Swapna Katta1 , Prabhishek Singh2
, Prabhishek Singh2 , Deepak Garg1
, Deepak Garg1  and Manoj Diwakar3,4
and Manoj Diwakar3,4
1School of Computer Science and Artificial Intelligence, SR University, Warangal, Telangana, India.
2School of Computer Science Engineering and Technology, Bennett University, Greater Noida, India,
3Department of CSE, Graphic Era Deemed to be University, Dehradun, Uttarakhand, India.
4Department of CSE, Department of CSE, Graphic Era Hill University, Dehradun, India.
Corresponding Author E-mail: prabhisheksingh1988@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/2991
Abstract
The presence of gaussian noise commonly weakens the diagnostic precision of low-dose CT imaging. A novel CT image denoising technique that integrates the non-subsampled shearlet transform (NSST) with Bayesian thresholding, and incorporates a modern method noise Deep Convolutional neural network (DCNN) based post-processing operation on denoised images to strengthen low-dose CT imaging quality. The hybrid method commences with NSST and Bayesian thresholding to mitigate the initial noise while preserving crucial image features, such as corners and edges. The novel aspect of the proposed approach is its successive application of a DnCNN on initial denoised image, which learns and removes residual noise patterns from denoised images, thereby enhancing fine detail preservation. This dual-phase strategy addresses both noise suppression and image-detail preservation. The proposed technique is evaluated through the use of metrics, such as PSNR, SNR, SSIM, ED, and UIQI. The results demonstrate that the hybrid approach outperforms standard denoising techniques in preserving image quality and fine details.
Keywords
DnCNN; LDCT imaging; Gaussian noise; Method Noise; NSST
Download this article as:| Copy the following to cite this article: Katta S, Singh P, Garg D, Diwakar M. A Hybrid Approach for CT Image Noise Reduction Combining Method Noise-CNN and Shearlet Transform. Biomed Pharmacol J 2024;17(3). |
| Copy the following to cite this URL: Katta S, Singh P, Garg D, Diwakar M. A Hybrid Approach for CT Image Noise Reduction Combining Method Noise-CNN and Shearlet Transform. Biomed Pharmacol J 2024;17(3). Available from: https://bit.ly/3BfPRum |
Introduction
Medical imaging refers to the different imaging techniques used in modern hospitals and clinics for medical diagnosis. X-rays, CT imaging, ultrasound scans, magnetic-resonance imaging (MRI) techniques are used to scan within the body to assess the cause of disease and provide appropriate treatment for medical conditions. In CT imaging , X-ray radiation is directed at the patient’s body from multiple projections to scan bone fractures, organs, fat, and blood vessels. Although repeated scans in patients provide invaluable information for the clinical diagnosis of various diseases, there is a potential risk of cancer threat1,2. Hence, it is essential to minimize radiation exposure. Lowering radiation exposure improves noise, blur and other minute artifacts in tomographical images3. Low dose CT images exhibit noise due to electrical interference, quantum effects and mathematical computations. An effective denoising technique is required to explicitly reduce the noise and artifacts from distorted CT images 4,5. CT image denoising techniques can be classified as classical CT image denoising, post-processing, and deep learning related methods.
Classical denoising techniques are classified as spatial and transform domain filtering methods. Spatial filtering methods manipulate the intensity values of the pixels directly based on spatial coordinates. Here, denoising is applied to the whole image. Spatial domain filtering methods,6,7,8,9 make use of low-pass filtering, suppressing noise to some extent and resulting in blurry images, for example, wiener, mean, bilateral and nonlocal means (NLM) filtering show the correlation between pixel intensities in their neighboring pixels around a given pixel. This results in average smoothing, and a loss of sharp features in an image.
Transform domain filtering methods analyze images in terms of their frequency levels, for example., Discrete fourier transform, Discrete cosine transform, Wavelet, and Shearlet transform domain10,11,12,13. In transform domain filtering, thresholding method is applied to denoise noisy CT images, and inverse transformation is used to reconstruct original images. Thresholding methods like SureShrink, VisuShrink, and BayesShrink. VisuShrink is a global thresholding method based on the pixel quantity in an image, where as SureShrink and BayesShrink based on each subband to evaluate the threshold values. The non-subsampled shearlet transform provides spatial localization and sparse representation of multiscale and multidirectional features to capture different directional features of an image to overcome the limitations such as isotropic features and the absence of multidirectionality of the wavelet transform. The shearlet transform is effectively used to capture and represent anisotropic features such as edges, corners, and fine details, and well-localized structures exhibit different features in different directions.
Unlike previous denoising techniques, post processing methods explicitly handle the reconstructed images in CT imaging, that is reconstruction without projectional data, which improves the performance of generalization. Traditional post-processing methods, Block-matching and 3D filtering (BM3D)14, adaptive nonlocal means (NLM)15, K means singular-valued decomposition (KSVD)16 methods effectively reduce noise and artifacts but the computational cost is high.
Hybrid methods are combinations of spatial and transform domain methods that provide improved image-denoising results, for example, denoising CT images through total variation using the shearlet domain17 with a multi-variate model, and method noise-based approach yield better results in suppressing noise, preserving the edges and structural details. The detection of noisy COVID-19 (SARS-CoV-2) virus on LDCT imaging using the nonlocal means filter in conjunction with method noise yields improved SSIM results compared to other existing methods18, for example, nonlocal means19 , total-variation methods20 using wavelets, to reduce noise while preserving image features in detail. However, images suffer from noise and artifacts owing to poor directionality, shift sensitivity, and a limited ability to capture directional information.
Recently, deep learning techniques have played a crucial role in image denoising.The progress in CNN-based methods21,22 has improved in CT image denoising in LDCT imaging. The deep denoising convolutional neural network with residual learning strategy (REDCNN) is a CNN architecture used in image denoising that includes residual mapping trained deeper networks to resolve the vanishing-gradient problem and allows deeper networks to be trained easily. The efficiency of the deep CNN differs at different CT radiation dose levels. For example, the Wasserstein distance-based generative adversarial network (W-GAN) provides superior GAN performance, and perceptual loss evaluates perceptual features with filtered output at ground truth level to maintain critical information and effectively reduce noise in CT images23. Currently, transformer models play a significant role in image- processing. The Transformer model is a (DL) deep learning architecture, which is a self-attention mechanism that captures long-range dependencies between input and output tokens24. A vision transformer (ViT) is a kind of neural-network framework, especially applied in domains like image recognition, recognition, and segmentation. Transformer-based Encoder-decoder Dilation network (TED) effectively preserves the structure and fine details while denoising images25.
Problem Statement
CT imaging is essential for accurate healthcare diagnostics because it provides precise images for the detection of health conditions. However, the existence of Gaussian noise in CT images significantly diminishes their quality, contributing to potential challenges in disease prediction and identification. This study aims to address the significant need for effective noise suppression method to improve CT image quality, thereby improving diagnostic precision and patient clinical judgement.
Major Contribution
The study presents a novel CT imaging denoising technique that combines method noise-based CNN with a non-subsampled shearlet transform to effectively mitigate Gaussian noise .The proposed technique improves noise suppression while retaining crucial image features by leveraging the multi-scale and multi-directional analysis of the shearlet transform. Additionally, the method noise-based CNN method intends to reduce residual noise patterns in denoised CT images. This hybrid strategy significantly improves image quality and diagnostic accuracy, providing an effective solution to noise-related challenges in LDCT imaging.
The rest of this paper is presented as follows. Section 2 introduces a brief literature review of CT image-denoising approaches. Section 3 outlines the major concepts of shearlet transform and DnCNN architecture. Section 4 discusses the proposed hybrid algorithm for CT image denoising with a method noise-based CNN method using a shearlet transform. The findings from the experiments and a comparison with various existing denoising methods are shown in Section 5. Finally, the conclusions and proposed future studies are presented in Section 6.
Literature Review
Clinical imaging plays a major role in diagnostic decision making using different modalities, to improve the accuracy of clinical diagnosis of the internal human body. In,26 Abhisheka proposed that the prominent medical imaging modalities include X-radiation, CT scan, MRI, Positron-Emission Tomographical (PET) imaging, and Ultrasound scans. These modalities effectively visualize a detailed image of inside the human body. For example, X-rays can be used to identify bone fractures, dislocations. Unlike X-ray, a CT scan provides a fast, more detailed image for medical diagnosis. CT scan is used to detect organ abnormalities, blood clots, subtle bone fractures, and internal bleeding. MRI scans provide highly detailed images of soft tissues. Ultrasound, or sonography, exploits high frequency sound waves to provide detailed imaging of human body, detect problems in the liver, kidney, heart, blood vessels, valvular regurgitation, and abdominal aorta etc. PET scans are used for detecting organ abnormalities, including soft tissue-related issues such as finding tumors, neurological (brain) diseases, cardiovascular (heart) diseases. These medical image modalities are used by healthcare professionals to diagnose various medical conditions in a timely, accurate, and non-invasive manner, thereby improving patient outcomes.
