Manuscript accepted on :01-05-2024
Published online on: 31-07-2024
Plagiarism Check: Yes
Reviewed by: Dr. Narasimha Murthy
Second Review by: Dr. Grigorios Kyriakopoulo
Final Approval by: Dr. Prabhishek Singh
Ajay Krishan Gairola 1,2* , Vidit Kumar2
, Vidit Kumar2 and Ashok Kumar Sahoo1
and Ashok Kumar Sahoo1
1Department of Computer Science and Engineering, Graphic Era Hill University, Dehradun, India.
2Department, of Computer Science and Engineering, Graphic Era deemed to be university, Dehradun, India.
Corresponding Author E-mail:ajaykrishangairola@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/2976
Abstract
The skin of an individual serves as the primary defense mechanism for safe guarding vital organs in the body. Although this barrier effectively protects internal organs from a variety of threats, it is still prone to damage from viral, fungal, or dust-related illnesses. Even minor skin injuries possess the potential to escalate into more severe and hazardous conditions. A prompt and precise skin disease diagnosis becomes crucial in expediting the healing process for individuals grappling with skin-related issues. The objective of this study is to develop a system based on Convolutional Neural Network (CNN) that can accurately identify various skin diseases. The proposed architecture, known as TFFNet (Two-Stream Feature Fusion Network), integrates two simultaneous modules featuring a Self-Attention (SA) block. We employ Self Attention-Convolutional Neural Networks (SACNNs) and Depthwise Separable Convolution (DWSC) to establish a diagnostic system for skin diseases. In this method, two separate CNN models are joined together, and two parallel modules (M1 and M2) are added. This greatly reduces the total number of trainable parameters. In comparison to other deep learning methods outlined in existing literature, the proposed CNN exhibits a notably lower number of learned parameters, specifically around 7 million for classification purposes. The skin disease classification was carried out on three datasets—ISIC2016, ISIC2017, and HAM10000. The model achieved testing accuracies of 89.70%, 90.52%, and 90.12% on each respective dataset.
Keywords
Convolution Layer; Deep Learning; Feature Fusions; Image Classification; Self-Attention; Skin Disease
Download this article as:| Copy the following to cite this article: Gairola A. K, Kumar V, Sahoo A. K. Enhancing Skin Disease Diagnosis with TFFNet: A Two-Stream Feature Fusion Network Integrating CNNs and Self Attention Block. Biomed Pharmacol J 2024;17(3). |
| Copy the following to cite this URL: Gairola A. K, Kumar V, Sahoo A. K. Enhancing Skin Disease Diagnosis with TFFNet: A Two-Stream Feature Fusion Network Integrating CNNs and Self Attention Block. Biomed Pharmacol J 2024;17(3). Available from: https://bit.ly/3LLYYFb |
Introduction
Melanoma, a highly aggressive skin cancer, accounts for only 1% of skin cancers, yet it is the leading cause of death 1. Computer-aided methods for skin cancer detection are necessary due to a shortage of dermatologists per capita. The American Cancer Society predicts 99,780 new melanoma cases (57,180 men and 42,600 women) and 7,650 deaths (5,080 men and 2,570 women) in 2022. As the field of computer vision and Artificial Intelligence has advanced, image analysis has become increasingly useful in a wide range of scene-parsing applications. Computer-assisted diagnosis and detection heavily rely on medical image analytics 2. Early disease detection and diagnosis are major challenges in healthcare. Only then can appropriate therapy begin. Millions of individuals around the world are affected by skin diseases today, which can be detrimental to both personal health and national economies if not addressed promptly 3. In 2021, according to the American Cancer Society, 7,180 individuals died from melanoma. Additionally, the American Cancer Society predicted in their 2022 annual report that there would be roughly 99,780 new instances of skin disease (melanoma), with an expected death rate of 7,650 people 4. Diseases that produce itching or pain, on the other hand, might lead to substantial damage and deformation. Damage to the skin from these disorders can also affect a person’s sense of well-being and confidence 5. The common perception is that some skin diseases are rather harmless. However, the vast majority of sufferers opt to treat their skin issues on their own. Medications for skin diseases can worsen the condition if they are not effective against the underlying cause. Perhaps the individual is unaware of the severity of their skin issue 6.
Examining dermoscopic images is the best standard for identifying skin diseases. Dermatologists utilize various dermoscopic tools, including the pigment network, dots/globules, and color regression, to make diagnoses from dermoscopy images. However, this method has several drawbacks, such as the need for advanced dermoscopic equipment and the time and effort required to train dermatologists in using these tools 7, 8. Additionally, the inflammatory nature of skin diseases and overlapping characteristics of infectious diseases result in considerable visual variation and irregularities in the overall appearance and feel of skin lesions. Inexperienced dermatologists often struggle to identify subtle variations using their eyes alone. Recent advancements in artificial intelligence, particularly in the healthcare industry, focusing on the analysis of medical images, have made it an attractive tool for developing algorithms for medical image interpretation. This is especially true in the context of the medical industry, where machine learning networks have proven to be very useful in image analysis due to their ability to independently learn image representations. Dermatologists have a critical need for computer-aided design (CAD) systems based on innovative problem-solving approaches 9. This would not only alleviate the strain on the nation’s healthcare infrastructure but also reduce the waiting time for medical dermoscopy. Convolutional neural networks (CNNs) and other forms of deep learning have demonstrated superiority over conventional methods in human disease diagnosis. The availability of powerful computational resources has led to the continuous development of more advanced deep learning systems. Nevertheless, due to the extensive training time required, these intricate systems could occasionally be wasteful. Due to their ability to achieve accurate results with fewer parameters and less effort, CNN models have gained popularity. In this study, the MobileNetV2 and NASNetMobile backbone architectures are employed to categorize skin diseases.
