Manuscript accepted on :06-02-2025
Published online on: 28-02-2025
Plagiarism Check: Yes
Reviewed by: Dr. Ananya Naha
Second Review by: Dr. Nishu Raina
Final Approval by: Dr. Prabhishek Singh
Abdulahi Mahammed Adem1 , Ravi Kant1
, Ravi Kant1 , Sonia2*
, Sonia2* , Karan Kumar3
, Karan Kumar3 , Vikas Mittal4
, Vikas Mittal4 , Pankaj Jain5
, Pankaj Jain5 and Kapil Joshi6
and Kapil Joshi6
1School of Core Engineering, Shoolini University, Solan, Himachal Pradesh, India.
2Yogananda School of AI Computers and Data Science, Shoolini University, Solan, Himachal Pradesh, India.
3Department of Electronics and Communication Engineering, MMEC, Maharishi Markandeshwar (Deemed to be University), Mullana, Ambala, India.
4Department of Electronics and Communication Engineering, Chandigarh University, Gharuan, Mohali, India.
5Department of Management, SJJTU University, Churu Rajasthan, India.
6Department of Computer Science and Engineering, Uttaranchal Institute of Technology (UIT), Uttaranchal University, Dehradun, Uttarakhand, India.
Corresponding Author E-mail: soniacsit@yahoo.com
DOI : https://dx.doi.org/10.13005/bpj/3073
Abstract
In digital image processing for disease categorization and detection, the introduction of neural networks has played a significant role. However, the need for substantial labelled data brings a challenge which often limits its effectiveness in pathology image interpretation. This study explores self-supervised learning’s potential to overcome the constraints of labelled data by using unlabeled or unannotated data as a learning signal. This study also focuses on self-supervised learning application in digital pathology where images can reach gigapixel sizes, requiring meticulous scrutiny. Advancements in computational medicine have introduced tools processing vast pathological images by encoding them into tiles. The review also explores cutting-edge methodologies such as contrastive learning and context restoration within the domain of digital pathology. The primary focus of this study centers around self-supervised learning techniques, specially applied to disease detection and classification in digital pathology. The study addresses the challenges associated with less labelled data and underscores the significance of self-supervised learning in extracting meaning full features from unlabelled pathology images. Using techniques like Longitudinal Self-supervised learning, the study provides a comparative study with traditional supervised learning approaches. The finding will contribute valuable insights and techniques by bridging the gap between digital pathology and machine learning communities.
Keywords
: Context restoration; Contrastive learning; Deep learning; Digital Pathology; Self-supervised learning
Download this article as:| Copy the following to cite this article: Adem A. M, Kant R, Sonia S, Kumar K, Mittal V, Jain P, Joshi K. Exploring Self-Supervised Learning for Disease Detection and Classification in Digital Pathology: A Review. Biomed Pharmacol J 2025;18(March Spl Edition). |
| Copy the following to cite this URL: Adem A. M, Kant R, Sonia S, Kumar K, Mittal V, Jain P, Joshi K. Exploring Self-Supervised Learning for Disease Detection and Classification in Digital Pathology: A Review. Biomed Pharmacol J 2025;18(March Spl Edition). Available from: https://bit.ly/3XlekXo |
Introduction
Advancements in deep learning, including image classification and object detection, are due to large and diversely labelled training data. Deep supervised learning requires many labelled data for good performance, but acquiring enough manually annotated data is expensive and requires technical expertise. These labelling efforts can be time-consuming and costly.1,3 Particularly, in the domain of digital pathology, numerous challenges have been impeded including the scarcity of annotated data and complexity of advancement in image analysis required for pathology image. Traditionally, supervised learning relies on extensive annotated dataset for training.
To mitigate this, various methods have been proposed, including unsupervised, self-supervised, and semi-supervised use of unlabelled or, in part, labelled data. Self-supervised learning is popular as it trains algorithms on data without needing large amounts of labelled examples. This makes it a promising approach for medical image analysis where labelled data is limited.4,6 Digital pathology images also present other challenges, the substantial image sizes and inherent variability in tissue appearance. The enormity of whole slide images reaching gigapixels coupled with intricate details and diverse structures in tissue samples complicates the interpretation and analysis of pathology images.
To tackle these challenges deep learning particularly CNN have been proven to tailored solutions. As mentioned above the advancements in deep learning, such as pathology image classification, are due to the full supervision of large and diverse labelled training data. Nevertheless, gathering enough labelled data for deep supervised learning needs technical know-how, is expensive, and takes much time. The neural network type known as a convolutional neural network (CNN) is optimized for image recognition and processing and comprises multiple layers trained using backpropagation.7,9
Figure 1 shows a CNN architecture with an input layer responsible for the input image. Then, there is the convolution layer to detect and learn features from the input data, such as images and max pooling, which is also used for down-streaming features. The complete connected layer, which serves as the final layer of the network, is used for transforming the high-level into predictions. The finally the layer which is responsible for producing the output. Animal vision, which detects light in discrete patches that overlap with one another across the visual field, served as an inspiration for CNN development. Layers will extract higher-level attributes with increasing accuracy as can be explored deeper into the network. Creating a correlation between the internal representation of pixel values and how those values are shown in the form of a two-dimensional matrix is a skill that CNN models are known to have. It works very well for data that can be connected to a specific area.
