba L.) leaves and their major flavonol quercetin 3-(6-malonylglucoside) attenuate atherosclerotic lesion development in LDL receptor-deficient mice. The Journal of nutrition 135, 729-734 (2005).
CrossRef - Valentova, K. et al. (Anti)mutagenic and immunomodulatory properties of quercetin glycosides. Journal of the science of food and agriculture 96, 1492-1499 (2016).
CrossRef - van der Woude, H., Boersma, M.G., Alink, G.M., Vervoort, J. & Rietjens, I.M. Consequences of quercetin methylation for its covalent glutathione and DNA adduct formation. Chemico-biological interactions 160, 193-203 (2006).
CrossRef - De Santi, C., Pietrabissa, A., Mosca, F. & Pacifici, G.M. Methylation of quercetin and fisetin, flavonoids widely distributed in edible vegetables, fruits and wine, by human liver. International journal of clinical pharmacology and therapeutics 40, 207-212 (2002).
CrossRef - Dell’Albani, P., Di Marco, B., Grasso, S., Rocco, C. & Foti, M.C. Quercetin derivatives as potent inducers of selective cytotoxicity in glioma cells. European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences 101, 56-65 (2017).
CrossRef - Bao, X.R. et al. Synthesis, Characterization and Cytotoxicity of Alkylated Quercetin Derivatives. Iranian journal of pharmaceutical research : IJPR 15, 329-335 (2016).
- Saija, A. et al. ‘In vitro’ antioxidant and photoprotective properties and interaction with model membranes of three new quercetin esters. European journal of pharmaceutics and biopharmaceutics : official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik e.V 56, 167-174 (2003).
- Jones, D.J., Jukes-Jones, R., Verschoyle, R.D., Farmer, P.B. & Gescher, A. A synthetic approach to the generation of quercetin sulfates and the detection of quercetin 3′-O-sulfate as a urinary metabolite in the rat. Bioorganic & medicinal chemistry 13, 6727-6731 (2005).
CrossRef - Daina, A., Michielin, O. & Zoete, V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific reports 7, 42717 (2017).
CrossRef - Masunov, A. ACD/I-Lab 4.5: an internet service review. Journal of chemical information and computer sciences 41, 1093-1095 (2001).
CrossRef - Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC bioinformatics 9, 40 (2008).
CrossRef - Ihlenfeldt, W.D., Bolton, E.E. & Bryant, S.H. The PubChem chemical structure sketcher. Journal of cheminformatics 1, 20 (2009).
CrossRef - Hur, J. & Wild, D.J. PubChemSR: a search and retrieval tool for PubChem. Chemistry Central journal 2, 11 (2008).
CrossRef - O’Boyle, N.M., Morley, C. & Hutchison, G.R. Pybel: a Python wrapper for the OpenBabel cheminformatics toolkit. Chemistry Central journal 2, 5 (2008).
CrossRef - Cousins, K.R. Computer review of ChemDraw Ultra 12.0. Journal of the American Chemical Society 133, 8388 (2011).
CrossRef - Chang, M.W., Ayeni, C., Breuer, S. & Torbett, B.E. Virtual screening for HIV protease inhibitors: a comparison of AutoDock 4 and Vina. PloS one 5, e11955 (2010).
CrossRef - Trott, O. & Olson, A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry 31, 455-461 (2010).
- Morris, G.M., Huey, R. & Olson, A.J. Using AutoDock for ligand-receptor docking. Current protocols in bioinformatics Chapter 8, Unit 8 14 (2008).
CrossRef - Lill, M.A. & Danielson, M.L. Computer-aided drug design platform using PyMOL. Journal of computer-aided molecular design 25, 13-19 (2011).
CrossRef - Chen, Y.L. et al. Inhibition of dengue virus by an ester prodrug of an adenosine analog. Antimicrobial agents and chemotherapy 54, 3255-3261 (2010).
CrossRef - Bhaskar, S. & Helen, A. Quercetin modulates toll-like receptor-mediated protein kinase signaling pathways in oxLDL-challenged human PBMCs and regulates TLR-activated atherosclerotic inflammation in hypercholesterolemic rats. Molecular and cellular biochemistry 423, 53-65 (2016).