The proposed literature review primarily concentrates on CT imaging using various deep learning approaches. In,27 Sehgal proposed novel CT image denoising algorithm, Political-Taylor Anti-coronavirus Optimization (PT-ACVO) combines deep learning and advanced optimization techniques to mitigate noise and enhance image qualities effectively. The method detects noisy pixels in images using a Deep Residual network (DRN) and reconstructs them using the Political Taylor-Anti-Coronavirus Optimization (Political Taylor -ACVO) algorithm and image-enhancement is achieved through Vectorial-Total-Variation approach. Image denoising was performed using Discrete Wavelet transform and NLM filtering, followed by image fusion to obtain final denoised image. However, the DRN and the Political-Taylor-ACVO Optimization algorithm might cause higher computational complexity and there is a requirement for extensive parameter-tuning.
The evolution of Deep learning methods has emerged as a rapid progress in CT image denoising28. To improve the quality of LDCT imaging,29 Zhang proposed an innovative denoising method using U-net and multi-attention mechanisms for effective feature extraction. This includes three attention modules. The local attention module provides localized surrounding pixel feature extraction based on feature mapping. The multi feature and channel-attention-module automatically acquire, extract, suppress noise, and contribute different weights to the existing feature-map based on different tasks. The hierarchical identification module enabled a deeper CNN for a substantial amount of feature extraction. Additionally, a study suggests that the enhanced learning-module increases the network depth by stacking a multi-layered CNN, activation layer and batch-normalization (BN) enabling the learning and maintenance of detailed image information. The Experimental quantitative analysis results show that the module effectively suppressed noise.
In,30 Huang proposed a deep cascade residual network (DCRN) that offers promising denoising results, combining attention mechanisms to enhance model performance, a hybrid loss function to provide better generalization ability of the model, and iterative refinement to iteratively refine the denoised image to obtain a better-quality image. In,31 Selig proposed a Dilated Residual U-Net (DRU-Net) for better LDCT image reconstruction and image enhancement to enhance image quality and performance. This involves two-stage process: Initially, filtered-back projection (FBP) was performed to improve image reconstruction. DRU-Net is pre-trained for denoising natural grayscale images, and mapping low-dose filtered back projection is applied to the reconstructed images to enhance the CT images. DRU-Net is fine-tuned and performs downstream image enhancement by leveraging LDCT imaging and appropriate normal-dose computed tomographical (NDCT) images. This method secured the topmost ranking in the low dose parallel beam CT-challenge (LoDoPaB), was computationally more efficient than Institute of Technology Network (ItNet), and increased the SSIM metric value. Here, the U-Net model was pretrained only for Gaussian denoising. If the pre-trained task and target CT image denoising differ, affects the performance of the model.
In,32 Song introduced a NeXtResUNet-CNN for industrial CT image denoising. It includes industrial CT image systems that operate on diverse energies and significantly affect distinct spatial resolutions. The proposed algorithm initiates an image fusion network that combines Con-vNeXt, ResNet, and U-Net, is assigned to a self-generated industrial tomographic denoising dataset. NeXtResUnet simulates a transformer model to acquire global features, and ResNet is used to extract the image details. CT-image noise reduction can be accomplished by downsampling the CNN. This results in an improved Peak signal-to-noise ratio (PSNR) and image-denoising, image segmentation, and contrast normalization. The NeXtResUNet network structure is concise and expandable, making it applicable to image denoising and image vision-based tasks, like CT image super-resolution and auto-segmentation involving industrial CT data.
In,33 Byeon proposed the use of a lightweight Deep CNN with multi-directional fuzzy non subsampled shearlet transformation (FNSST) for better image decomposition and suppression of noisy patterns and artifacts in LDCT imaging. FNSST is a multi-scale and multi-directional localization technique used for the decomposition of low and normal dose images to produce high and low-resolutional subimages. High-resolutional subimages with varying noise levels in a fuzzy setting and other artifacts were given as input to the CNN to establish an association between the LDCT high-resolution subimages and the residual subimages generated throughout the training process. FNSST-CNN discriminates low and high-frequency subimages while testing process to suppress noise and other relevant artifacts. In LDCT imaging, FNSST-CNN effectively reduces noisy patterns while preserving the edges and structural features. The main limitation is that the cost of implementing fuzzy-based methods is high.
In,34 Li proposed a multistage noise reduction framework for LDCT images. The framework was mainly trained using un-paired data. The (PCCNN) Progressive-Cyclical-CNN performs latent -space utilization from CT images to suppress noisy areas and other artifacts. PCCNN, a multistage denoising framework, suggests a noise transfer model that enables the transfer of noise from low-dose to normal dose CT images. The PCCNN also has a progressive module that includes a multi-stage wavelet transform to extract high frequency coefficients to reduce noisy coefficients and preserve contours of the image. The main constraint is that there is a need for pairs of perfectly matched low and normal-dose images to elevate model performance.
In,35 Çalişkan introduced, effective method for identifying and handling noisy pixels in 2D images using to enhance the quality of CT images, specifically focusing on accurate detection of noisy pixels in 2 Dimensional CT images using hidden resource decomposition approach. The hidden resource decomposition approach with extreme learning Machines (ELM) to improve efficiency in training and high learning speed suitable for handling large volumes of the CT images to preserve critical structural information in detail. The ELM method markedly improved noise suppression and imaging quality, attaining peak performance with 250 hidden layer neurons. he ELM method significantly reduced mean- squared-error (MSE) and peak-signal-to-noise ratio (PSNR).The incorporation of hidden resource decomposition with ELM may result in complexity in implementation.In,36 Çalişkan suggested, deep learning-driven hybrid approach to categorize seven mineral types, with precision, employing refined feature selection and the complement rule for clustering. The method utilizes deep learning models for feature-extraction and applied metaheuristic optimization to identify key features, and complement rule for grouping ineffective features for achieving exceptional classification precision. The image denoising can be obtained using metaheuristic optimization algorithm due to their capability to explore vast and complex search spaces.
Major Concepts
The following section presents some essential concepts that were exploited to implement the proposed methodology.
Non-subsampled Shearlet Transform
The NSST transform is constitutes an extended variant form of wavelet transformation and is a mathematical tool,37 used in image processing, particularly for image denoising and feature extraction. The nonsubsampled shearlet transform is a multi-directional, multi-dimensional, shift-invariant, and well-localized analysis that combines multiscale and directional analysis separately. NSST coefficients are related to sparse and capture the mathematical and geometric properties of an image. NSST is depicted in conjunction with Nonsubsampled-Laplacian pyramid (NS-LP) and shearing-filters. Initially, (NSLP) is used to analyze the images into different low (approximation) and high (detail) frequency components, and directional filtering helps to generate various subbands and extract shearlet components. The shear matrix accomplishes directional filtering and provides an analysis across various directions.
For image data with dimensions of n=2, and for j>0, k ∈ R, l ∈ R2
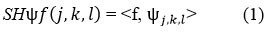
Where ψf(j, k, l) are called shearlets. Here, j > 0, k ∈ R, l ∈ R2, the shearlets are evaluated as:
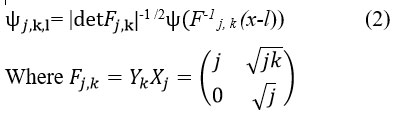
The anisotropic-dilation is represented as:
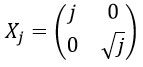
where j>0, manages the shearlet’s scale, and provides the frequency to obtain finer scales. The shearing transformation matrix is obtained as follows:
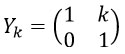
The shearing matrix manages only the shearlet direction.
Hence, the shearlet transform is determined by the three variables, includes scale j, orientation k and location l.
Each f ∈ L2 (R2)is restored using:
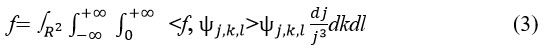
The Discrete shearlet transform is used to represent multi-dimensional functions. Here j= 2-2m ,k=-L with m, l=k∈Z2 and L ∈ Z.
The Discrete shearlet transformation can be denoted as:
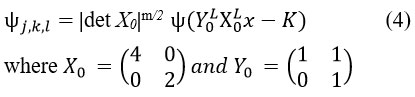
From each function f ∈ L2 (R2),the given method is reconstructed using the characteristics of as described below:
The fundamental framework of DnCNN is depicted in Fig. 1.