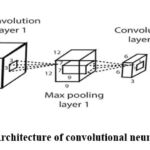 |
Figure 1: Architecture of convolutional neural network10Click here to view Figure |
Transfer learning is a machine learning system that lets a model qualified on one job be adjusted and used in another related job. It is a valuable approach because it can save time and resources by leveraging the knowledge gained from previously solving a similar problem.11,12
Medical image analysis requires time-consuming data labelling, which has led to methods that use unlabelled or partially labelled data to boost performance. Self-supervised learning, which assigns surrogate supervisory signals to unlabelled data without human annotation, is a promising solution for scenarios where labelled data is limited or expensive to obtain. This approach benefits from using substantial quantity of unnamed data, which is simpler and cheaper to acquire.13,16
Digital pathology offers new opportunities in analysis medical image with high-quality WSIs of histopathology scans available. Self-supervised learning is popular in histopathology as it trains models on large amounts of unlabelled data, saving time and effort in manual labelling. A common tactic to self-supervise in histopathology is to use a masked language model pre-training technique, where the model predicts a masked region of an image given the rest as context. The model can then be fine-tuned on a labelled dataset for specific tasks like image segmentation or classification.17-19
Additionally, self-learning can enhance the functioning of a model on a specific task by providing it with additional training data. For example, a self-supervised learning model that is trained on a substantial quantity of text can learn valuable features of the language that can then be used to increase the ability of the model on a specific processing task, for instance, sentiment analysis or machine conversion.20
In contrastive self-supervised learning, the model is given two samples and must predict whether they are from the same distribution. For example, the model might be given two images and must predict whether they are both images of the same object or not. To perform this, the model must learn to obtain meaningful elements from the inserted data and then use them to make accurate predictions.13,21,22
Non-contrastive self-supervised learning trains models to perform specific tasks using input data, unlike contrastive self-supervised learning. Digital pathology and (WSIs) have opened new opportunities for medical image analysis. Due to its capacity to train deep learning models on substantial volumes of unlabelled data, self-supervised learning is popular in histopathology. This makes it advantageous for histopathology images, where labelling is laborious.23,25
Self-supervised learning is a common approach, using masked language model pre-training to predict masked regions of an image. The model learns to identify features and patterns and then fine-tunes labelled data for a specific task like image segmentation or classification. One application of self-supervised learning is using CNNs to analyze tissue images for disease recognition.26
Once trained, a CNN can automatically analyze new tissue images and identify any abnormalities or signs of disease. This could speed up the diagnostic process and improve its accuracy. Additionally, because CNNs can learn to recognize patterns and features that may not be obvious to a human observer, they could potentially identify diseases that a human pathologist might miss.27,28
In the domain of digital pathology, different learning paradigms, including supervised and unsupervised approaches play critical roles. Table 1 provides summary of the different machine learning approaches used in digital pathology.
Table 1: Summary of different machine learning approaches
| Approach | Methodology | Limitation | Application in digital pathology |
| Supervised learning | · Requires large, labeled data. | · Challenges in acquiring extensive annotated data. | · Hindered by the scarcity of precisely labeled pathology image. |
| Unsupervised learning | · Operates without labeled data but may lack the ability to capture meaningful representation | · It may struggle to capture intricate patterns and features in complex images. | · Limited efficiency in cases where detailed understanding is needed. |
| Self-supervised learning | · Uses unlabeled or partially labeled data, relay on inherent data structure. | · Mitigate the need for labeled data.· Exploit unlabeled data.· Allows efficient training on diverse and comprehensive dataset. | · Especially useful in overcoming challenges related to scarcity of annotated data. |
Objective
The purpose of this work is to thoroughly investigate and assess the application of self-supervised learning methods in the field of digital pathology, with an emphasis on tasks involving disease identification and classification. In order to increase accuracy and efficiency in illness detection and classification using digital pathology images, the study intends to present a comprehensive overview of self-supervised learning approaches, their strengths, limits, and effectiveness. The purpose of this work, which involves a thorough review of recent studies, is to
Examine and classify the several self-supervised learning approaches used in digital pathology, such as contrastive learning, generative adversarial networks (GANs), and others.
Examine digital pathology applications: Review the literature to comprehend how self-supervised learning methods have been used in tasks involving the detection and classification of diseases in digital pathology. This study includes identifying the diseases that have been targeted and the results that have been obtained.
Evaluation of accuracy, robustness, and generalization: In comparison to conventional supervised methods, evaluate the accuracy, robustness, and generalization of self-supervised learning models across a variety of datasets and illness categories.
Identify best practices: To provide academics and practitioners with useful information, identify the best practices, structures, and techniques for self-supervised learning that have demonstrated promise in the identification and categorization of diseases.
Bridge the gap: Provide information on how these cutting-edge methods can be modified and incorporated into real-world clinical applications to bridge the gap between the digital pathology community and the self-supervised learning research community.
Other than the above listed objectives the study provides a substantial contribution to the field of digital pathology and self-supervised learning by identifying best practices. When it comes to digital pathology, this study enhances the accuracy and efficiency of disease detection and classification by providing detailed discussion on advanced methodologies. The significant impact of this study extends positively influencing medical communities and practitioners. Improved accuracy in disease detection ensures timely intervention contributing to patient outcomes. By achieving these goals and others, the paper hopes to advance the understanding of how cutting-edge machine-learning methods can revolutionize disease diagnosis and classification through the analysis of digital pathology images and contribute to the expanding body of knowledge in self-supervised learning and digital pathology.