CrossRef - Weckerle, B., Michel, K., Balazs, B., Schreier, P. & Toth, G. Quercetin 3,3′,4′-tri-O-beta-D-glucopyranosides from leaves of Eruca sativa (Mill.). Phytochemistry 57, 547-551 (2001).
CrossRef - Miean, K.H. & Mohamed, S. Flavonoid (myricetin, quercetin, kaempferol, luteolin, and apigenin) content of edible tropical plants. Journal of agricultural and food chemistry 49, 3106-3112 (2001).
CrossRef - Saito, A., Sugisawa, A., Umegaki, K. & Sunagawa, H. Protective effects of quercetin and its metabolites on H2O2-induced chromosomal damage to WIL2-NS cells. Bioscience, biotechnology, and biochemistry 68, 271-276 (2004).
CrossRef - Haskins, A.H. et al. Data for induction of cytotoxic response by natural and novel quercetin glycosides. Data in brief 6, 262-266 (2016).
CrossRef - Shimoda, K. et al. Glycosylation of quercetin with cultured plant cells and cyclodextrin glucanotransferase. Natural product communications 9, 647-648 (2014).
- Kato, K., Ninomiya, M., Tanaka, K. & Koketsu, M. Effects of Functional Groups and Sugar Composition of Quercetin Derivatives on Their Radical Scavenging Properties. Journal of natural products 79, 1808-1814 (2016).
CrossRef - Haas, M.J. et al. Induction of hepatic apolipoprotein A-I gene expression by the isoflavones quercetin and isoquercetrin. Life sciences 110, 8-14 (2014).
CrossRef - Wang, Y. & Wang, X. Binding, stability, and antioxidant activity of quercetin with soy protein isolate particles. Food chemistry, 188, 24-29 (2015).
CrossRef - Mehranfar, F., Bordbar, A. K., & Parastar, H. A combined spectroscopic, molecular docking and molecular dynamic simulation study on the interaction of quercetin with beta-casein nanoparticles. Journal of photochemistry and photobiology. B, Biology, 127, 100-107 (2013).
CrossRef - Dobrikova, A. G. & Apostolova, E. L. Damage and protection of the photosynthetic apparatus from UV-B radiation. II. Effect of quercetin at different pH. Journal of plant physiology, 184, 98-105 (2015).
CrossRef - Wu, J. et al. Quercetin alters energy metabolism in swimming mice. Applied physiology, nutrition, and metabolism = Physiologie appliquee, nutrition et metabolisme, 37(5), 912-922 (2012).
- Maciej, J. et al. Bioavailability of the flavonol quercetin in neonatal calves after oral administration of quercetin aglycone or rutin. Journal of dairy science, 98(6), 3906-3917 (2015).
CrossRef - Fitzenberger, E., Deusing, D. J., Marx, C., Boll, M., Luersen, K., & Wenzel, U. The polyphenol quercetin protects the mev-1 mutant of Caenorhabditis elegans from glucose-induced reduction of survival under heat-stress depending on SIR-2.1, DAF-12, and proteasomal activity. Molecular nutrition & food research, 58(5), 984-994 (2014).
CrossRef - Pfeuffer, M. et al. Effect of quercetin on traits of the metabolic syndrome, endothelial function and inflammation in men with different APOE isoforms. Nutrition, metabolism, and cardiovascular diseases : NMCD, 23(5), 403-409 (2013).
CrossRef - Zhu, X. The effects of quercetin-loaded PLGA-TPGS nanoparticles on ultraviolet B-induced skin damages in vivo. Nanomedicine : nanotechnology, biology, and medicine, 12(3), 623-632 (2016).
CrossRef - Xie, Y. et al. Phytic acid enhances the oral absorption of isorhamnetin, quercetin, and kaempferol in total flavones of Hippophae rhamnoides L. Fitoterapia, 93, 216-25 (2014).
CrossRef - Nogara, P. A. et al. Virtual screening of acetylcholinesterase inhibitors using the Lipinski’s rule of five and ZINC databank. BioMed research international 2015, 870389 (2015).
CrossRef