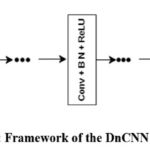 |
Figure 1: Framework of the DnCNN network |
Architecture of DnCNN
The DnCNN architecture,28 has been extensively utilized in image restoration and artifact removal. DnCNN is a popular neural network denoising framework that is intended to denoise additive white Gaussian noise, image restoration, single image super-resolution, and deblocking JPEG images. For a given noisy input image, the noise observation is denoted as x = y + v, and the discriminative model for denoising attempts to acquire the mapping-function, F(x) = y. The DnCNN approach leverages the residuals or skip connection learning process to train the residual transformation R(x) = v; subsequently, y = x – R(x), and DnCNN applies the loss function (l) to enhance model parameters that are R (∂) specific to the DnCNN framework.
The Loss function is calculated as the average mean-squared error among the residual and the predicted image based on noisy images.
The training process of the DnCNN was performed using a loss function.
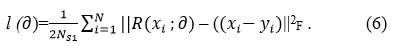
where,
∂ represents trainable parameters of the DnCNN network.
N represents the pairs of clean and distorted training image patches.
The architecture of the DnCNN was accustomed to reduce boundary artifacts. It includes
Deep Architecture
DnCNN architecture with depth D, includes 3 types of layers.
Layer 1
Convolutional_layers + ReLu (Rectified Linear Unit) activation function, including 64 filters with each aspect of existed dimensions [3 × 3 × c] (total channels) to generate 64 feature representations and Rectified Liner Units. Here, non linearity is obtained by ReLu ((max (0,.)). Here, the channel quantity for grayscale image is 1; The number-of-channels for a color image is 3 (R-red, G-green, B-blue).
Layer 2
Convolutional_layer + ReLu + Batch normalization (BN) through the addition of 64 convolutional_filters, the size of each dimension is 3 × 3 × 64, batch normalization was performed between each aspect of the convolutional_layer and the (ReLu ) activation function, and the depth of the layers was 2 ~ (D – 1).
Layer 3
The convolutional_layer is mainly applied for image restoration via 64 filters of size 3 × 3 × 64.
Reducing Boundary Artifacts
In general, core vision techniques require the size of resulting image to be consistent with the given input image. Here, DnCNN applies simple-zero padding and does not exhibit any artifacts. DnCNN directly pads the zeroes before convolution; thus, each feature mapping relating to existing middle layers exhibit a size equivalent to that of the specified image.
The main contribution of DnCNN in noise reduction approach is that, it effectively utilizes the residual network strategy and batch normalization process to expedite the training and regularize the learning problem. The image Denoising approach is represented as a Discriminative-learning challenge to separate distortion from latent images.
Method noise refers to residual noise or artifacts introduced by the denoising algorithm used in image processing. The residual noise retains pixel information in the image after applying a noise suppression or denoising algorithm. This shows the discrepancy among a noisy input and a (filtered) denoised image.
Method noise = noisy input image – denoised (processed) image.
Method noise was applied to assess the effectiveness of the denoising techniques while preserving image structure and fine details.
Proposed Methodology
In this portion, firstly describe the flowchart and proposed methodology in detail.
Mathematically, image denoising can be represented as:
Y1 (i, j) = X1 (i, j) + n1(i, j).
where X1 (i, j) denotes a clean image, Y1 (i, j) denotes a noisy or distorted CT image, and n1 is supposed to be (AWGN) Additive-White-Gaussian-Noise in conjunction with a standard deviation (σ), and (i, j) represents pixel’s locations in an image.
The comprehensive summary of the suggested approach is depicted in Fig. 2. The proposed hybrid approach combining the preprocessing approach of NSST and postprocessing approach of method noise with CNN to achieve superior denoising results, i.e., effectively combining the structural features of NSST, the statistical features of Bayesian thresholding,38 and the learning capability of method noise-based CNN ,39 will further enhance the image denoising approach.
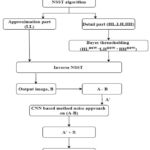 |
Figure 2: Flowchart of proposed CT image denoising using NSST with method noise-based CNN. |
Proposed Algorithm
Input: A Pre-processed, noisy CT image Ai,j.
Output: Final denoised CT image A”i,j.
Step 1: Perform NSST transform to decompose gaussian noisy input CT image Ai,j to acquire low- frequency components NSSTAl and high-frequency components NSSTAl (HL new, LH new and HH new).
Step 2: Implement CT image denoising using the following steps:
Evaluate noise variability.
Determine thresholding value.
Implement empirical Bayesian thresholding method on noisy coefficients to obtain thresholded NSST coefficients.
Evaluate Noise Variance
Estimate noise variance σ ̃noisy from noisy shearlet coefficients using robust median estimation 40 of CT image noise level in (HH) high-high shearlet diagonal coefficients.
Estimate the noise standard deviation σ ̃noisy

med= median (|ck (:)|), ∈HH subbands
where
ck represents set of high frequency (HH) coefficients of NSST decomposition of noisy CT image.
med represents median absolute deviation of all coefficients.
The constant 0.6745 is used for robust estimation of the noise level.
Calculate noise variance σ ̃noisy2
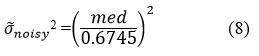
Implement Bayesian Thresholding Process
The threshold value is calculated for retaining the image’s intricate details of an image and perform noise suppression effectively.
The threshold value is selected as:
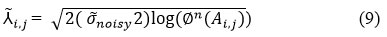
where,
⅄ ̃i,j denotes the threshold value used for empirical Bayes thresholding.
σ ̃noisy2 denotes the estimated noise variation.
∅n (Ai,j) denotes the amount of elements of image Ai,j.
log signifies the natural logarithm function.
Perform thresholding of coefficients tk using empirical Bayes thresholding.
Bayesian soft thresholding process:
For each coefficient|Ai,j|, find the absolute value of Ai,j.
Compute the sign of the coefficients.
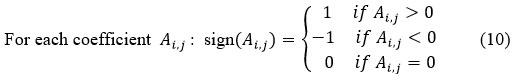
Apply Bayesian soft thresholding approach for each coefficient, is described as:
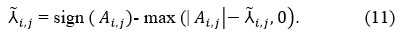
This function works as follows for thresholding:
If |Ai,j| is less than or equal to (<=), the given thresholding A ̃ i,j is set to 0. This procedure effectively removes small coefficients assumed to be dominated by gaussian noise. Then replace the original noisy coefficients Ai,j with thresholded coefficients tk
Step 3: Perform inverse NSST transform on thresholded NSSTAh coefficients (HLnew, LH new and HH new) to reconstruct the semi denoised CT image, Bi,j.
Calculate the residual image A’i,j.
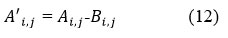
Here A’i,j represents the discrepancy between a noisy image Ai,j and its reconstructed semi denoised image counterpart Bi,j.
When A’i,j approaches 0, the noise in the original signal is successfully eliminated through denoising. A’i,j is residual image that also has some noisy coefficients that affect the quality of image.
Step 4: Calculate C’i,j by applying method noise CNN on residual image A’ i.e., DnCNN on (A’i,j).
Step 5: calculate final denoised image
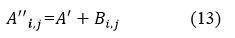
By combining inverse thresholded NSST coefficients (Bi,j) with the denoised method noise-based CNN on the residual image (Ci,j) to get the final denoised image (A’’i,j).
A Brief Explanation of Proposed Methodology
During the experiments , noisy CT images are commonly contaminated by Gaussian noise, The hybrid approach, non-subsampled shearlet transform is applied for decomposition of noisy image into an approximation (LL new) and detail part (LH new, HL new and HH new). The (approximation) high frequency components are further decomposed into multidirectional subbands to represent image features in a detailed manner. The noise variance was estimated in high frequency components using the median-absolute-deviation (MAD).The Bayesian thresholding function, which selects all noisy NSST coefficients, then calculates optimal threshold values to obtain the denoised coefficients. The inverse NSST transform is used to reconstruct denoised CT images from high frequency thresholded NSST coefficients, resulting in a denoised CT image. The denoised image, that is, the reconstructed image, preserves fine details but retains some residual noise. To solve this problem, calculated the method noise to visualize the discrepancies among the noisy and denoised NSST coefficients. Here, the method noise process was applied to capture residuals or any distortions introduced during the CT image denoising process. The deep CNN,28 network architecture is applied to method noise to learn complex noise patterns in order to capture intricate details effectively and reduce noise components. Finally, residual learning denoised image was combined with the NSST high frequency thresholded denoised image to acquire the final denoised CT-image. The final restored image preserves fine details and structural information while simultaneously removing noise and other artifacts.