Materials and Methods
Self-supervised learning can be applied to digital pathology for disease detection by training a model on large amounts of unlabelled pathology images. The model may be made to learn for the purpose of identifying elements and patterns in the images that indicate specific diseases. As an example, detecting abnormal cells, tissue or patterns (anomaly detection) using autoencoders and self-supervised anomaly detection techniques done by.29-31 Organ or tissue segmentation, which is Segmenting different organs or tissues in pathology images, is another application of self-supervised learning.32,33
Additionally, self-learning can be applied to teach a model to identify cancerous cells in pathological images. The model can be taught on an extensive dataset of unannotated pathology images and then fine-tuned on a smaller dataset of labelled images to improve its performance in identifying cancerous cells. Once trained, using this model new images can be analyzed, and predictions can be made about the presence of cancer in those images. This could increase the precision and effectiveness of diagnostic processes and also enable the discovery of new insights in pathology research.34,36
Another example of the application of self-supervised learning in computer-assisted pathology is to detect the severity of disease. The model can be trained on different stages of pathology images of the same disease and fine-tune on labelled data to predict the severity of the disease in new images. This can help in making the diagnosis and treatment plan more accurate.37,39 Other than this, self-supervised learning is also used in checking the quality of pathology images that will help identify images unsuitable for diagnosing the disease.40,43 Grading pathology and multiclass classification of diseases using deep learning and self-supervised multiclass classification is also one area of application of self-supervised, as indicated in the studies done by.44,50
Overall, self-supervised learning can be a powerful tool for computers, enabling the creation of models that can produce precise predictions based on the analysis of vast amounts of data about the presence and severity of diseases in pathology images, thus improving the diagnostic process and enabling new insights into pathology research. Table 2 summarizes applications of self-supervised learning with the disease being applied to and the technique/method being used.
Table 2: Methods used for Applications of Unsupervised pre-training
| Application | Disease | Method | Article |
| Cancer cell identification | Identifying cancerous cells in pathology images of breast cancer | Convolutional Neural Networks (CNN), Self-supervised pre-training | [34], [35], [36] |
| Disease severity prediction | Predicting the severity of Alzheimer’s disease in pathology images of brain tissue | Recurrent Neural Networks (RNN), Self-supervised representation learning | [37], [38], [39] |
| Organ or tissue segmentation | Segmenting different organs or tissues in pathology images of lungs | U-Net, Self-supervised instance segmentation | [32], [51], [52], [53] |
| Anomaly detection | Detecting abnormal cells, tissues or patterns in pathology images of liver | Autoencoders, Self-supervised anomaly detection | [29], [30], [31] |
| Grading of pathology | Classifying pathology images of skin lesions as melanoma or non-melanoma | Deep learning, Self-supervised multi-class classification | [44], [45], [46], [47] |
| Multi-class classification | Classifying pathology images of skin lesions as melanoma or non-melanoma | Deep learning, Self-supervised multi-class classification | [47], [48], [49] |
Results
Based on Longitudinal Self-Supervised Learning
Longitudinal SSL is a machine learning approach designed for scenarios involving untagged longitudinal data, especially in the context of disease forecasting. Longitudinal Unsupervised pre-training is a method for studying from untagged longitudinal data, effective when there is a large number of image sets from the same subject over time, and the desired downstream task (e.g., disease forecasting) changes over time.54 Several research have been done using longitudinal self-supervised learning. This method is effective in scenarios where the data captures the progression of a disease over time. By continuously extracting meaningful features and temporal dependencies from untagged data.
Longitudinal scans can be used to train a CNN without labelled data. By using MRI images, the network can be trained with a contrastive loss, which is instrumental in guiding the model to embed similar longitudinal instances closely together while pushing dissimilar instances which means to identify scans from the same individual and a classification loss which plays a role when the self-supervised task involves predicting specific temporal aspects or attributes within the longitudinal data, in general to predict vertebral levels.55 Figure 2 Shows the structure of the LSSL Network. In which encoder networks are represented by orange blocks and decoder networks by blue blocks.56 Both encoders and decoders capture temporal dependencies and learn representation. The encoder’s role is transforming the input data into meaningful latent space representation. On the other hand, the decoder in tandem with encoder aiming to reconstruct the original input sequence. Its task is to produce an output that is similar to the input.57
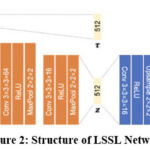 |
Figure 2: Structure of LSSL NetworkClick here to view Figure |
Based on contrastive learning
Contrastive learning is a machine learning method that learns comparable and unrelated information in a dataset. It can also be viewed as a classification method that sorts data based on similarity.58
Medical image interpretation using trained ML models often outperforms specialists using relevant data. A self-supervised model trained with chest X-rays without annotations performs similarly to radiologists in pathology classification validated on external chest X-ray dataset.59, 60
The unsupervised pre-training job’s objective is to investigate a representation that delineates the target classes in the feature space, ensuring that the predicted logits are very confidently assigned to the right class. According to equation (1), by lowering the negative log-probability of the supplied predicted class probabilities, the NT-Xent loss function promotes this. The Normalized Temperature-scaled Cross Entropy Loss, also called the NT-Xent, is a loss function applied in self-supervised learning. As given in Equation 1, the cosine similarity, a gauge of how similar two vectors are, forms the basis of the loss function. The cosine similarity is calculated from two vectors, u and v, by dividing the dot product of the two vectors by the product of their magnitudes. This can be mathematically stated as sim(u, v) = uT v / ||u|||||v||. Then
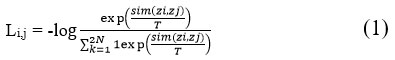
where T indicates a temperature parameter and 1[k≠i]∈{0,1} is an indicator function that evaluates to 1 iff k ≠ i. The final loss is calculated in a mini batch over all positive pairs, including (I, j) and (j, I).