Below is the outline for the remainder of the paper. We reviewed the studies that have been conducted on the topic of skin disease classification in Section 2. The methodological approach and overall structure of the model are detailed in Section 3. In Section 4, we examine the training and validation processes for the model. The proposed work concludes with some last notes and an outline of potential future work in Section 5.
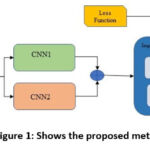 |
Figure 1: Shows the proposed method’s workflow. |
Literature Review
Skin diseases are a major health risk for humans. The diagnosis might be impacted by factors such as high sensitivity, the need for laborious laboratory procedures, considerable time investment, and intricate physical manipulation. Furthermore, the similarity among many skin lesions often leads to frequent misidentifications 10. This work aimed to construct a unified CAD model for segmenting and classifying skin lesions using a deep learning architecture. At the outset of this procedure, source dermoscopic images are pre-processed using a variant of a bio-inspired multiple exposure fusion method that emphasizes contrast enhancement. The second step is to create a bespoke CNN architecture with 26 layers specifically for the task of identifying and isolating skin lesions. Finally, four CNN models are learned from the segmented lesion images (ResNet-50, Xception, VGG16, and ResNet-101). Finally, a convolutional sparse image decomposition fusion method is used to combine the deep feature vectors that were obtained from each CNN model. In the last stage, the ideal features are chosen for classification using univariate measurements and the Poisson distribution feature selection method. The final step in the classification process involves employing a multi-class support vector machine with the selected features.
The first steps involve using a lightweight attention module to identify feature correlations, fine-tuning a pre-trained model (ResNet-50) on the HAM10000 dataset to extract latent high-level features, increasing the number of samples from underrepresented groups using synthetic minority class oversampling, and feeding those into an XGBoost model for training and prediction 11. This combination of high-level attributes and generic statistics will be employed. A hybrid network with multi-scale Gaussian difference preprocessing, dual-stream convolutional neural networks, and transformers 12 is used to reliably separate skin lesions found with dermoscopy. To cautiously improve the lesion area and edge information and eliminate noisy features like hair, three Gaussian difference convolution kernels were trained. By utilizing multi-scale Gaussian convolution, the model can effortlessly extract and incorporate edge and lesion information while simultaneously reducing noise. Secondly, for accurate alignment, a dual-stream network is employed to extract features from both the original image and the Gaussian difference image separately. Then, these features are fused in the feature space. Combining models from vision transformers with convolutional neural networks enhances data consumption on a local and global scale. Lastly, self-attention and coordination techniques are utilized to make important aspects more noticeable. A densely connected Res2Net and feature fusion attention module-based approach is proposed for gesture image recognition 13. Using dense connections and group convolution, they propose the densely connected Res2Net to improve upon Res2Net. By using SK-Net to choose features, densely connected Res2Net is made more adaptable to the receptive field. The resulting network is used to extract features from high- and low-level gesture images. The FFA was created to combine high-level and low-level features and eliminate superfluous data from features.
The AlexNet model 14 was modified by changing the activation function to detect skin tumors in the HAM10000 dataset. F-score, accuracy, and recall all reached a new high of 98.20%. To classify skin lesions, an ensemble model 15 was introduced, combining stacked ensemble techniques from Inceptionv3, Xception, DenseNet121, DenseNet201, and InceptionResNet-V2, based on fine-tuning and transfer learning. Compared to state-of-the-art approaches, the proposed model achieved a higher accuracy of 97.93%. They initiated the image classification process by introducing their innovative ESRGAN preprocessing strategy for ISIC 2018 images. Subsequently, they applied different deep learning models 16, achieving an overall accuracy of 83.2%. Notably, CNN, Resnet-50 (83.7%), InceptionV3 (85.8%), and InceptionResnet (84%) contributed to this success. The pre-trained versions of the deep learning models MobileNetV2 and DenseNet201 were improved by adding more convolution layers, which allowed for more accurate diagnosis of skin cancer 17. In the most recent iteration, both models have three convolutional layers stacked on top of one another. The approach that has been presented has the potential to distinguish between benign and malignant types. With an accuracy of 95.50 percent, the modified version of the DenseNet201 model that was proposed beats both the state-of-the-art baselines and the state-of-the-art approaches from the most recent literature review. Additionally, the enhanced sensitivity of the DenseNet201 model is 93.96%, while its specificity is 97.03%.
A lightweight model capable of accurately diagnosing skin lesions was proposed 18. Even though this results in a very small number of trainable parameters, the employment of dynamically scaled kernels in the layers is what allows for the achievement of optimal outcomes. Within the framework of the suggested paradigm, the activation functions ReLU and leaky ReLU are both put to use. The model correctly categorized every class included in HAM-10000, with a success percentage of 97.85% overall. Employing HAM-10000 19, researchers tested 11 distinct CNN models by using seven skin disease classes. They addressed the problem of imbalance and the striking similarities between images of different skin diseases by employing transfer learning, fine-tuning, and data augmentation. DenseNet169 emerged as the top-performing algorithm out of 12 different CNN architecture variants. The system achieved 92.25% accuracy, 93.59% sensitivity, and a 93.27% F1-score. The framework developed for automating the SLC process of dermoscopy is named “Dermo-Expert” 20. The preprocessing step and the convolutional stage are both included in the hybrid CNN’s processing pipeline. To develop more precise lesion feature maps, the hybrid CNN presented uses three distinct feature extractor modules. After categorizing the various feature maps using a variety of completely linked layers, the resulting maps are assembled to provide a prediction regarding the type of lesion. Lesion segmentation, augmentation (based on geometry and intensity), and class rebalancing are all components of their proposed preprocessing step for their methodology. These features include imposing a cost on the decline of every class and combining additional graphics with the underrepresented groups. After being put through its paces on the ISIC-2017, ISIC-2018, and HAM-10000 datasets, Dermo-Expert earned an AUC of 0.96, 0.95, and 0.97, respectively.