Results and Discussion
The exploratory evaluation is carried out on noisy grayscale CT images with pixel’s size 512×512. Initially, CT scan test images are obtained from “large COVID-19 CT-scan slice dataset” to determine the efficacy of the suggested denoising method. The Noise-free or clean CT images are required as a reference image to analyse the denoising method’s performance. Fig. 3. is considered as CT images 1, 2,3, and 4 respectively. Fig. 4. depicts the addition of additive gaussian with a noise variance of 10. To test the experimental results, Additive-white-gaussian-noise is added at different noise levels(𝜎 = 5, 10, 15, 20) to analyze the effectiveness of various denoising techniques and assess the qualitative performance of noisy CT images.
Quantitative Evaluation Metrics
The qualitative result analysis of the suggested methodology uses diverse quality factors , i.e., PSNR, SNR, SSIM, ED, and UIQI,10,41.
PSNR (Peak Signal-to-Noise ratio) is used to assess the quality of restored images relative to the original images. PSNR evaluates the ratio between the given maximum signal strength and the power of distorted noise, i.e., the difference between the clean image and filtered image. For the input CT image X and the denoised CT image Y.
PSNR is denoted as
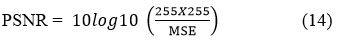
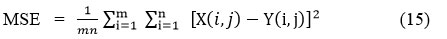
Where MSE represents the mean-square-error between the original image and the denoised CT image.
X (i, j) depicts the clean CT image.
Y (i, j) depicts the filtered CT image.
m x n represents the pixel’s size of clean CT image and a denoised CT image.
(SNR) Signal-to-Noise ratio is a qualitative metric to measure and quantify the desired signal strength related to the distortion or noise level. It is measured in the form of decibels, and it is also used to analyze the image quality.
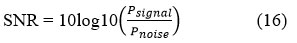
SSIM (Structural-Similarity-Index-Measure) is a metric serves as a measure to evaluate the similarity among two images. It is mainly relying on three parameters: luminance, contrast, and structural features, and the SSIM values vary between -1 and 1, where 1 denotes absolute similarity and -1 denotes discrepancy between two images.
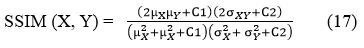
X represents clean CT image.
Y represents denoised or filtered CT image.
µX and µY are denoted as the local means, σX, σY are denoted as standard deviations, and σXY is image’s covariance of X and Y Here, C1=(k1D)2, C2= (k2D)2 are constant values to stabilize division with zeros, where D is the variation in pixel values between 2bits-per-pixel -1 and 1. Here, k1= 0.01 & k2 = 0.03.
The Entropy Difference is the statistical measure of randomness present in an image suitable for analyzing the texture of the given source images. Shannon entropy is estimated between the clean image (Xi) and the denoised-CT image (Yi). The dissimilarity in the mean value is denoted as ED.
ED is computed as:
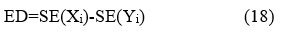
Where, SE denotes Shannon Entropy.
Shannon Entropy is computed as:
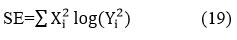
UIQI (Universal-Image-Quality-Index) is a benchmark used to determine the quality of a denoised CT image and its corresponding reference image.
The UIQI between two images original image distorted image is defined as:
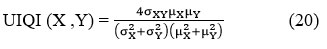
μX μY represents the average values of the given CT images X and Y.
σx2 and σy2denotes the variances of X and Y.
σXY represents the covariance of the images X and Y.
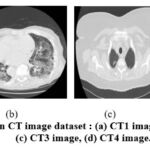 |
Figure 3: Clean CT image dataset : (a) CT1 image, (b) CT2 image, (c) CT3 image, (d) CT4 image. |
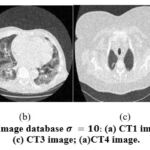 |
Figure 4: Noisy CT image database (a) CT1 image; (b) CT2 image; (c) CT3 image; (a)CT4 image. |
Table 1: PSNR Comparison for the proposed methodology with wiener, median, bilateral, DWT, curvelet transform, contourlet transform, and DnCNN for CT1, CT2, CT3, and CT4 images at various gaussian noise variances (𝜎=5, 10, 15, 20 of noise intensity).
|
CT Image |
𝜎 |
PSNR |
|
|
|
CT Image |
PSNR |
|
|
|
|
|
|
5 |
10 |
15 |
20 |
|
5 |
10 |
15 |
20 |
|
CT 1 |
Wiener filter9 |
26.9227 |
21.0561 |
17.1016 |
15.9510 |
CT 2 |
28.8351 |
22.3409 |
20.0957 |
18.0717 |
|
512×512 |
Median filter6 |
25.4818 |
21.2815 |
18.0291 |
16.1741 |
512×512 |
26.8228 |
22.6641 |
19.3134 |
17.7229 |
|
|
Bilateral filter7 |
25.8456 |
20.9348 |
18.3621 |
15.5553 |
|
28.2931 |
22.3078 |
19.6007 |
17.4489 |
|
|
NSST with Bivariate17 |
35.0639 |
29.4426 |
26.1123 |
23.7630 |
|
34.9416 |
29.2023 |
25.8483 |
23.5604 |
|
|
DWT11 |
36.1510 |
31.7197 |
28.9528 |
27.1211 |
|
36.8484 |
32.4378 |
29.6757 |
27.8361 |
|
|
Curvelet transform12 |
24.7381 |
17.8315 |
15.2633 |
15.2232 |
|
24.4989 |
17.8109 |
15.2434 |
15.3776 |
|
|
Contourlet transform13 |
31.1441 |
28.4641 |
27.7560 |
27.4756 |
|
31.9356 |
29.3965 |
28.6934 |
28.5351 |
|
|
DnCNN28 |
26.9654 |
21.3395 |
17.8176 |
16.1083 |
|
27.1848 |
23.4148 |
19.8904 |
17.8606 |
|
|
Proposed |
38.6875 |
35.4432 |
33.5507 |
32.0938 |
|
39.3028 |
35.7841 |
33.6865 |
32.2141 |
|
CT 3 |
Wiener filter9 |
27.2958 |
21.5341 |
18.9843 |
15.5837 |
CT 4 |
30.4803 |
24.1262 |
20.8647 |
19.7389 |
|
512×512 |
Median filter6 |
27.6440 |
20.3498 |
17.9301 |
16.7986 |
512×512 |
29.9581 |
24.2495 |
20.9963 |
19.8384 |
|
|
Bilateral filter7 |
27.4662 |
21.0894 |
18.3180 |
16.2724 |
|
31.6511 |
24.4505 |
20.8721 |
19.4807 |
|
|
NSST with Bivariate17 |
34.6530 |
28.8408 |
25.4758 |
23.1329 |
|
34.5294 |
28.9670 |
25.8372 |
23.6112 |
|
|
DWT11 |
37.7380 |
32.8812 |
30.1778 |
27.9877 |
|
35.9373 |
31.3968 |
28.6151 |
26.5497 |
|
|
Curvelet transform12 |
24.1199 |
17.7353 |
15.3748 |
15.5832 |
|
24.6752 |
18.2605 |
15.7387 |
15.7257 |
|
|
Contourlet transform13 |
31.2001 |
28.7470 |
28.1473 |
27.9475 |
|
32.1699 |
30.3563 |
30.0802 |
30.2322 |
|
|
DnCNN28 |
25.5178 |
21.4493 |
19.0634 |
16.3844 |
|
30.2987 |
25.0755 |
22.3263 |
19.9453 |
|
|
Proposed |
40.0415 |
37.0541 |
35.2108 |
33.7444 |
|
38.7044 |
35.1286 |
33.1462 |
31.7048 |
Table 2: SNR comparison for the suggested approach with wiener, median, bilateral, DWT, curvelet transform, contourlet transform, and DnCNN for CT1, CT2, CT3, and CT4 images at various gaussian noise variances (𝜎=5, 10, 15, 20 of noise intensity).