CS-CO is a mixed unsupervised pre-training technique for histopathological image representation learning. It is a novel approach that distinguishes itself by integrating both generative discriminative and discriminative models. In this approach the generative model focuses on generating diverse augmented views of input data and the discriminative model is responsible for comparing the augmented views and this approach does not require human supervision.61 It includes cross-stain (CS) and contrastive learning (CO) and uses a technique called stain vector discomposure to promote contrastive learning technique. This method is domain-specific, requires no additional input, and is adaptable. WSI classification, with only slide-level labels, becomes a multiple instance learning (MIL) issue.62
Autonomous MM detection system in the eyelid with minimal annotation using ResNet50 and the PCam dataset. AUC 0.981, accuracy 90.9%, sensitivity 85.2%, specificity 96.3% for patch-level categorization of ZJU-2 dataset. AUC 0.974, accuracy 93.8%, sensitivity 75%, specificity 100% for WSI-level diagnosis. Heatmap for malignancy likelihood for each WSI.63
Improved self-supervised models using primary site data from 6000+ WSIs from TCGA. The self-supervised model outperforms the fully supervised model in detecting 3/8 pathologies in the external chest X-ray dataset. The SASSL model uses domain-adversarial training for robust detection across multiple tasks, datasets, and institutions.45, 64
Figure 3 shows an overview contrastive learning method, it utilizes a consistent CNN block shared among image patches, in which each image is augmented twice. The features are taken using CNN and transformed into a lower dimensional space by the MLP. The objective of this pretraining is to bring together augmented image patches from the same source in the feature space emphasizing their similarity, while pushing apart image patches from different sources.65
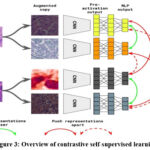 |
Figure 3: Overview of contrastive self-supervised learningClick here to view Figure |
Based on Generative adversarial network
A type of learning known as “adversarial learning” involves putting different networks into competition. The Generative Adversarial Network is the form in which adversarial learning is often applied.
To include semantic guidance into a GAN (generative adversarial network)-based framework for stain normalization and maintain intricate structural details, integrates semantic information between a semantic network that has already been trained and a network that normalizes stain colours at multiple levels.66
Equation 2, shows the generator loss, which can evaluate how well the generator network performs in providing instances that are representative of the actual data.67
![]()
where:
The generator network, or G
The discriminator network is D.
A random noise vector called z is fed into the generator as input.
The expectation operator is shown by E.
Discriminator loss measures (Equation 3) the performance of the discriminator network in correctly classifying the actual data and the produced samples, and can be represented as:
![]()
where x is actual data.
The sum of the generator and discriminator losses is the overall loss function for a GAN as shown in the following equation, Equation 4:
![]()
The goal is to find the Nash equilibrium of this minimax game, in which the discriminator attempts to categorize the actual and created samples, and the generator accurately strives to produce samples that are indistinguishable from actual data. In adversarial learning, the process establishes a competitive dynamic in which the generator seeks to produce increasingly realistic data to deceive the discriminator, while the discriminator’s goal is to improve its ability to identify between real and generated data.68
Table 3 provides summary of the techniques used in most of the papers reviewed in this study, the datasets used in those articles and the accuracy of their findings or techniques. As stated in the table, using the longitudinal self-supervised learning technique on datasets of NRG Oncology, ADNI dataset and chest CT dataset is done by.57,69,70. Using self-supervised contrastive learning techniques, a number of research have been done on different datasets and diseases some of them are on padChest dataset, PCam & ZJU-2, (TCGA), NCT-CRC, Bioimaging and Databiox, TCGA-CRC, INbreast and CBIS-DDSM by.71,73
Table 3: Techniques of self-supervised learning
| Technique | Paper | Dataset | Accuracy |
| Self-supervised SSL | [69] | NRG Oncology | 97.6% |
| Self-supervised SSL | [57] | ADNI dataset | 87% |
| Self-supervised SSL | [70] | chest CT dataset | 91% |
| Self-supervised Contrastive learning | [60] | PadChest test dataset | 95% |
| Self-supervised Contrastive learning | [63] | PCam & ZJU-2 | 90.9% & 97.4% |
| Self-supervised Contrastive learning | [45] | The Cancer Genome Atlas Program (TCGA) | 88% |
| Self-supervised Contrastive learning | [71] | NCT-CRC | AUC 0.99 |
| Self-supervised Contrastive learning | [72] | Bioimaging andDatabiox | 94.28% and80.44% |
| Self-supervised Contrastive learning | [73] | TCGA-CRC | 0.87 of AUROC |
| Self-supervised discriminative learning | [74] | NCT-CRC-HE-100K | 82.9% |
| Self-supervised pretext Training | [75] | (PPMI) | 87.50% |
| Self-supervised | [76] | TCGA | AUCs of 0.97 |
| Partial self-supervised | [77] | Colorectal Adenocarcinoma Gland (CRAG) | 93.91 |
| Self-supervised learning | [78] | MoNuSeg dataset | AJI of 70.63% |
| Self-supervised learning | [79] | TCGA | 89% |
The other technique mentioned in this paper is the self-supervised discriminative technique. Many research works have been done using discriminative techniques, one of them done by.74 Using pretext learning technique which is also one of the self-supervised learning techniques several works have been done to mention some which is done by.75 Using partial self-supervised in which full self-supervision is not done,77 and full self-supervised learning is applied is the following research works.76, 78, 79
Based on Context restoration
Context restoration is a vital concept in self-supervised learning, emphasizing the preservation of contextual information during training a neural network. This method is straightforward and operates by randomly selecting two distinct tiny patches from the target image and then changing the context of those patches. To keep the intensity distribution but change the spatial information, repeat these steps a total of T times.80
Figure 4 shows the generic CNN architecture of self-supervised learning for context restoration. The blue, green, and orange strides in the figure stand for convolutional, down-sampling, and up-sampling units, respectively. CNN structures in the reconstruction phase could change depending on the type of ensuing work. Simple designs, like certain deconvolution layers in the second row, are recommended for further classification tasks. The complicated structures (1st row) compatible with the segmentation CNNs are selected for further segmentation jobs.81
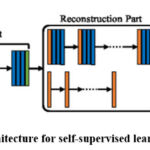 |
Figure 4: General CNN architecture for self-supervised learning with context restorationClick here to view Figure |
The context restoration technique effectively restores semantic image features and has broad applicability in image analysis. It is easy to implement and has been shown to excel in classifying, localizing and segmenting images, such as detecting scan planes in fetal ultrasound, localizing organs in CT, and segmenting brain tumors in MR images.82
Based on Learning Pretext Task and Discriminative leaning
The term “Pretext Training” refers to the process of training a model for a task different from the one for which it will ultimately be taught and deployed. This Pretext Training is carried out before the model is put through its training. Because of this, Pretext Training can also be referred to as Pre-Training in some circles.