These points summarize the problems with existing studies. To begin with, most of the research currently available relies on unprocessed visualizations for the purpose of identifying skin diseases, which is inefficient and inaccurate. Second, the importance of combining multiple features for skin disease detection is often under-researched. To overcome the limitations of prior research, we propose a TFFNet (Two-Stream Feature Fusion Network) for skin disease diagnosis that can classify a wide range of skin diseases. The proposed approach takes both color and grayscale images and extracts the most relevant features from each.
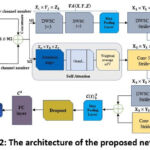 |
Figure 2: The architecture of the proposed network TFFNet |
Table 1: Fused Layers with SAConv and DWSC modules.
|
Stage |
M1 |
M2 |
stride |
Layers |
|
0 |
DWSC1 3×3 |
– |
1 |
1 |
|
1 |
DWSC2 5×5 |
– |
1 |
2 |
|
2 |
DWSC3 3×3 |
Conv1 5×5 |
2 |
4 |
|
3 |
DWSC4 5×5 |
Conv2 7×7 |
2 |
6 |
|
4 |
DWSC5 3×3 |
Conv3 5×5 |
2 |
8 |
Research gaps and motivation
In terms of skin disease identification, the following research gaps have been identified
The majority of research has been conducted using small datasets 5 6. Therefore, there is a need for analysis using large datasets to enhance the performance of trained models.
Using conventional image processing methods to identify and extract disease-specific traits from skin examinations is a daunting task.
As every skin disease has its own unique characteristics, automatic feature extraction is necessary to improve classification accuracy. However, this incurs a considerable amount of computation. Although the vast majority of deep learning models offer automated feature learning, tailored deep architectures are required to resolve the trade-off between complexity and accuracy.
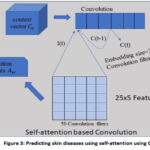 |
Figure 3: Predicting skin diseases using self-attention using CNN. |
Research contributions
The primary research contributions of the proposed effort are as follows:
This study establishes a new architecture known as “TFFNet” by modifying the conventional CNN design and creating two parallel modules: the CNN with a Self-Attention (SA) block 21 and the Depthwise Separable Convolution (DWSC) module. After implementing these changes, the overall number of trainable parameters dropped significantly. The proposed approach learned more than seven million characteristics to identify diseases, surpassing other deep learning approaches detailed in the literature.
The proposed architecture achieved good classification accuracy while utilizing minimal processing resources. In contrast to CNN models such as MobileNetV2 and NASNetMobile, these networks employ appropriate customization to address the complexity versus accuracy trade-off.
We used three datasets and 13,894 images to depict various skin diseases in this work. After developing six data pre-processing correction methods for image enhancement, we progressively merged the unique information from each modality.
Table 2: Specifics of the data set used.
|
Data-set |
Classes |
Test set |
Training set |
|
ISIC 2016 |
Benign, Malignant |
379 |
900 |
|
ISIC 2017 |
Benign nevi, Melanoma, Seborrheic keratosis |
600 |
2000 |
|
HAM10000 |
Basal cell carcinoma, Actinic keratoses, Dermatofibroma, Benign keratosis, Melanocytic nevi, Melanoma, Vascular lesions |
3004 |
7011 |
Methodology
The system’s proposed workflow is illustrated in Figure 1, providing an overview of the entire process.
Proposed network’s architecture
The system’s proposed workflow is illustrated in Figure 1, providing an overview of the entire process. Skin disease classification has recently utilized various cutting-edge CNN models. This investigation incorporates MobileNetV2 and NASNetMobile as foundational architectures. These models have demonstrated superior performance with reduced computational complexity in addressing a range of computer vision challenges compared to alternative methods. The key components include SACNNs and DWSC modules. The newly proposed network, named ‘TFFNet,’ integrates existing CNN architectures through the incorporation of two-stream feature fusion modules. Figure 2 illustrates the comprehensive design of the proposed model. In contrast to traditional CNN networks, it aims to provide accurate categorization while minimizing the number of parameters involved. Table 1 provides a breakdown of the 21 layers. Here, we will discuss the importance of each module in the proposed architecture.
DWSC Module
Initially, Sifre 22 proposed DWSC, which found application in image classification. The concept entails decomposing the convolution operation, a strategy known as DWSC. This approach converts a conventional convolution operation into a blend of depthwise separable and pointwise convolution operations. In the depthwise separable convolution process, each input channel undergoes filtration independently, and the resultant linear input channels are subsequently merged. This convolutional method substitutes a solitary convolutional layer with two distinct layers — one for spatial filtering and the other for merging purposes. Consequently, depthwise separable convolution effectively reduces both the model’s size and the parameter count. By combining depthwise separable and pointwise convolution, this method turns a regular convolution into something new. Prior to merging the linear input channels, the separable convolution procedure applies a separate filter to each channel input. Rather than using a single convolution layer, this convolutional method splits the processing into two distinct layers: one to perform spatial filtering and another to combine results. Both the size of the model and the number of parameters can be successfully reduced using depthwise separable convolution. When it comes to input feature maps, however, a typical convolution kernel just requires three parameters: the height (H), width (W), and input channel (Ic ). With Oc standing for the number of output channels, the resulting convolution layer (h × w × Ic ) is depicted as K x K x Ic x Oc. Two crucial operations are involved in depthwise separable convolution: the depthwise separable convolution operation and the pointwise convolution operation. To put it mathematically, the operation of depthwise separable convolution is:
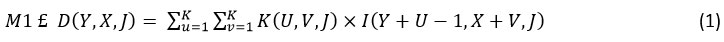
In the context of the depthwise separable convolution operation, K denotes the kernels characterized by dimensions K x K x Ic. The input feature map (I) is used to regenerate the G output feature map by applying the nth filters of the K kernels to the nth set of channels. Using pointwise convolution is a part of learning new features. In mathematical terms, this can be expressed as the following:
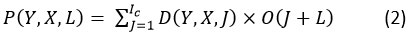
In pointwise convolution, the kernel’s dimension is K x K x Ic x Oc .