|
CT Image |
𝜎 |
SNR |
|
|
|
CT Image |
SNR |
|
|
|
|
|
|
5 |
10 |
15 |
20 |
|
5 |
10 |
15 |
20 |
|
CT 1 |
Wiener |
23.1806 |
17.3140 |
13.3595 |
12.2089 |
CT 2 |
24.3356 |
17.8414 |
15.5962 |
13.5722 |
|
512×512 |
Median |
21.7397 |
17.5394 |
14.2870 |
12.4320 |
512×512 |
22.3233 |
18.1646 |
14.8139 |
13.2234 |
|
|
Bilateral |
22.1035 |
17.1927 |
14.6200 |
11.8132 |
|
23.7936 |
17.8083 |
15.1012 |
12.9494 |
|
|
NSST with Bivariate17 |
31.3218 |
25.7005 |
22.3702 |
20.0209 |
|
30.4421 |
24.7028 |
21.3488 |
19.0609 |
|
|
DWT11 |
32.4093 |
27.9826 |
25.2195 |
23.3900 |
|
32.3488 |
27.9392 |
25.1814 |
23.3493 |
|
|
Curvelet transform12 |
20.8339 |
13.7838 |
11.1153 |
10.9738 |
|
19.9783 |
13.2432 |
10.6259 |
10.6641 |
|
|
Contourlet transform13 |
21.0065 |
14.0935 |
11.5272 |
11.3866 |
|
19.9740 |
13.2892 |
10.8254 |
10.8066 |
|
|
DnCNN28 |
23.2233 |
17.5974 |
14.0755 |
12.3663 |
|
22.6853 |
18.9153 |
15.3909 |
13.3611 |
|
|
Proposed |
34.9454 |
31.7011 |
29.8086 |
28.3517 |
|
34.8033 |
31.2846 |
29.1870 |
27.7154 |
|
|
|
|
|
|
|
|
|
|
|
|
|
CT 3 |
Wiener |
24.3152 |
18.5534 |
16.0036 |
12.6031 |
CT 4 |
24.7123 |
18.3583 |
15.0967 |
13.9709 |
|
512×512 |
Median |
24.6634 |
17.3691 |
14.9494 |
13.8179 |
512×512 |
24.1901 |
18.4815 |
15.2284 |
14.0705 |
|
|
Bilateral |
24.4855 |
18.1087 |
15.3373 |
13.2917 |
|
25.8831 |
18.6825 |
15.1041 |
13.7127 |
|
|
NSST with Bivariate17 |
31.6724 |
25.8601 |
22.4952 |
25.1522 |
|
28.7614 |
23.1990 |
20.0692 |
17.8432 |
|
|
DWT11 |
34.7583 |
29.9035 |
27.2033 |
25.0127 |
|
30.1727 |
25.6366 |
22.8592 |
20.7994 |
|
|
Curvelet transform12 |
21.1356 |
14.6744 |
12.1857 |
12.2720 |
|
18.9421 |
12.6007 |
10.0722 |
09.9956 |
|
|
Contourlet transform13 |
21.0577 |
14.7171 |
12.4321 |
12.5608 |
|
18.8854 |
12.5286 |
09.9644 |
09.9185 |
|
|
DnCNN28 |
22.5371 |
18.4687 |
16.0827 |
14.4038 |
|
24.5307 |
19.3075 |
16.5583 |
14.1773 |
|
|
Proposed |
37.0609 |
34.0742 |
32.2301 |
30.7637 |
|
32.9364 |
29.6594 |
27.3780 |
25.9118 |
Table 3: SSIM comparison for the proposed ensembled approach with wiener, median, bilateral, DWT, curvelet transform, contourlet transform, and DnCNN for CT1, CT2, CT3, and CT4 images at various gaussian noise variances (𝜎=5, 10, 15, 20 of noise intensity).
|
CT Image |
𝜎 |
SNR |
|
|
|
CT Image |
SNR |
|
|
|
|
|
|
5 |
10 |
15 |
20 |
|
5 |
10 |
15 |
20 |
|
CT 1 |
Wiener |
23.1806 |
17.3140 |
13.3595 |
12.2089 |
CT 2 |
24.3356 |
17.8414 |
15.5962 |
13.5722 |
|
512×512 |
Median |
21.7397 |
17.5394 |
14.2870 |
12.4320 |
512×512 |
22.3233 |
18.1646 |
14.8139 |
13.2234 |
|
|
Bilateral |
22.1035 |
17.1927 |
14.6200 |
11.8132 |
|
23.7936 |
17.8083 |
15.1012 |
12.9494 |
|
|
NSST with Bivariate17 |
31.3218 |
25.7005 |
22.3702 |
20.0209 |
|
30.4421 |
24.7028 |
21.3488 |
19.0609 |
|
|
DWT11 |
32.4093 |
27.9826 |
25.2195 |
23.3900 |
|
32.3488 |
27.9392 |
25.1814 |
23.3493 |
|
|
Curvelet transform12 |
20.8339 |
13.7838 |
11.1153 |
10.9738 |
|
19.9783 |
13.2432 |
10.6259 |
10.6641 |
|
|
Contourlet transform13 |
21.0065 |
14.0935 |
11.5272 |
11.3866 |
|
19.9740 |
13.2892 |
10.8254 |
10.8066 |
|
|
DnCNN28 |
23.2233 |
17.5974 |
14.0755 |
12.3663 |
|
22.6853 |
18.9153 |
15.3909 |
13.3611 |
|
|
Proposed |
34.9454 |
31.7011 |
29.8086 |
28.3517 |
|
34.8033 |
31.2846 |
29.1870 |
27.7154 |
|
|
|
|
|
|
|
|
|
|
|
|
|
CT 3 |
Wiener |
24.3152 |
18.5534 |
16.0036 |
12.6031 |
CT 4 |
24.7123 |
18.3583 |
15.0967 |
13.9709 |
|
512×512 |
Median |
24.6634 |
17.3691 |
14.9494 |
13.8179 |
512×512 |
24.1901 |
18.4815 |
15.2284 |
14.0705 |
|
|
Bilateral |
24.4855 |
18.1087 |
15.3373 |
13.2917 |
|
25.8831 |
18.6825 |
15.1041 |
13.7127 |
|
|
NSST with Bivariate17 |
31.6724 |
25.8601 |
22.4952 |
25.1522 |
|
28.7614 |
23.1990 |
20.0692 |
17.8432 |
|
|
DWT11 |
34.7583 |
29.9035 |
27.2033 |
25.0127 |
|
30.1727 |
25.6366 |
22.8592 |
20.7994 |
|
|
Curvelet transform12 |
21.1356 |
14.6744 |
12.1857 |
12.2720 |
|
18.9421 |
12.6007 |
10.0722 |
09.9956 |
|
|
Contourlet transform13 |
21.0577 |
14.7171 |
12.4321 |
12.5608 |
|
18.8854 |
12.5286 |
09.9644 |
09.9185 |
|
|
DnCNN28 |
22.5371 |
18.4687 |
16.0827 |
14.4038 |
|
24.5307 |
19.3075 |
16.5583 |
14.1773 |
|
|
Proposed |
37.0609 |
34.0742 |
32.2301 |
30.7637 |
|
32.9364 |
29.6594 |
27.3780 |
25.9118 |
Table 4: ED Comparison for the proposed approach with wiener, median, bilateral, DWT, curvelet transform, contourlet transform, and DnCNN for CT1, CT2, CT3, and CT4 images at various gaussian noise variances (𝜎=5, 10, 15, 20 of noise intensity).