Discriminative learning uses encoders to group similar instances and separate dissimilar ones by predicting anatomical placements as features, eliminating manual annotation. Five annotated subjects increased the mean Dice metric from 0.811 to 0.852 for image segmentation compared to U-net. A generative model was built to expand the visual field and was evaluated on CAMELYON17 and CRC datasets, outperforming self-supervised and pre-trained digital pathology methods.83
Categorization of Parkinson’s disease (PD) involves feeding successive frames of preprocessed magnetic resonance imaging (MRI) data into the classifier module of 3D ResNet18. In particular, the attention mechanism is utilized in order to study the distinctive qualities. A regression task is created, as well as a self-supervised learning pretext task, to make training easier and strengthen the validity of the suggested model respectively. The results of the experiments show that the strategy that was suggested has achieved an accuracy of 87.50 percent when classifying PD, exceeding the most advanced deep learning systems.75
Discussion
Self-supervised learning in digital pathology disease detection faces some current challenges, including:
Limited annotated data: Digital pathology images are often limited in annotated data, making it challenging for self-supervised models to learn effectively.84 To solve this problem methods like jigsaw which was introduced to help children learn as a pretext.85
Heterogeneity of tissue: Digital pathology images can have high variability in tissue appearance, texture, and structure, leading to difficulties in developing generalizable models.86
High dimensional data: Digital pathology images can have high resolution and multiple channels, making them computationally intensive to process and requiring large amounts of memory and computing power.87
Inter-observer variability: Annotating digital pathology images can be subject to inter-observer variability, leading to potential annotation inaccuracies and affecting the performance of self-supervised models.88
Robustness to artefacts: Digital pathology images can contain various artefacts, such as staining variations or tissue folding, that can influence the functioning of self-supervised techniques.89, 90
The studies have limitations, such as the need for a large and diverse dataset to represent a variety of genres, topics and the reliance on ground-truth labels, which can be unavailable or expensive to acquire. Further research is needed to address these limitations and improve the effectiveness of self-supervised learning in digital pathology disease detection.
Conclusion
Machine learning, especially deep learning, needs a vast quantity of high-quality labelled images for a better performance of the models. Based on the papers or articles reviewed, the availability of annotated or labelled data/images is a big problem or difficulty that researchers face. Considering self-supervised learning methods will alleviate the difficulty in annotated data.
Self-supervised learning presents a promising avenue for overcoming challenges in digital pathology, particularly in scenarios with limited annotated data, leveraging techniques such as pretext task, contrastive learning and LSSL. The field has witnessed advancement in disease detection and classification.
The application of self-supervised learning extends across different domains in digital pathology such as detection of abnormal cells, tissue segmentation and disease severity. However, it is not without its limitations or challenges. Issues like limited annotated data, tissue heterogeneity and others present ongoing hurdles that need further exploration. As the field grows addressing the mentioned challenges is pivotal for unlocking its full potential.
This review covers self-supervised learning methods, which have recently been done in the area of analysis of medical images, specifically on digital histopathology. The review is classified based on the categories of contrastive learning, Longitudinal Self-Supervised Learning, discriminative learning, Generative adversarial network, Context restoration and Learning Pretext Task.
Acknowledgement
The authors would like to express their gratitude to Maharishi Markandeshwar (Deemed to be University), Mullana-Ambala, for supporting and facilitating this research work.
Funding Sources
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
The author(s) do not have any conflict of interest.
Data Availability Statement
This statement does not apply to this article.
Ethics Statement
This research did not involve human participants, animal subjects, or any material that requires ethical approval.
Informed Consent Statement
This study did not involve human participants, and therefore, informed consent was not required.
Clinical Trial Registration
This research does not involve any clinical trials
Author Contributions
Conceptualization & Methodology: Abdulahi Mahammed Adem
Visualization & Supervision: Ravi Kant and Sonia
Analysis and Writing – Original Draft: Pankaj Jain, Karan Kumar and Kapil Joshi
Visualization & Reviewing: Vikas Mittal
All authors made a significant and equal contribution to this work.
References
- Gazda M, Plavka J, Gazda J and Drotar P. Self-supervised deep convolutional neural network for chest X-ray classification. IEEE Access, 2021; 9:151972-151982.
CrossRef - Rawat S, Solomon D.D, Kanwar K, Garg S, Kumar K, Mijwil M.M and Beňova E. Indian sign language recognition system for interrogative words using deep learning. In International Conference on Advances in Communication Technology and Computer Engineering, 2023; 383-397.
CrossRef - Gunduz H. Deep learning-based Parkinson’s disease classification using vocal feature sets. IEEE Access, 2019; 7:115540-115551.
CrossRef - Zhao L, Luo W, Liao Q, Chen S and Wu J. Hyperspectral image classification with contrastive self-supervised learning under limited labeled samples. IEEE Geosci Remote Sens Lett, 2022; 19: 1-5.
CrossRef - Luo Y, Pan J, Fan S, Du Z and Zhang G. Retinal image classification by self-supervised fuzzy clustering network. IEEE Access, 2020; 8: 92352-92362.
CrossRef - Wang D, Pang N, Wang Y and Zhao H. Unlabeled skin lesion classification by self-supervised topology clustering network. Biomed Signal Process Control, 2021; 66, 102428.
CrossRef - Aggarwal K, Bhamrah M.S and Ryait H.S. The identification of liver cirrhosis with modified LBP grayscaling and Otsu binarization. SpringerPlus, 2016; 5:1-15.
CrossRef - Srinidhi C.L, Kim S.W, der Chen F and Martel A.L. Self-supervised driven consistency training for annotation efficient histopathology image analysis. Med Image Anal., 2022; 75:102256.