One advantage of this module (DWSC) over regular convolutions is the reduced number of parameters. Both ReLU and global average pooling are connected to the three 3 × 3 depth-wise convolutions and the two 5 × 5 DWS convolutions. Results showed that DWSC improved training speed in stages 1-3 with little overhead on parameters, while in stages 4–8, it vastly increased. An optimal trade-off between training time and parameters was achieved.
SACNN Module
Firstly, let’s establish the meanings of the terms “convocation-layer” and “SA”. Subsequently, we delve into an elucidation of the mechanics underlying the SA-CNN.
Convolution neural network
Features are extracted using the two models from both color and grayscale images. To reduce the overall cross-entropy loss between multi-label predictions, we employ a method that is co-trained by pairs of images from two different modalities. IRGB‘s loss function is denoted by LFrgb , Igray‘s by LFgray, and LFfusion‘s by Ifusion . The cross-entropy loss function is denoted by CrEnt (). We train the RGB branch, the grayscale branch, and the fusion branch jointly using a loss function that combines the below three loss functions.
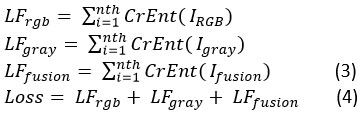
We have developed the proposed method to obtain the multi-receptive field of skin disease in order to gain insight into a wider variety of skin diseases. The multi-receptive fields are made up of nested convolution layers with different kernel sizes, like 5 × 5 and 7 × 7. In order to obtain more diseased areas multi-receptive fields are used to cover a broader area of skin disease. The lesser convolutional kernel and the larger convolutional kernel work together to train layers with varying weights that correspond to their respective receptive fields. They probe a wider diseased area, which ultimately enhances the model’s precision. In order to execute channel-integrated and non-linear processing, we combine the feature maps of all convolutions and the ReLU + 5 × 5 convolution layer. Before this, we used the 7 × 7 and 5 × 5 convolutional layers+Max pooling layer combination. The 5 × 5 convolution is sliding a filter across the image to create feature maps. ReLU is then applied to the feature maps to keep only the positive values, aiding the neural network in recognizing important patterns in the data. The filter calculates the feature map region’s average using two average and max pooling layers. Thus, max pooling returns the most prominent feature in a feature map patch, while average pooling returns the average of all features.
In the TFFNet method, the diseases identified by their characteristics are given equal weight through the use of a maximum and average pooling process. When we pool features, we start with a feature vector F and end up with a vector V. In the instance where one makes use of maximum pooling, this vector Vmp is given by:
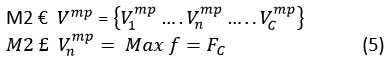
where C is the feature map’s channel count. For feature map n = (1, C), let be the set. The network produces a total of C similar feature maps as its output. All the features included in make up the f, and the mp denotes the max pooling operation. And when we consider the use of average max pooling, this vector Vavg is given by
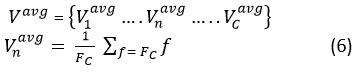
Self-attention mechanism
Employing the attention mechanism across the sequence of hidden states obtained from convolution allows us to compute the vector for convolved features. Here at the convolution layer, our model made use of the intra-layer convolution connection that we just described. Therefore, because of the convolution link, Hu hidden units will influence numerous nearby units through the employment of right context HuR and left contexts HuL, as defined as:
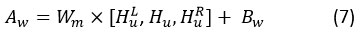
The vectors are obtained in self-attention by taking into consideration all hidden states, which are represented by convolve features. Convolutional features will yield a context vector Cv. Hu in the following way, using a weighted sum of all convolve features:
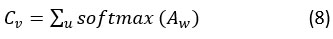
The attention weight (A)w and weight matrix ( Wm) are used here. With convolution training, Cv , which stand for vectors, learn together. Focusing on convolve characteristics that significantly impact patient disease prediction is the goal of these attention vectors.
SACNN
We combined the context vector that was computed before with the convolution in the following way for the SACNN (Fig. 3):
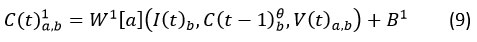
Where represents a weight matrix with dimensions defined by the product of a=1, 2, …. i and b= 1, 2, …. j, where i and j are specific parameters. The term I(t) corresponds to the input text representation, and B1 is the bias term. C (t-1)θ denotes a previously computed convolution at time (t-1). Va signifies an attention vector, and the result of a convolution obtained from C1 (t) is considered the feature-map.
Rectified Linear Unit (ReLU) activation functions capture non-linear correlations in feature maps. ReLU is mathematically defined as:
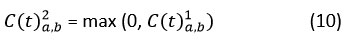
The attention method is incorporated into the convolution that follows a non-linear operation in the convolution that has been developed. When compared to some established approaches that are considered to be state-of-the-art, the experimental results reveal that the convolution that was developed achieves a higher level of accuracy.