|
CT Image |
𝜎 |
ED |
|
|
|
CT Image |
ED |
|
|
|
|
|
|
5 |
10 |
15 |
20 |
|
5 |
10 |
15 |
20 |
|
CT 1 |
Wiener |
0.2614 |
0.4414 |
0.5418 |
0.6709 |
CT 2 |
0.2177 |
0.3527 |
0.4904 |
0.5725 |
|
512×512 |
Median |
0.2170 |
0.4034 |
0.5029 |
0.5559 |
512×512 |
0.2219 |
0.3135 |
0.3555 |
0.4014 |
|
|
Bilateral |
0.1795 |
0.4045 |
0.5949 |
0.7027 |
|
0.1673 |
0.3201 |
0.4543 |
0.5704 |
|
|
NSST with Bivariate17 |
0.4774 |
0.7059 |
0.8858 |
1.0016 |
|
0.5220 |
0.7635 |
0.9367 |
1.0175 |
|
|
DWT11 |
0.1864 |
0.0496 |
0.0314 |
0.0524 |
|
0.7585 |
0.6040 |
0.5585 |
0.4996 |
|
|
Curvelet transform12 |
0.5713 |
0.5397 |
0.5113 |
0.5491 |
|
0.5713 |
0.5397 |
0.5113 |
0.5491 |
|
|
Contourlet transform13 |
0.5716 |
0.5356 |
0.5147 |
0.5532 |
|
0.0596 |
0.0004 |
0.0272 |
0.0219 |
|
|
DnCNN28 |
0.2900 |
0.1760 |
0.1220 |
0.0996 |
|
0.2331 |
0.1782 |
0.1261 |
0.1137 |
|
|
Proposed |
0.0383 |
0.0583 |
0.0719 |
0.0841 |
|
0.0330 |
0.0550 |
0.0699 |
0.0793 |
|
CT 3 |
Wiener |
0.0944 |
0.2976 |
0.4661 |
0.4913 |
CT 4 |
0.0019 |
0.0415 |
0.0594 |
0.0611 |
|
512×512 |
Median |
0.1303 |
0.2915 |
0.3970 |
0.4959 |
512×512 |
0.0165 |
0.1158 |
0.1584 |
0.1983 |
|
|
Bilateral |
0.0648 |
0.2603 |
0.4550 |
0.6012 |
|
0.0449 |
0.0429 |
0.0566 |
0.1164 |
|
|
NSST with Bivariate17 |
0.5307 |
0.8390 |
1.0659 |
1.2341 |
|
0.3086 |
0.4871 |
0.5824 |
0.6565 |
|
|
DWT11 |
0.0734 |
0.0207 |
0.0020 |
0.0052 |
|
0.0342 |
0.0760 |
0.1391 |
0.2021 |
|
|
Curvelet transform12 |
0.2521 |
0.2080 |
0.2045 |
0.2320 |
|
0.0490 |
0.1310 |
0.1386 |
0.0825 |
|
|
Contourlet transform13 |
0.2514 |
0.2189 |
0.2108 |
0.2303 |
|
0.0511 |
0.1296 |
0.1394 |
0.0881 |
|
|
DnCNN28 |
0.0544 |
0.0314 |
0.0292 |
0.0307 |
|
0.0521 |
0.1397 |
0.1617 |
0.1828 |
|
|
Proposed |
0.0361 |
0.0544 |
0.0772 |
0.0771 |
|
0.0204 |
0.0360 |
0.0484 |
0.0587 |
Table 5: UIQI Comparison for the proposed method with comparison of UIQI values with wiener, median, bilateral, DWT, curvelet transform, contourlet transform, and DnCNN for CT1, CT2, CT3, and CT4 images at various gaussian noise variances (𝜎=5, 10, 15, 20 of noise intensity).
|
CT Image |
𝜎 |
UIQI |
|
|
|
CT Image |
UIQI |
|
|
|
|
|
|
5 |
10 |
15 |
20 |
|
5 |
10 |
15 |
20 |
|
CT 1 |
Wiener |
0.9945 |
0.9776 |
0.9412 |
0.9209 |
CT 2 |
0.9958 |
0.9803 |
0.9655 |
0.9430 |
|
512×512 |
Median |
0.9922 |
0.9792 |
0.9543 |
0.9277 |
512×512 |
0.9935 |
0.9824 |
0.9602 |
0.9408 |
|
|
Bilateral |
0.9930 |
0.9700 |
0.9567 |
0.9130 |
|
0.9953 |
0.9802 |
0.9613 |
0.9339 |
|
|
NSST with Bivariate17 |
0.9989 |
0.9959 |
0.9910 |
0.9845 |
|
0.9983 |
0.9949 |
0.9890 |
0.9812 |
|
|
DWT11 |
0.9991 |
0.9976 |
0.9955 |
0.9931 |
|
0.9991 |
0.9976 |
0.9955 |
0.9932 |
|
|
Curvelet transform12 |
0.9874 |
0.9363 |
0.8822 |
0.8774 |
|
0.9874 |
0.9363 |
0.8822 |
0.8774 |
|
|
Contourlet transform13 |
0.9874 |
0.9364 |
0.8824 |
0.8746 |
|
0.9848 |
0.9275 |
0.8696 |
0.8644 |
|
|
DnCNN28 |
0.9947 |
0.9800 |
0.9525 |
0.9272 |
|
0.9940 |
0.9854 |
0.9656 |
0.9431 |
|
|
Proposed |
0.9995 |
0.9989 |
0.9984 |
0.9977 |
|
0.9995 |
0.9989 |
0.9982 |
0.9975 |
|
CT 3 |
Wiener |
0.9955 |
0.9824 |
0.9667 |
0.9212 |
CT 4 |
0.9963 |
0.9834 |
0.9631 |
0.9510 |
|
512×512 |
Median |
0.9960 |
0.9779 |
0.9600 |
0.9462 |
512×512 |
0.9958 |
0.9839 |
0.9645 |
0.9527 |
|
|
Bilateral |
0.9957 |
0.9807 |
0.9614 |
0.9348 |
|
0.9972 |
0.9847 |
0.9633 |
0.9479 |
|
|
NSST with Bivariate17 |
0.9979 |
0.9917 |
0.9820 |
0.9693 |
|
0.9985 |
0.9946 |
0.9888 |
0.9812 |
|
|
DWT11 |
0.9989 |
0.9967 |
0.9939 |
0.9899 |
|
0.9989 |
0.9969 |
0.9941 |
0.9906 |
|
|
Curvelet transform12 |
0.9751 |
0.8941 |
0.8200 |
0.8228 |
|
0.9853 |
0.9346 |
0.8800 |
0.8759 |
|
|
Contourlet transform 3 |
0.9747 |
0.8934 |
0.8291 |
0.8207 |
|
0.9853 |
0.9350 |
0.8798 |
0.8747 |
|
|
DnCNN28 |
0.9936 |
0.9833 |
0.9703 |
0.9429 |
|
0.9962 |
0.9869 |
0.9744 |
0.9540 |
|
|
Proposed |
0.9993 |
0.9989 |
0.9981 |
0.9973 |
|
0.9994 |
0.9986 |
0.9979 |
0.9971 |
Visual Evaluation of the Experimental Results with Additive White Gaussian Noise
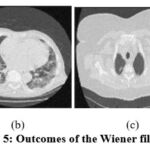 |
Figure 5: Outcomes of the Wiener filter [9]. |
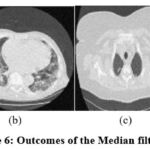 |
Figure 6: Outcomes of the Median filter [6]. |
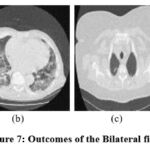 |
Figure 7: Outcomes of the Bilateral filter [7]. |
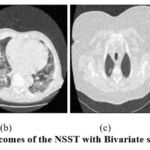 |
Figure 8: Outcomes of the NSST with Bivariate shrinkage [17]. |
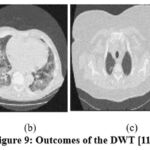 |
Figure 9: Outcomes of the DWT [11]. |
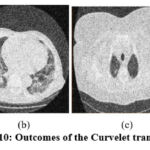 |
Figure 10: Outcomes of the Curvelet transform [12]. |
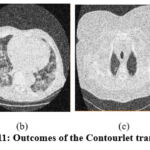 |
Figure 11: Outcomes of the Contourlet transform [13]. |
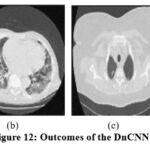 |
Figure 12: Outcomes of the DnCNN [28] |
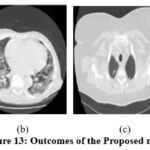 |
Figure 13: Outcomes of the Proposed method. |
For strong comparison, the noisy CT images are denoised using various approaches, like wiener, median, bilateral, DWT, curvelet transform, contourlet transform, DnCNN, and the proposed method. The performance criteria, including PSNR, SNR, SSIM, ED, and UIQI, are assessed across different noise variances, as presented in Table no. from 1 to 5. The results of the proposed method are highlighted in bold. It is clear from, comparing Tables 1 to 5 that the proposed method outperforms the mentioned standard methods.Table no.1, illustrates comparative analysis of different denoising methods based on PSNR, while Table 2, based on SNR, while Table 3 on SSIM, while Table 4 on ED and Table 5 on UIQI.
Table no.1 shows PSNR results for different denoising approaches applied to four CT images (CT1, CT2, CT3 and CT4) at various gaussian noise levels (𝜎 = 5,10,15,20). The PSNR refers how efficiently the signal is preserved in relation to the level to which its representation has been distorted by noise. The higher PSNR values generally indicates better imaging quality. The proposed method proves to be consistently achieving the highest PSNR values among all mentioned methods. For PSNR, a 0.5% increase in noise results in a considerable decrease of 2-5 points in the PSNR value. SNR measures the ratio between intensity of the desired signal and amount of the noise present in the image. For SNR, increase in noise causes substantial reduction of 1-6 points in SNR value as shown in Table no.2. The method consistently delivers the highest SNR value in comparison with other standard denoising methods.