CrossRef - Alsharef A, Sonia, Arora M and Aggarwal K. Predicting time-series data using linear and deep learning models—an experimental study. In: Data, Engineering and Applications: Proceedings of IDEA 2021. Singapore: Springer Nature Singapore; 2022;505-516.
CrossRef - Sarker I.H. Deep cybersecurity: A comprehensive overview from neural network and deep learning perspective. SN Comput Sci., 2021; 2(3), 154.
CrossRef - Mehrotra R, Ansari M.A, Agrawal R and Anand RS. A transfer learning approach for AI-based classification of brain tumors. Mach Learn Appl., 2020; 2, 100003.
CrossRef - Abbet C, Zlobec I, Bozorgtabar B and Thiran J.P. Divide-and-rule: Self-supervised learning for survival analysis in colorectal cancer. Lect Notes Comput Sci. 2020; 12265: 480-489.
CrossRef - Güldenring R, Nalpantidis L. Self-supervised contrastive learning on agricultural images. Comput Electron Agric., 2021; 191, 106510.
CrossRef - Abbood ZA, Khaleel I and Aggarwal K. Challenges and future directions for intrusion detection systems based on AutoML. Mesopotamian J CyberSecurity, 2021; 16-21.
CrossRef - Wang Q, Chen K, Dou W and Ma Y. Cross-attention based multi-resolution feature fusion model for self-supervised cervical OCT image classification. IEEE/ACM Trans Comput Biol Bioinform., 2023; 1-14.
CrossRef - Solomon DD, Sonia, Kumar K, Kanwar K, Iyer S and Kumar M. Extensive review on the role of machine learning for multifactorial genetic disorders prediction. Archives of Computational Methods in Engineering, 2024; 31(2): 623-40.
CrossRef - Ashrafi Fashi P. A. self-supervised contrastive learning approach for whole slide image representation in digital pathology. 2022. Accessed 2023.
- Chen C, Lu M.Y, Williamson D.F.K, Chen T.Y, Schaumberg A.J and Mahmood F. Fast and scalable search of whole-slide images via self-supervised deep learning. Nat Biomed Eng., 2022; 6(12): 1420-1434.
CrossRef - Bashab A, Ibrahim A.O, Tarigo Hashem I.A, et al. Optimization techniques in university timetabling problem: Constraints, methodologies, benchmarks, and open issues. Comput Mater Continua., 2023; 74(3): 1-10.
CrossRef - Gao W, Wu M, Lam S. K, Xia Q and Zou J. Decoupled self-supervised label augmentation for fully-supervised image classification. Knowl Based Syst., 2022; 235: 107605.
CrossRef - Pöppelbaum J, Chadha G.S and Schwung A. Contrastive learning-based self-supervised time-series analysis. Appl Soft Comput., 2022; 117:108397.
CrossRef - Wei C.T, Hsieh M.E, Liu C.L and Tseng V.S. Contrastive Heartbeats: Contrastive learning for self-supervised ECG representation and phenotyping. ICASSP IEEE Int Conf Acoust Speech Signal Process Proc. 2022; 2022:1126-1130.
CrossRef - Tran V.N, Liu S.H, Li Y.H and Wang J.C. Heuristic attention representation learning for self-supervised pretraining. Sensors. 2022; 22(14): 5169.
CrossRef - Rani P, Lamba R, Sachdeva R.K, Kumar K and Iwendi C. A machine-learning model for Alzheimer’s disease prediction. IET Cyber Phys Syst Theory Appl. 2024;9(2):125-134.
CrossRef - Anand D, Annangi P and Sudhakar P. Benchmarking self-supervised representation learning from a million cardiac ultrasound images. Proc Annu Int Conf IEEE Eng Med Biol Soc EMBC. 2022;2022: 529-532
CrossRef - Ciga O, Xu T and Martel A.L. Self-supervised contrastive learning for digital histopathology. Mach Learn Appl. 2022; 7: 100198.
CrossRef - Yang P, Hong Z, Yin X, Zhu C and Jiang R. Self-supervised visual representation learning for histopathological images. Lect Notes Comput Sci. 2021;12902: 47-57.
CrossRef - Zhang R, Isola P and Efros A.A. Colorful image colorization. Lect Notes Comput Sci. 2016; 9907: 649-666.
CrossRef - Bogatinovski J, Nedelkoski S, Cardoso J and Kao O. Self-supervised anomaly detection from distributed traces. Proc IEEE ACM Int Conf Util Cloud Comput UCC. 2020; 2020: 342-347.
CrossRef - Liu Y, Li Z, Pan S, Gong C, Zhou C and Karypis G. Anomaly detection on attributed networks via contrastive self-supervised learning. IEEE Trans Neural Netw Learn Syst. 2022; 33(6): 2378-2392.
CrossRef - Zhang X, Mu J, Zhang X, Liu H, Zong L and Li Y. Deep anomaly detection with self-supervised learning and adversarial training. Pattern Recognit., 2022; 121: 108234.
CrossRef - Wang B, Li Q and You Z. Self-supervised learning based transformer and convolution hybrid network for one-shot organ segmentation. Neurocomputing. 2023;527: 1-12.
CrossRef - Taleb A, Rohrer C, Bergner B, De Leon G, Rodrigues JA, Schwendicke F, Lippert C and Krois J. Self-supervised learning methods for label-efficient dental caries classification. Diagnostics. 2022;12(5): 1237.
CrossRef - Özen Y, Aksoy S, Kösemehmetoğlu K, Önder S and Üner A. Self-supervised learning with graph neural networks for region of interest retrieval in histopathology. Proc Int Conf Pattern Recognit. 2020; 6329-6334.
CrossRef - Li T, Feng M, Wang Y and Xu K. Whole slide images based cervical cancer classification using self-supervised learning and multiple instance learning. Proc IEEE Int Conf Big Data Artif Intell Internet Things Eng (ICBAIE). 2021;192-195.