Max pooling layer
The DWSC and SACNN modules are concatenated before the max pooling layer. In order to carry out the pooling procedure, the feature map C(t)2a,b derived from convolution in order to identify, P(Y, X, L) from DWSC and select large-granular features from images of diseases. Assuming that image-level granular features are obtained, the pooling process is anticipated to yield phrase-level granular features, as shown below:
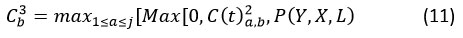
Where C(t) 3b illustrates the feature-map that was produced as a result of the max-pooling.
Fully connected layer
Furthermore, at this particular layer, a fully connected operation is carried out on the feature map C3 , which is produced from the max-pooling layer, as illustrated in Figure 2:
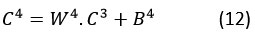
Here, C3 and C4 denote the feature maps derived from pooling and full connection operations, respectively. B4 and W4 represent the bias and weight parameters of the full connection layer.
Each of the feature maps that are derived from full connection and pooling procedures is denoted by C3and C4 in this context. The parameters of the fully connected layer are denoted by W4 and B4, which stands for weight and bias.
The pseudo-code of the skin disease feature detection based two stream fusion algorithms is shown in Algorithm 1.
Algorithm 1: Skin disease feature detection based two-stream fusion
|
Input: Two-stream model two input images Irgb and Igray., the proposed model includes MobileNetV2 and NASNetMobile models, Maximum epochs Mepoch = 120 with batch size of Bs = 16 for the Network, and LR (learning rate) = 0.001. Output: The optimized feature detection based two-stream feature fusion network 1: While M1 ≤ Mn × Bs do 2: Two-stream images batch as Irgb and Igray. . 3: Apply Pre-processing (image resize, shift range, rescale, rotation, shear & zoom, flip) to both RGB and Grayscale images. 4: Apply fusion on CNN models and add DWSC by Equations (1, (2), and (3). 5: Proposed approach jointly using a loss function by Equation (4). 6: Extract gradient images using gradient operator by Equation (5), and (6). 7: Employing the Self-attention-based convolution by Equation (9), and (10). 8: Jointly optimized feature detection based dual-scale fusion network by Equation (11), and (12). 9: End |
Results and Discussion
This section offers a comprehensive explanation of the dataset, along with the techniques for experimentation, model training, and validation. The final sub-section of the paper provides an analysis of performance utilizing several cutting-edge models.
Implementation Details
In this Google Colab Keras experiment, we utilize the K80, P100, and T4 GPUs. To maximize the effectiveness of segmentation networks, we use a batch size of 16 with the Adam optimizer. We employed a learning rate of 0.001. The optimal values for the maximum number of epochs are 80 for transfer learning and 120 for the fused model. Images are randomly downsized to 256 × 224 for CNN training and then horizontally flipped.
Dataset description
Images utilized in this study were sourced from the HAM10000 23, ISIC2017 24, and ISIC2016 25 databases. All three datasets contain 13,894 images, each depicting various skin diseases across different areas of the body. Table 2 displays the distribution of images across the different datasets. The effectiveness of the proposed method in identifying skin diseases is demonstrated using each dataset. The models are divided into training and testing sets in a 70:30 ratio.
Data Preprocessing
In this section, we provide a full description of the preprocessing conducted on the datasets. The preprocessing involves resizing the images and augmenting the data.
Resize
Images of high resolution are found in the HAM10000, ISIC2016, and ISIC2017 datasets. When used directly for training, images of skin lesions have a resolution of 600 × 450 pixels, which results in a significant increase in the amount of calculation required. Therefore, to comply with the specifications of the model, we reduced the dimensions of each image from 600 × 450 pixels to 224 × 224 pixels. We allocate 70% for training and 30% for testing.
Data augmentation
Despite the fact that the three datasets contain 13894 images between them, this amount of information is insufficient to satisfy the requirements of deep learning algorithms. As a result, we apply six different data augmentation procedures to the training samples. These include randomly rotating the samples, shifting them horizontally and vertically, randomly zooming, randomly twisting, flipping, and resizing them.
Evaluation Metrics
Accuracy, precision, recall, and F1 score are some of the evaluation measures we utilize based on recommendations from our dataset. Definitions for these quantitative measures can be found in (13)-(16). Following is the formula for their computation:

Findings and comparison
This segment provides a thorough perspective on experimentation, comparison, and discussion. In the concluding subsection, an evaluation of performance is showcased through the analysis of various cutting-edge models.
The findings from single models
Table 3 displays the classification results for multiclass skin diseases using the HAM10000, ISIC2016, and ISIC2017 datasets with two single-model frameworks: MobileNetV2 and NASNetMobile. The NASNetMobile model combined with the HAM10000 dataset achieves the best results, with an accuracy of 81%. It also attains a 70 F1-score, 81 recall rate, and 68 precision rates. On the ISIC2017 dataset, the NASNetMobile model performs second-best, achieving an accuracy rate of 80% with an F1-score of 69, recall of 80, and precision of 74. The highest-performing metrics are bolded for emphasis. Additionally, the NASNetMobile model has approximately 4.1 million trainable parameters, while MobileNetV2 has around 3.2 million.