SSIM is commonly employed to evaluate the imaging quality after compression, denoising, or other processing approaches. It plays a vital role in assessing how well various image denoising algorithms preserve structural information. The SSIM range extends between 0 and 1, with values closer to 1 indicating better denoising performance. For instance, the SSIM values of CT image no. 3 (0.9400) and CT image no. 4 (0.9632) at 𝜎 = 5 are somewhat inferior to those of the proposed method, as shown in Table no.3. CT images with SSIM values greater than 0.80 are considered to be of high quality. It is evident that the proposed method yields superior results compared to existing standard methods in terms of SSIM values.
Table no.4 presents ED values of various denoising methods. A lower ED indicates that the denoising method has well preserved the features of the clean image. The ED values of the Wiener filter for CT image 2 at noise levels 10, 15, and 20 exhibit inconsistent performance, with the ED11 values changing markedly. In CT image 3, the ED7 values of the DWT approach are also quite a bit lower compared to the proposed method at noise levels 10, 15, and 20. However, the difference between the proposed method and the outcomes of other standard approaches is quite small. The proposed method consistently outperforms the other denoising techniques in terms of minimizing entropy difference and preserving the original image’s details across various noise intensity levels.
Table no.5 shows a detailed comparison of various denoising methods applied on 4 CT images (CT1, CT2, CT3 and CT4) including proposed method based on UIQI values at different gaussian noise levels (σ = 5,10,15,20). Here, UIQI is used to analyze the quality of denoised-CT images, where higher UIQI value indicates superior image quality. The proposed method performs well at (𝜎 =5) lower noise levels. The proposed technique achieves the highest UIQI values among all standard methods, with DWT and proposed method exhibiting slightly better performance.
The exploratory results of, 9 as depicted in Fig. 5 are evaluated, and it is noted that the denoising scheme is performed well but does not effectively preserve structural details at higher noise levels. In the experimental data of ,6 as illustrated in Fig. 6, it is observed that the noise suppression is performed effectively, but at higher noise levels, it is unable to preserve the image’s smoothness and edge information in detail. According to the experimental results of,7 as demonstrated in Fig. 7, the noise suppression is done effectively. The SSIM of CT3 and CT4 images shows better outcomes at noise variance 5. The experimental findings of,17 as depicted in Fig. 8, show that noise suppression and other artifacts are reduced successfully, and it is observed that as more noise is added, resulting blurry images.
In the experimental results of,11 as shown in Fig. 9, the noise suppression is performed well, but as the noise increases, it affects image clarity and quality. The experimental outcomes of,12 as shown in Fig. 10, give superior noise suppression but fail to preserve the image’s structural and fine details at higher noise levels. The ED of the CT2 image at gaussian noise level 10, 20 gives better outcomes. In the experimental results of,13 as shown in Fig. 11, the suppression of noise is performed well. If noise variance has increased, the resulting image overall looks blurry. The experimental results of the proposed technique as depicted in Fig. 12, illustrate that the proposed study mitigates noise effectively and also retain edges, other structural, and fine details of an image. The experimental outcomes are tested on different noisy intensities; however, the images are displayed only at noise variance 10.
The proposed algorithm combines NSST with a thresholding function and its noise-based CNN approach. This approach exploits NSST with Bayes thresholding to get denoised NSST coefficients. Here, the NSST domain provides various features of an image depicts in different dimensions and different directional subbands. The main benefit of recommended methodology is, applying the method noise-based CNN approach gives better noise reduction and preserves edge’s information. In high textured, noisy CT images, some residuals or the image’s structural and fine details may get damaged during denoising using the NSST domain. To overcome that, the proposed method noise-based approach using CNN gives better performance in order to improve image quality. Performance metrics like PSNR, SNR, SSIM, ED, and UIQI have proven that the proposed method remarkably reduces noise at different noise intensities and also provides better images and fine detail preservation as compared to other denoising techniques. Here, the ED values in the proposed method are near zero. Hence, it has been proven this novel hybrid approach gives improvised outcomes in case of visual clarity, quality, and performance benchmarks.
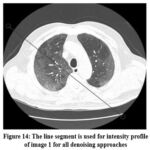 |
Figure 14: The line segment is used for intensity profile of image 1 for all denoising approaches |
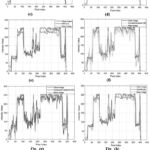 |
Figure 15: Intensity profiles of clean image, noisy image and proposed approach, respectively; |
(a). Intensity profile of clean image against noisy image and proposed filtered image9; (b) Intensity profile of clean image ,noisy image and proposed filtered image6; (c) Intensity profile of clean image ,noisy image and proposed filtered image7; (d) Intensity profile of clean image ,noisy image and proposed filtered image17; (e) Intensity profile of clean image ,noisy image and proposed filtered image11; (f) Intensity profile of clean image ,noisy image and proposed filtered image12; (g) Intensity profile of clean image ,noisy image and proposed filtered image13; (h) Intensity profile of clean image, noisy image and proposed filtered image28; (i) Intensity profile of clean image, noisy image and proposed filtered image.
Another critical assessment for addressing variations among noisy CT image, clean CT image, and denoised or filtered CT image, is obtaining the pixel’s intensity’s profile. The outcome shows the clean image, a noisy image (noise variance 10), and a denoised or filtered CT image’s intensity profile as shown in Fig. 14, the lowest difference has been figured out between an original or clean image and proposed denoised image i.e, the ensembled method provides effective noise suppression as well as preserving edges and fine details.
Conclusion and Future work
The CT images are extensively used in the medical and healthcare domain, as they precisely recognise the abnormality information of the patient. In the proposed work, initially gaussian noise was added at various levels, ranging in noise variance from 5 to 20. These noise variances are used to estimate the efficacy of various denoising techniques. The proposed study includes NSST and a noise-based CNN method to remove Gaussian noise in CT images. Here, NSST is used as a preprocessing operation to resolve the noisy CT image into various frequency subbands. Bayesian thresholding is applied to denoise noisy NSST coefficients. After denoising NSST coefficients, a postprocessing approach is used to get residuals that were preserved in denoised CT images. The DnCNN was applied to method noise to extract structural information and fine details of CT images. The experimental study employed four CT scan images, i.e., CT2, CT3, and CT4. Standard denoising filters were applied to noisy CT images. All experimental outcomes in proposed method are evaluated against standard methods. So, the ensembled method gives benchmarked performance in case of PSNR, SNR, SSIM, ED, and UIQI. The proposed study has proven that the experimental results from Table.no. from 1 to 5 and Fig.no. from 5 to 15, shows better results over the CT imaging from the perspective of visual quality.
For future work, must investigate the integration of advanced deep learning models to further improve denoising effectiveness and enhance computational efficiency. Moreover, broadening the study to encompass a larger and more varied collection of CT images can verify the reliability of the proposed method. Analyzing the use of the technique in various medical imaging modalities such as X-rays or MRI or could expand its applicability. Implementing real-time denoising features would improve clinical practices. Finally, analyzing the effect of denoising on diagnostic precision and patient outcomes would reveal valuable insights and understanding of its practical value.
Acknowledgment
I would like to express my deepest gratitude to my PhD supervisor, Prof. Deepak Garg, for his invaluable guidance, support, and encouragement throughout the course of this research. His insightful feedback and unwavering mentorship have been instrumental in shaping this work. I am equally grateful to my co-supervisor, Dr. Prabhishek Singh, for his constant advice, suggestions, and support, which have been crucial in navigating the complex challenges of this research work. A special thanks to Prof. Manoj Diwakar for sharing his expert knowledge and providing specific insights on CT image denoising, which significantly contributed to the refinement and depth of this research.
Funding Sources
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
The authors do not have any conflict of interest
References
- Albeshan, S., Algamdi, S., Alkhybari, E., Alhailiy, A., Fisal, N., Alsufyan, M., … & Abuhaimed, A. Radiation exposure and cancer risk of pediatric head CT scans: impact of age and scanning parameters. Radiation Physics and Chemistry.2024;216:111459.
CrossRef - Bagherzadeh, S., MirDerikvand, A., & MohammadSharifi, A. Evaluation of radiation dose and radiation-induced cancer risk associated with routine CT scan examinations. Radiation Physics and Chemistry.2024;217:111521.
CrossRef - Zhang, J., Gong, W., Ye, L., Wang, F., Shangguan, Z., & Cheng, Y. A review of deep learning methods for denoising of medical low-dose CT images. Computers in Biology and Medicine. 2024;108112.
