Manuscript accepted on :22-02-2023
Published online on: 07-05-2024
Plagiarism Check: Yes
Reviewed by: Dr. Narasimha Murthy
Second Review by: Dr. Yuvaraj Mandavkar
Final Approval by: Dr. H Fai Poon
Sowmya Priya Manoharan , Sangilimuthu Alagar Yadav*
, Sangilimuthu Alagar Yadav* , Gnanaselvan Suvathika
, Gnanaselvan Suvathika , Priyadharshini Anandhan, Balamurugan Pandiyan
, Priyadharshini Anandhan, Balamurugan Pandiyan
Department of Biotechnology, Karpagam Academy of Higher Education, Coimbatore, Tamil Nadu, India.
Corresponding Author E-mail: smuthu.al@gmail.com
Abstract
Background: Phosphoinositide 3 kinase belongs to the enzyme family which is responsible for the development of cellular trafficking and uncontrolled cellular division which in turn to cancer metastasis. Activation of the PI3k-Akt-mTOR pathway tends to promote oncogenesis in Lung Cancer and its mutation leads to resistance in EGFR tyrosine kinase inhibitors. Objective: Inhibition of the initial PI3k receptor through natural bioactive phytocompounds from Argemone mexicana that may prevent the side effects caused by the synthetic medication and improve the patient's life quality as natural medicine. Method: According to the previous research we did the Bioactive phytochemical screening from the methanol extract (AME) and powder (AMP) of A. mexicana leaf by analysing the GC-MS (Agilent), UPLC-QTOF-ESI-MS (Waters India Pvt Limited). Analyzed phytochemicals from the studies were subjected to molecular docking analysis using SeeSAR 9.2 software against PI3K (PDP ID: 4FA6) receptor. Result: GC-MS revealed 40 compounds including Cryptopine (17.5), beta-sitosterol (9.57), and Protopine (8.6) were found as major compounds. LC-MS analysis showed the presence of major compounds with 13 elements compared to the literature survey in both positive and negative ESI ranges. 29 compounds from the GC-MS and LC-MS were selected for analyzing the interaction between the cancer receptor-ligand complex according to the binding strategy. Conclusion: Phytoconstituents such as Galactitol, Succinic acid and N-feruloyltryamine from A. mexicana showed good binding affinity range and maximised hydrogen bonding that may assure possessing anticancer effect by controlling the PI3K pathway. This study would lead a major role in the preliminary screening of Drug Targets against the PI3K receptor.
Keywords
Argemone mexicana; GC-MS; Lung Cancer; Molecular docking; PI3Kreceptor; UPLC-Q-TOF-MS
| Copy the following to cite this article: Manoharan S. P, Yadav S. A, Suvathika G, Anandhan P, Pandiyan B. Screening of Phosphoinositide-3 Kinase Inhibitors from Argemone mexicana Leaves. Biomed Pharmacol J 2024;17(2). |
| Copy the following to cite this URL: Manoharan S. P, Yadav S. A, Suvathika G, Anandhan P, Pandiyan B. Screening of Phosphoinositide-3 Kinase Inhibitors from Argemone mexicana Leaves. Biomed Pharmacol J 2024;17(2). Available from: https://bit.ly/4afwOvT |
Introduction
Phosphoinositide 3 kinase (PI3K) is also known as Phosphatidylinositol-3-kinase is a breakthrough receptor that responsible for the regulation of the pathway PI3K/Akt/mTOR (mechanistic target of rapamycin). Activation of PI3K involves numerous hallmarks of cancer such as apoptosis inhibition, invasion of tissue at an increased level, sustained angiogenesis, insensitivity to anti-growth signals and acquiring growth signal anatomy 1. Impairment in the downstream signaling of the Phosphoinositide 3-kinase (PI3K/Akt/mTOR) pathway causes an instant signal transduction cascade in the lung carcinoma cell growth. Activation of such PI3K pathway results in anti-apoptotic cancer cell proliferation and cell differentiation 2. Copansilib is such a pan inhibitor standard drug of PI3Kα inhibiting the autophosphorylation in the tyrosine residues of the receptor. This is also effective against PI3Kδ form by inducing apoptosis in tumor cells as well as primary inhibition in malignant B cell proliferation. Although side effects symptoms also persist in Copansilib usage as the drug for the malignant types 3.
Cells are the structural and functional unit of a healthy being. Several mutations or genetic impairment develops the proliferation of abnormal aberrated cells that leads to cancer prognosis. There are more than 200 types of cancer effects unisexually both men and women worldwide. Each cancer type differs in its location and exhibits diverse causes 4. Among the other types of cancer, Non-Small Cell Lung Cancer (NSCLC) remains the second most predominant case and the first in deaths worldwide. NSCLC is caused mainly due to direct and passive smoking, disordered unhealthy diet, pollution, radioactive and stress at last. These external, as well as internal effects, cause changes in the PI3K/Akt/mTOR pathway which are responsible for the abnormal growth of squamous cell carcinoma (NSLC) 1.