Table 3: The classification results of two single models on different datasets.
|
Single network – ISIC -2016 Dataset |
||||
|
Model |
Precision |
Recall |
F1-Score |
Testing Accuracy |
|
MobileNetV2 |
77 |
79 |
78 |
79 |
|
NASNetMobile |
73 |
78 |
74 |
78 |
|
Single network – ISIC -2017 Dataset |
||||
|
MobileNetV2 |
74 |
79 |
65 |
79 |
|
NASNetMobile |
74 |
80 |
69 |
80 |
|
Single network – HAM10000 Dataset |
||||
|
MobileNetV2 |
72 |
79 |
62 |
79 |
|
NASNetMobile |
68 |
81 |
70 |
81 |
The findings from fused model
Table 4 displays the classification outcomes of a fused model (MobileNetV2 + NASNetMobile) for multiscale skin diseases using the HAM10000, ISIC2016, and ISIC2017 datasets. With the fused model architecture, the best results are achieved by the MobileNetV2 + NASNetMobile model on the ISIC2017 dataset, attaining an accuracy rate of 84%. The NASNetMobile model achieves an F1-Score, recall, and precision of 81, 76, and 81, respectively. On the HAM10000 dataset, the MobileNetV2 + NASNetMobile fused model performs second-best, with an accuracy rate of 83%. For this dataset, MobileNetV2 + NASNetMobile achieve an F1-Score, recall, and precision of 81, 80, and 76, respectively. This fused model comprises approximately 8.6 million trainable parameters.
Table 4: The classification results of fused models on different datasets.
|
Late fusion – ISIC -2016 Dataset |
||||
|
Model |
Precision |
Recall |
F1-Score |
Testing Accuracy |
|
MobileNetV2 + NASNetMobile |
77 |
79 |
80 |
82 |
|
Late fusion – ISIC -2017 Dataset |
||||
|
MobileNetV2 + NASNetMobile |
81 |
76 |
81 |
84 |
|
Late fusion – HAM10000 Dataset |
||||
|
MobileNetV2 + NASNetMobile |
76 |
80 |
81 |
83 |
The findings from proposed model
Table 5 presents the classification results obtained by applying the TFFNet model to the HAM10000, ISIC2016, and ISIC2017 datasets for the analysis of multiclass skin diseases. Testing the TFFNet model architecture on the HAM10000 dataset yielded a remarkable accuracy rate of 90%. The proposed model achieved an F1-score of 81, a recall of 80, and a precision of 83, respectively. Similarly, the TFFNet model demonstrated strong performance on the ISIC2017 dataset, achieving a 90% accuracy rate. It received scores of 81, 82, and 78 for F1 score, recall, and precision, respectively. This model comprises approximately 7.5 million trainable parameters.
Table 5: The classification results of proposed models on different datasets.
|
Proposed method – ISIC -2016 Dataset |
||||
|
Model |
Precision |
Recall |
F1-Score |
Testing Accuracy |
|
TFFNet |
82 |
78 |
80 |
89 |
|
Proposed method – ISIC -2017 Dataset |
||||
|
TFFNet |
78 |
82 |
81 |
90 |
|
Proposed method – HAM10000 Dataset |
||||
|
TFFNet |
83 |
80 |
81 |
90 |
Analysis of TFFNet architecture
The training accuracy of the TFFNet model proposed in this study demonstrated a discernible increase, characterized by a swift initial ascent followed by a gradual levelling off. In contrast, the validation accuracy exhibited a consistent upward trajectory, as depicted in Fig. 4c, albeit with fluctuations throughout the training period. Unlike the CNN models, the validation curves of TFFNet displayed comparatively less fluctuation. Particularly noteworthy is the fact that signs of saturation started to emerge during the 80th epoch. A significant decrease in the discrepancy between validation and training accuracy was obtained compared to the CNN model. TFFNet consistently displayed stable performance in terms of validation accuracy, suggesting that the model’s fitting capabilities could potentially result in superior generalization on unseen test data when compared to CNN models.
Comparison on our single, fused, and proposed models with three datasets
Compared to the single and fused approaches, the proposed framework outperforms them consistently across all metrics, including F1-score, precision, accuracy, and recall, as depicted in Figure 4. In this experiment, we utilized three distinct datasets: HAM10000, ISIC-2017, and ISIC-2016. Testing on the HAM10000 and ISIC2017 datasets yielded an impressive accuracy rate of 90% for the proposed TFFNet model architecture. The results of the study are presented in Table 6, which indicates that TFFNet achieved the highest accuracy among all models. Additionally, TFFNet had fewer parameters compared to MobileNetV2, NASNetMobile, and the combination of the two models.
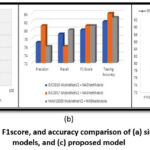 |
Figure 4: Precision, recall, F1score, and accuracy comparison of (a) single models; (b) fused models, and (c) proposed model |
Table 6: Analysis of TFFNet with different models on trainable parameters.
|
Models |
Training parameters |
|
MobileNetV2 |
3,257,895 |
|
NASNetMobile |
4,125,468 |
|
MobileNetV2 + NASNetMobile |
8,684,985 |
|
TFFNet |
7,572,286 |
Comparison of proposed model with state of the art on HAM10000 dataset
Among state-of-the-art methods, our proposed TFFNet method surpasses them all, with an average improvement of 1.4% over the second-best method. Table 7 presents the results comparing the accuracy of the HAM10000 dataset to that of the previous equivalent reference.
Table 7: Accuracy comparison with a previous related reference for the HAM10000 dataset.
|
Authors |
Year |
Accuracy (%) |
|
Gouda 26 |
2022 |
83.2 |
|
Hoang 27 |
2022 |
86.33 |
|
Kim 28 |
2023 |
88.6 |
|
Ours |
2023 |
90.12 |
Comparison of proposed model with state of the art on ISIC 2017 dataset
We conducted a comparative analysis to assess the efficiency of our proposed TFFNet approach against the most recent and innovative classification strategies. The results of these comparisons were analyzed and evaluated. Table 8 presents the comparison between the ISIC2017 dataset and a relevant previous reference, aimed at assessing the accuracy of the ISIC2017 dataset. Our proposed method demonstrates superior performance compared to state-of-the-art methods, with an average accuracy that is 2.5% higher than the method currently considered to be in second place.