CrossRef - Fan L, Sowmya A, Meijering E and Song Y. Cancer survival prediction from whole slide images with self-supervised learning and slide consistency. IEEE Trans Med Imaging. 2022; 42(5): 1401-12.
CrossRef - Fang F, Yao Y, Zhou T, Xie G and Lu J. Self-supervised multi-modal hybrid fusion network for brain tumor segmentation. IEEE J Biomed Health Inform. 2021; 26(11): 5310-20.
CrossRef - Hu C, Li C, Wang H. Self-supervised learning for MRI reconstruction with a parallel network training framework. Lect Notes Comput Sci. 2021; 12906:382-391.
CrossRef - Ouyang J, Zhao Q, Adeli E, Sullivan EV, Pfefferbaum A, Zaharchuk G and Pohl K.M. Self-supervised longitudinal neighbourhood embedding. Lect Notes Comput Sci. 2021; 12902: 80-89.
CrossRef - Haghighi F, Ghosh S, Ngu H, et al. Self-supervised learning for segmentation and quantification of dopamine neurons in Parkinson’s disease. arXiv. 2023.08141.
- Li B, Keikhosravi A, Loeffler A.G and Eliceiri K.W. Single image super-resolution for whole slide image using convolutional neural networks and self-supervised color normalization. Med Image Anal. 2021;68: 101938.
CrossRef - Baykaner K, Xu M, Bordeaux L, et al. Image model embeddings for digital pathology and drug development via self-supervised learning. bioRxiv. 2021; 2021-09.
CrossRef - Chen C, Lu MY, Williamson DF, Chen TY, Schaumberg AJ, Mahmood F. Fast and scalable search of whole-slide images via self-supervised deep learning. Nature Biomedical Engineering. 2022; 6(12):1420-34.
CrossRef - Grullon S, Spurrier V, Zhao J, Chivers C, Jiang Y, Motaparthi K, Lee J, Bonham M and Ianni J. Using Whole Slide Image Representations from Self-supervised Contrastive Learning for Melanoma Concordance Regression. In European Conference on Computer Vision 2022; 442-456.
CrossRef - Fashi P.A, Hemati S, Babaie M, Gonzalez R and Tizhoosh H.R. A self-supervised contrastive learning approach for whole slide image representation in digital pathology. J Pathol Inform. 2022;13: 100133.
CrossRef - Chen R.J. and Krishnan R.G. Self-supervised vision transformers learn visual concepts in histopathology. arXiv. 2022.
- Aryal M and Soltani N.Y. Context-aware graph-based self-supervised learning of whole slide images. Proc IEEE Int Conf Acoust Speech Signal Process (ICASSP). 2022; 3553-3557.
CrossRef - Kwasigroch A, Grochowski M, Mikołajczyk A. Self-supervised learning to increase the performance of skin lesion classification. Electronics. 2020; 9(11): 1930.
CrossRef - Sriram A. COVID-19 prognosis via self-supervised representation learning and multi-image prediction. arXiv. 2021.
- Hayashi T and Fujita H. OCFSP: Self-supervised one-class classification approach using feature-slide prediction subtask for feature data. Soft Comput. 2022;26(19):10127-10149.
CrossRef - Taleb A, Lippert C, Klein T, Nabi M. Multimodal self-supervised learning for medical image analysis. Lect Notes Comput Sci. 2021;12729: 661–673.
CrossRef - Haghighi F, Taher MRH, Zhou Z, Gotway MB, Liang J. Transferable visual words: Exploiting the semantics of anatomical patterns for self-supervised learning. IEEE Trans Med Imaging. 2021; 40(10): 2857–2868.
CrossRef - Tang YT. Self-supervised pre-training of Swin transformers for 3D medical image analysis. Published online 2022. Accessed 2023. Available at: https://www.synapse.org/#!Synapse:syn3193805/
CrossRef - Ouyang J. Self-supervised longitudinal neighbourhood embedding. Lect Notes Comput Sci. 2021; 12902: 80–89.
CrossRef - Jamaludin A, Kadir T, Zisserman A. Self-supervised learning for spinal MRIs. Lect Notes Comput Sci. 2017; 10553: 294–302.
CrossRef - Zhao Q, Liu Z, Adeli E, Pohl KM. Longitudinal self-supervised learning. Med Image Anal. 2021; 71: 102051.
CrossRef - Ke J, Shen Y, Liang X, Shen D. Contrastive learning-based stain normalization across multiple tumor types in histopathology. Lect Notes Comput Sci. 2021; 12908: 571–580.
CrossRef - Ciga O, Xu T, Martel AL. Self-supervised contrastive learning for digital histopathology. Mach Learn Appl. 2022; 7: 100198.
CrossRef - Tiu E, Talius E, Patel P, Langlotz CP, Ng AY, Rajpurkar P. Expert-level detection of pathologies from unannotated chest X-ray images via self-supervised learning. Nat Biomed Eng. 2022; 6(12): 1399-1406.
CrossRef - Yang P, Hong Z, Yin X, Zhu C, Jiang R. Self-supervised visual representation learning for histopathological images. Lect Notes Comput Sci. 2021;12902: 47–57.
CrossRef - Yang P, Yin X, Lu H. CS-CO: A hybrid self-supervised visual representation learning method for H&E-stained histopathological images. Med Image Anal. 2022;81: 102539.
CrossRef - Wang L, Self-supervised learning mechanism for identification of eyelid malignant melanoma in pathologic slides with limited annotation. Front Med (Lausanne). 2022; 9: 2761.
CrossRef - Ye H-L, Wang D-H. Stain-adaptive self-supervised learning for histopathology image analysis. 2022.