CrossRef - Diwakar, M., Singh, P., & Garg, D. Edge-guided filtering-based CT image denoising using fractional order total variation. Biomedical Signal Processing and Control. 2024;92:106072.
CrossRef - Sadia, R. T., Chen, J., & Zhang, J. CT image denoising methods for image quality improvement and radiation dose reduction. Journal of Applied Clinical Medical Physics. 2024;25(2).
CrossRef - Diwakar, M., & Kumar, M. A review on CT image noise and its denoising. Biomedical Signal Processing and Control. 2018;42:73-88.
CrossRef - Zhang, M., & Gunturk, B. K.Multiresolution bilateral filtering for image denoising. IEEE Transactions on image processing. 2008; 17(12):2324-2333.
CrossRef - Li, Z., Yu, L., Trzasko, J. D., Lake, D. S., Blezek, D. J., Fletcher, J. G., … & Manduca, A. Adaptive nonlocal means filtering based on local noise level for CT denoising. Medical physics. 2014;41(1):011908.
CrossRef - Siddiqi, A. A Filter selection for removing noise from CT scan images using digital image processing algorithm. Biomedical Engineering: Applications, Basis and Communications. 2024;36(01):2350038.
CrossRef - Sagheer, S. V. M., & George, S. N. A review on medical image denoising algorithms. Biomedical signal processing and control. 2020;61:102036.
CrossRef - Mallat, S. G.A theory for multiresolution signal decomposition: the wavelet representation. IEEE transactions on pattern analysis and machine intelligence. 1989;11(7):674-693.
CrossRef - Candes, E., Demanet, L., Donoho, D., & Ying, L.Fast discrete curvelet transforms. multiscale modeling & simulation. 2006;5(3): 861-899.
CrossRef - Do, M. N., & Vetterli, M. The contourlet transform: an efficient directional multiresolution image representation. IEEE Transactions on image processing. 2005;14(12):2091-2106.
CrossRef - Kim, K., & Lee, Y. Block-matching and 3D filtering algorithm in X-ray image with photon counting detector using the improved K-edge subtraction method. Nuclear Engineering and Technology. 2024;56(6):2057-2062.
CrossRef - Li, Z., Yu, L., Trzasko, J. D., Lake, D. S., Blezek, D. J., Fletcher, J. G., … & Manduca, A. Adaptive nonlocal means filtering based on local noise level for CT denoising. Medical physics. 2014;41(1):011908.
CrossRef - Yang, W., Hong, J. Y., Kim, J. Y., Paik, S. H., Lee, S. H., Park, J. S., … & Jung, Y. J. A novel singular value decomposition-based denoising method in 4-dimensional computed tomography of the brain in stroke patients with statistical evaluation. Sensors. 2020;20(11):3063.
CrossRef - Diwakar, M., & Singh, P. CT image denoising using multivariate model and its method noise thresholding in non-subsampled shearlet domain. Biomedical Signal Processing and Control. 2020;57:101754.
CrossRef - Diwakar, M., Singh, P., Swarup, C., Bajal, E., Jindal, M., Ravi, V., … & Singh, T. Noise suppression and edge preservation for low-dose COVID-19 CT images using NLM and method noise thresholding in shearlet domain. Diagnostics. 2022; 12(11):2766.
CrossRef - Diwakar, M., & Kumar, M. CT image denoising using NLM and correlation‐based wavelet packet thresholding. IET Image Processing. 2018;12(5):708-715.
CrossRef - Chen, W., Shao, Y., Wang, Y., Zhang, Q., Liu, Y., Yao, L., … & Gui, Z. A novel total variation model for low-dose CT image denoising. IEEE Access. 2018;6:78892-78903.
CrossRef - Jifara, W., Jiang, F., Rho, S., Cheng, M., & Liu, S. Medical image denoising using convolutional neural network: a residual learning approach. The Journal of Supercomputing. 2019;75:704-718.
CrossRef - Trung, N. T., Trinh, D. H., Trung, N. L., & Luong, M. Low-dose CT image denoising using deep convolutional neural networks with extended receptive fields. Signal, Image and Video Processing. 2022; 16(7):1963-1971.
CrossRef - Yang, Q., Yan, P., Zhang, Y., Yu, H., Shi, Y., Mou, X., … & Wang, G. Low-dose CT image denoising using a generative adversarial network with Wasserstein distance and perceptual loss. IEEE transactions on medical imaging. 2018;37(6):1348-1357.
CrossRef - Yuan, J., Zhou, F., Guo, Z., Li, X., & Yu, H. HCformer: hybrid CNN-transformer for LDCT image denoising. Journal of Digital Imaging. 2023;36(5):2290-2305.
CrossRef - Han, K., Wang, Y., Chen, H., Chen, X., Guo, J., Liu, Z., … & Tao, D. A survey on vision transformer. IEEE transactions on pattern analysis and machine intelligence. 2022;45(1):87-110.
CrossRef - Abhisheka, B., Biswas, S. K., Purkayastha, B., Das, D., & Escargueil, A. Recent trend in medical imaging modalities and their applications in disease diagnosis: a review. Multimedia Tools and Applications. 2024;83(14): 43035-43070.
CrossRef - Sehgal, R., & Kaushik, V. D. Deep Residual Network and Wavelet Transform-Based Non-Local Means Filter for Denoising Low-Dose Computed Tomography. International Journal of Image and Graphics. 2024;2550072.
CrossRef - Zhang, K., Zuo, W., Chen, Y., Meng, D., & Zhang, L. Beyond a gaussian denoiser: Residual learning of deep cnn for image denoising. IEEE transactions on image processing. 2017;26(7):3142-3155.
CrossRef - Zhang, J., Niu, Y., Shangguan, Z., Gong, W., & Cheng, Y. A novel denoising method for CT images based on U-net and multi-attention. Computers in Biology and Medicine. 2023;152:106387.
CrossRef - Huang, Z., Chen, Z., Quan, G., Du, Y., Yang, Y., Liu, X., … & Hu, Z. Deep Cascade Residual Networks (DCRNs): Optimizing an Encoder–Decoder Convolutional Neural Network for Low-Dose CT Imaging. IEEE Transactions on Radiation and Plasma Medical Sciences. 2022; 6(8):829-840.
CrossRef - Selig, T., März, T., Storath, M., & Weinmann, A. Low-Dose CT Image Reconstruction by Fine-Tuning a UNet Pretrained for Gaussian Denoising for the Downstream Task of Image Enhancement. arXiv preprint arXiv:2403.03551. 2024.
- Song, G., Xu, W., & Qin, Y. NeXtResUNet: A neural network for industrial CT image denoising. Journal of Radiation Research and Applied Sciences. 2024;17(1):100822.
CrossRef - Byeon, H., Patel, R. K., Vidhate, D. A., Kiyosov, S., Rahin, S. A., Keshta, I., & Lakshmi, T. V. Non-sample fuzzy based convolutional neural network model for noise artifact in biomedical images. Discover Applied Sciences.2024;6(1):16.
CrossRef - Li, Q., Li, R., Li, S., Wang, T., Cheng, Y., Zhang, S., … & Wang, L. Unpaired low‐dose computed tomography image denoising using a progressive cyclical convolutional neural network. Medical Physics.2024; 51(2):1289-1312.
CrossRef - Çalişkan, A., & Çevik, U. An efficient noisy pixels detection model for CT images using extreme learning machines. Tehnički vjesnik. 2018;25(3):679-686.
CrossRef - Çalışkan, A. Finding complement of inefficient feature clusters obtained by metaheuristic optimization algorithms to detect rock mineral types. Transactions of the Institute of Measurement and Control. 2023;45(10):1815-1828.
CrossRef - Diwakar M, Lamba S, Gupta H. CT image denoising based on thresholding in shearlet domain. Biomed Pharmacol J. 2018;11(2):671-677.
CrossRef - Silverman BW. Wavelets in statistics: beyond the standard assumptions. Philos Trans R Soc Lond A. 1999;357(1760):2459-2473.
CrossRef - Singh, P., Diwakar, M., Gupta, R., Kumar, S., Chakraborty, A., Bajal, E., … & Paul, R. A method noise-based convolutional neural network technique for CT image Denoising. Electronics, 2022;11(21): 3535.
CrossRef - Abramovich, F., Angelini, C., & De Canditiis, D. Pointwise optimality of Bayesian wavelet estimators. Annals of the Institute of Statistical Mathematics. 2007;59(3):425-434.
CrossRef - Diwakar, M., & Kumar, M. CT image denoising using NLM and correlation‐based wavelet packet thresholding. IET Image Processing. 2018;12(5):708-715.
CrossRef