Table 8: Accuracy comparison with existing methods for the ISIC 2017dataset.
|
Authors |
Year |
Accuracy (%) |
|
Al-masni 29 |
2020 |
81.57 |
|
Yilmaz 30 |
2021 |
82 |
|
Kim 28 |
2023 |
87.5 |
|
Ours |
2023 |
90.52 |
Comparison of proposed model with state-of-the-art on ISIC2016 dataset
According to Table 9, our proposed TFFNet method achieves the highest average accuracy of 89.70% among all compared approaches. This accuracy is 0.81% and 1.4% higher than the previous two methods, namely Dahou 33 and Wei 32, respectively.
Table 9: Accuracy compared with previous methods for the ISIC 2016dataset.
|
ss |
Year |
Accuracy (%) |
|
Al-masni 29 |
2020 |
80 |
|
Yu 31 |
2020 |
86.8 |
|
Wei 32 |
2020 |
87.6 |
|
Dahou 33 |
2023 |
88.19 |
|
Ours |
2023 |
89.70 |
Conclusion
Skin diseases affect a significant number of individuals and rank among the most widespread categories of ailments globally. Among these, acne stands out at the top of the list alongside various other skin diseases, each posing its own set of risks, ranging from minor discomforts to life-threatening conditions. The integration of computer-aided diagnosis 34 has greatly facilitated the medical community in the identification and categorization of skin diseases, addressing a substantial challenge. The field of skin disease classification 35 has seen the emergence of several deep learning models. However, there remains room for improvement in areas such as computational efficiency and dataset-specific accuracy. To truly enhance the effectiveness of computer-aided diagnosis, it must be capable of accurately discerning specific skin diseases from an extensive list. However, as the number of skin classes increases, the complexity and parameter count of a model naturally escalate. This is where the TFFNet model comes into play, offering a solution with fewer parameters while maintaining satisfactory accuracy. Contrary to the assumption that augmenting parameter numbers enhances accuracy, the results of our study challenge this notion. The incorporation of two modules with the SA block demonstrates a reduction in parameters without compromising accuracy. The proposed TFFNet model exhibited total accuracies of 90.12%, 90.52%, and 89.70% on test datasets. Further evaluation using microscopic and histological images can shed light on potential challenges in disease categorization for this innovative network.
Acknowledgment
We thank Graphic Era (Deemed to be University) Dehradun for the use of their research facilities.
Conflict of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Funding Sources
This research received no external funding.
Data availability
Not applicable.
Reference
- Zhou, S.K.; Greenspan, H.; Davatzikos, C.; Duncan, J.S.; Van Ginneken, B.; Madabhushi, A.; Prince, J.L.; Rueckert, D.; Summers, R.M. A Review of Deep Learning in Medical Imaging: Imaging Traits, Technology Trends, Case Studies with Progress Highlights, and Future Promises. Proc. IEEE 2021, 109, 8s20–838.
CrossRef - Bai, W.; Suzuki, H.; Huang, J.; Francis, C.; Wang, S.; Tarroni, G.; Guitton, F.; Aung, N.; Fung, K.; Petersen, S.E.; et al. A population-based phenome-wide association study of cardiac and aortic structure and function. Nat. Med. 2020, 26, 1654.
CrossRef - Shanthi, T.; Sabeenian, R.S.; Anand, R. Automatic Diagnosis of Skin Diseases Using Convolution Neural Network. Microprocess. Microsyst. 2020, 76, 103074.
CrossRef - American Cancer Society. Key Statistics for Melanoma Skin Cancer. 2022. Available online: https://www.cancer.org/cancer/ melanoma-skin-cancer/about/key-statistics.html.
- Wei, L.-S.; Gan, Q.; Ji, T. Skin Disease Recognition Method Based on Image Color and Texture Features. Comput. Math. Methods Med. 2018, 2018, 8145713.
CrossRef - Amarathunga, A.A.L.C.; Ellawala, E.P.W.C.; Abeysekara, G.N.; Amalraj, C.R.J. Expert System for Diagnosis of Skin Diseases. Int. J. Sci. Technol. Res. 2015, 4, 174–178.
- Bajwa, M.N.; Muta, K.; Malik, M.I.; Siddiqui, S.A.; Braun, S.A.; Homey, B.; Dengel, A.; Ahmed, S. Computer-Aided Diagnosis of Skin Diseases Using Deep Neural Networks. Appl. Sci. 2020, 10, 2488.
CrossRef - Monisha, M.; Suresh, A.; Rashmi, M.R. Artificial Intelligence Based Skin Classification Using GMM. J. Med. Syst. 2019, 43, 3.
CrossRef - Kassem, M.A.; Hosny, K.M.; Damaševiˇcius, R.; Eltoukhy, M.M. Machine Learning and Deep Learning Methods for Skin Lesion Classification and Diagnosis: A Systematic Review. Diagnostics 2021, 11, 1390.
CrossRef - Maqsood, S., & Damaševičius, R. (2023). Multiclass skin lesion localization and classification using deep learning-based features fusion and selection framework for smart healthcare. Neural Networks, 160, 238-258.
CrossRef - Cai, H., Brinti Hussin, N., Lan, H., & Li, H. (2023). A Skin Cancer Detector Based on Transfer Learning and Feature Fusion. Current Bioinformatics, 18(6), 517-526.
CrossRef - Zhao, X., & Ren, Z. (2023, January). Multi-scale Gaussian Difference Preprocessing and Dual Stream CNN-Transformer Hybrid Network for Skin Lesion Segmentation. In International Conference on Multimedia Modeling (pp. 671-682). Cham: Springer Nature Switzerland.
CrossRef - Tian, Q., Sun, W., Zhang, L., Pan, H., Chen, Q., & Wu, J. (2023). Gesture image recognition method based on DC-Res2Net and a feature fusion attention module. Journal of Visual Communication and Image Representation, 95, 103891.