- Mahapatra D, Bozorgtabar B, Thiran JP, Shao L. Structure-preserving stain normalization of histopathology images using self-supervised semantic guidance. Lect Notes Comput Sci. 2020; 12265: 309–319.
CrossRef - Goodfellow I.J. Generative adversarial networks. Advances in neural information processing systems, 2014; 27.
CrossRef - Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: A review. Med Image Anal. 2019;58: 101552.
CrossRef - Esteva A. Prostate cancer therapy personalization via multi-modal deep learning on randomized phase III clinical trials. NPJ Digit Med. 2022; 5(1): 1–8.
CrossRef - Yan K. SAM: Self-supervised learning of pixel-wise anatomical embeddings in radiological images. IEEE Trans Med Imaging. 2022; 41(10):2658–2669.
CrossRef - Yan J, Chen H, Li X, Yao J. Deep contrastive learning-based tissue clustering for annotation-free histopathology image analysis. Comput Med Imaging Graph. 2022; 97: 102053.
CrossRef - Gong R. Self-distilled supervised contrastive learning for diagnosis of breast cancers with histopathological images. Comput Biol Med. 2022; 146: 105641.
CrossRef - Schirris Y, Gavves E, Nederlof I, Horlings HM, Teuwen J. DeepSMILE: Contrastive self-supervised pre-training benefits MSI and HRD classification directly from H&E whole-slide images in colorectal and breast cancer. Med Image Anal. 2022; 79: 102464.
CrossRef - Li J, Lin T, Xu Y. SSLP: Spatial guided self-supervised learning on pathological images. Lect Notes Comput Sci. 2021; 12902: 3–12.
CrossRef - Zhang Y, Lei H, Huang Z. Parkinson’s disease classification with self-supervised learning and attention mechanism. In: 2022 26th International Conference on Pattern Recognition (ICPR). IEEE; 2022; 4601-4607.
CrossRef - Saillard C, Dehaene O, Marchand T. Self-supervised learning improves dMMR/MSI detection from histology slides across multiple cancers. arXiv preprint arXiv:2109.05819. 2021.
- Bukhari SUK, Syed A, Bokhari SKA. The histological diagnosis of colonic adenocarcinoma by applying partial self-supervised learning. MedRxiv. 2020; 2020-08.
CrossRef - Xie X, Chen J, Li Y, Instance-aware self-supervised learning for nuclei segmentation. In: Medical Image Computing and Computer Assisted Intervention – MICCAI 2020: 23rd International Conference, Lima, Peru, 2020, Proceedings, Part V. Springer International Publishing; 2020; 341-350.
CrossRef - Lai ZF, Zhang G, Zhang XB, Liu HT. High-resolution histopathological image classification model based on fused heterogeneous networks with self-supervised feature representation. BioMed Res Int. 2022; 2022(1): 8007713.
CrossRef - Chen L, Bentley P, Mori K, Misawa K, Fujiwara M, Rueckert D. Self-supervised learning for medical image analysis using image context restoration. Med Image Anal. 2019; 58: 101539.
CrossRef - Bai W, Chen C, Tarroni G. Self-supervised learning for cardiac MR image segmentation by anatomical position prediction. In: Medical Image Computing and Computer Assisted Intervention – MICCAI 2019: 22nd International Conference, Shenzhen, China, 2019, Proceedings, Part II. Springer International Publishing; 2019; 541-549.
CrossRef - Koohbanani NA, Unnikrishnan B, Khurram SA, Krishnaswamy P, Rajpoot N. Self-path: Self-supervision for classification of pathology images with limited annotations. IEEE Trans Med Imaging. 2021; 40(10): 2845-2856.
CrossRef - Noroozi M, Favaro P. Unsupervised learning of visual representations by solving jigsaw puzzles. Lect Notes Comput Sci. 2016; 9910: 69–84.
CrossRef - Al-Thelaya K, Gilal NU, Alzubaidi M. Applications of discriminative and deep learning feature extraction methods for whole slide image analysis: A survey. J Pathol Inform. 2023; 14:100335.
CrossRef - Meirelles ALS, Kurc T, Kong J, Ferreira R, Saltz JH, Teodoro G. Building efficient CNN architectures for histopathology images analysis: A case-study in tumor-infiltrating lymphocytes classification. Front Med (Lausanne). 2022; 9:894430.
CrossRef - Wong ANN, He Z, Leung K.L. Current developments of artificial intelligence in digital pathology and its future clinical applications in gastrointestinal cancers. Cancers (Basel). 2022; 14(15): 3780.
CrossRef - Patil A, Diwakar H, Sawant J. Efficient quality control of whole slide pathology images with human-in-the-loop training. J Pathol Inform. 2023; 14:100306.
CrossRef - Brixtel R, Bougleux S, Lézoray O. Whole slide image quality in digital pathology: Review and perspectives. IEEE Access. 2022;10: 131005–131035.
CrossRef - Bhakuni M, Kumar K, Sonia, Iwendi C, Singh A. Evolution and evaluation: Sarcasm analysis for twitter data using sentiment analysis. Journal of Sensors. 2022;2022(1):6287559.
CrossRef - Alsharef A, Aggarwal K, Sonia, Kumar M, Mishra A. Review of ML and AutoML solutions to forecast time-series data. Archives of Computational Methods in Engineering. 2022;29(7): 5297-311.
CrossRef - Alsharef A, Kumar K, Iwendi C. Time series data modeling using advanced machine learning and AutoML. Sustainability. 2022;17:14(22): 15292.
CrossRef - Verma R, Garg S, Kumar K, Gupta G, Salehi W, Pareek PK, Kniežova J. New Approach of Artificial Intelligence in Digital Forensic Investigation: A Literature Review. In International Conference on Advances in Communication Technology and Computer Engineering 2023; 399-409. Cham: Springer Nature Switzerland.
CrossRef
Abbreviations
CNN: Convolutional Network