CrossRef - Rajput, G.; Agrawal, S.; Raut, G.; Vishvakarma, S.K. An accurate and noninvasive skin cancer screening based on imaging technique. Int. J. Imaging Syst. Technol. 2022, 32, 354–368.
CrossRef - Raza, R.; Zulfiqar, F.; Tariq, S.; Anwar, G.B.; Sargano, A.B.; Habib, Z. Melanoma Classification from Dermoscopy Images Using Ensemble of Convolutional Neural Networks. Mathematics 2022, 10, 26.
CrossRef - Gouda, W.; Sama, N.U.; Al-Waakid, G.; Humayun, M.; Jhanjhi, N.Z. Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning. Healthcare 2022, 10, 1183.
CrossRef - Rehman, M.Z.U.; Ahmed, F.; Alsuhibany, S.A.; Jamal, S.S.; Ali, M.Z.; Ahmad, J. Classification of Skin Cancer Lesions Using Explainable Deep Learning. Sensors 2022, 22, 6915.
CrossRef - Aldhyani, T.H.H.; Verma, A.; Al-Adhaileh, M.H.; Koundal, D. Multi-Class Skin Lesion Classification Using a Lightweight Dynamic Kernel Deep-Learning-Based Convolutional Neural Network. Diagnostics 2022, 12, 2048.
CrossRef - Kousis, I.; Perikos, I.; Hatzilygeroudis, I.; Virvou, M. Deep Learning Methods for Accurate Skin Cancer Recognition and Mobile Application. Electronics 2022, 11, 1294.
CrossRef - Hasan, K.; Elahi, T.E.; Alam, A.; Jawad, T.; Martí, R. DermoExpert: Skin lesion classification using a hybrid convolutional neural network through segmentation, transfer learning, and augmentation. Inform. Med. Unlocked 2022, 28, 100819.
CrossRef - Tan, L., Wu, H., Xia, J., Liang, Y., & Zhu, J. (2024). Skin lesion recognition via global-local attention and dual-branch input network. Engineering Applications of Artificial Intelligence, 127, 107385.
CrossRef - Muhammad, W., Aramvith, S., & Onoye, T. (2021). Multi-scale Xception based depthwise separable convolution for single image super-resolution. Plos one, 16(8), e0249278.
CrossRef - P. Tschandl, C. Rosendahl, and H. Kittler, ‘‘The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions,’’ Scientific Data, vol. 5, no. 1, pp. 1–9, Aug. 2018.
CrossRef - Hasan MK, Elahi MTE, Alam MA, Jawad MT, MartíR. DermoExpert: skin lesion classification using a hybrid convolutional neural network through segmentation, transfer learning, and augmentation. Inform Med Unlock. (2022) 28:100819. doi: 10.1016/j.imu.2021.100819.
CrossRef - D. Gutman, N.C. Codella, E. Celebi, B. Helba, M. Marchetti, N. Mishra, A. Halpern, Skin lesion analysis toward melanoma detection: a challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC), arXiv preprint arXiv:1605.01397, https://doi.org/10. 48550/arXiv.1605.01397, 2016.
- Gouda, W.; Sama, N.U.; Al-Waakid, G.; Humayun, M.; Jhanjhi, N.Z. Detection of skin cancer based on skin lesion images using deep learning. Healthcare 2022, 10, 1183.
CrossRef - Hoang, L., Lee, S. H., Lee, E. J., & Kwon, K. R. (2022). Multiclass skin lesion classification using a novel lightweight deep learning framework for smart healthcare. Applied Sciences, 12(5), 2677.
CrossRef - Kim, C., Jang, M., Han, Y., Hong, Y., & Lee, W. (2023). Skin Lesion Classification Using Hybrid Convolutional Neural Network with Edge, Color, and Texture Information. Applied Sciences, 13(9), 5497.
CrossRef - Al-masni, A.M.; Kim, D.; Kim, T. Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput. Methods Programs Biomed. 2020, 190, 105351.
CrossRef - Yilmaz, A., Kalebasi, M., Samoylenko, Y., Guvenilir, M. E., & Uvet, H. (2021). Benchmarking of Lightweight Deep Learning Architectures for Skin Cancer Classification using ISIC 2017 Dataset. arXiv preprint arXiv:2110.12270.
- Yu, Z.; Jiang, F.; Zhou, F.; He, X.; Ni, D.; Chen, S.; Wang, T.; Lei, B. Convolutional descriptors aggregation via cross-net for skin lesion recognition. Appl. Soft Comput. 2020, 92, 106281.
CrossRef - Wei, L.; Ding, K.; Hu, H. Automatic skin cancer detection in dermoscopy images based on ensemble lightweight deep learning network. IEEE Access 2020, 8, 99633–99647.
CrossRef - Dahou, A., Aseeri, A. O., Mabrouk, A., Ibrahim, R. A., Al-Betar, M. A., & Elaziz, M. A. (2023). Optimal Skin Cancer Detection Model Using Transfer Learning and Dynamic-Opposite Hunger Games Search. Diagnostics, 13(9), 1579.
CrossRef - Ding, H., Huang, Q., & Alkhayyat, A. (2024). A computer aided system for skin cancer detection based on Developed version of the Archimedes Optimization algorithm. Biomedical Signal Processing and Control, 90, 105870.
CrossRef - Akilandasowmya, G., Nirmaladevi, G., Suganthi, S. U., & Aishwariya, A. (2024). Skin cancer diagnosis: Leveraging deep hidden features and ensemble classifiers for early detection and classification. Biomedical Signal Processing and Control, 88, 105306.
CrossRef