Medicinal plants remain in debate for cancer treatment and thus they have been used since the ancient period as traditional medications against several ailments. Phytocompounds such as withaferin, vinblastine, Camptothecin, Taxol and diosgenin have anticancer effects as already reported 5. Still, most of the anticancer potential of phytomedicine and medicinal plants remains unexplored. Argemone mexicana also known as the Mexican poppy belongs to the Papaveraceae family and grows up to 1.2 meters alongside agricultural and wasteland weed areas. This plant is found widely in Bangladesh and all parts of India. A. mexicana has potential medicinal properties such as anti-oxidant, analgesic, anti-malarial, antispasmodic, chemo sterilant, wound healing, nematocidal and remedy for snake poisoning. In Ayurveda and Yunani, the plant parts were used as purgative and sedative with skin disease treatment properties 6. Phytochemicals such as Phenolics, Alkaloids, Tannins, Carotenoids, Flavonoids, Terpenoids, Saponins, and Pectin were reported from A. mexicana 7. In silico screening of medicinally important phytocompounds of this plant against cancer, receptor remains less explored.
Artificial Intelligence (AI) plays important role in discovering the lead molecules by molecular interaction with specific receptors through Machine Learning (ML). This trending method adds more advantages to the primary selection of the drug targets and receptors of a particular disease that would help at the cellular level 8. Preliminary screening of the drugs which hits the molecular pathways should be focussed celestially to develop a medication for the predominant disease. The detailed study of functional and structural prospects of drug properties is so important in the drug development field that is carried out by pharmaceutical as well as research industries9. Investigation of the essential phytocompounds mainly secondary metabolites can be acquired in mass with the help of analytical instruments. Automated screening of volatile and non-polar phytocompounds such as alkaloids, lipids, fatty acids and volatile essential oil is majorly achieved by Gas Chromatography-Mass Spectrum Analysis (GC-MS). Phenolic phytoconstituents are detected with the help of LC-QTOF- ESI-MS in terms of positive and negative ionization. In this study, we attempted the integrated approach with both the volatile and phenolic phytocompound screening to fulfill the research gap in the binding mechanism against the PI3K receptor which is responsible for lung cancer proliferation.
Materials and Methods
Plant collection
Fresh and healthy plant of the A. mexicana was collected from Rangareddy District, Telangana state and authenticated in Botanical Survey of India, Coimbatore (vide No: BSI/SRC/5/23/2013-14/Tech./1855). Separated leaves were washed under tap water to free them from sand and dust. This is made to shade dry for a continuous week. Thus, dried leaves were course powdered in an electric grinder and weighed. This was stored in an air-tight container at room temperature for further studies.
Sample Preparation
For GC-MS analysis, Soxhlet extraction was performed with 50 ± 0.8g, of course, powdered A. mexicana Leaves. Analytical grade reagent of Carbinol was purchased from Himedia and used as the solvent for extraction. Up to 8 suctions were undergone to obtain the extract yield from A. mexicana (AME) in 24 hrs continuous extraction 10. Preparing the plant sample is a bit tedious process. The dried coarse powder of A. mexicana Leaves is mixed with a 1:2 proportion of carbinol and made centrifuged for 5 mins at 3000rpm. Thus, the supernatant was carefully removed without disturbing the pellet surface and sterile-filtered in a 0.22 µm PAL syringe filter. Followed by the process of sonication and degassing until 5 mins thoroughly in ultra sonicator – A. mexicana Powder (AMP). Respective extracts (AME & AMP) were taken for GC-MS and LC-MS studies.
Gas Chromatography-Mass Spectrum Analysis
AME has been checked for the Bioactive volatile phytoconstituents utilizing Agilent technologies GC Systems with GC7890A/ MS-59795C model (Agilent Technologies, Santa Clara, CA, USA). Capillary column – DB5MS was allocated with a length of 30 meters and diameter of 0.25mm with a film thickness of about 0.25mm for the screening of phytochemicals. High electron energy of 70eV was used to perform the spectroscopic detection of the electron ionization system with a mass scan range of 45-380 m/z. The initial temperature range was set to 30ºC- 350ºC and maintained at 10ºC for 4min as standard. Pure helium gas of 99.995% was used and the flow rate derivates as 1ml/min. The fragmentation pattern of A. mexicana Leaf extract for the Retention time of 25mins has been determined using the stored phytocompound in the NIST Library (National Institute of Science and Technology). Thus, retrieved data are separated and noted down for the Tentative name, structure, Molecular weight, Molecular Formula and Retention time 11.
Conditions and Sampling in UPLC-ESI-Q-TOF-MS analysis
Tentative identification of phenolic compounds present in the AMP was performed using Waters ACQUITY UPLC-ESI-Q-TOF-MS (Waters, USA). C18 Reverse Phase column has been used for the chromatographic separation and maintained the temperature of 40ºC. The gradient solvent A contains 100% Milliq water (HPLC Grade) and 0.1% of Formic acid whereas solvent B consisted of 100% of acetonitrile. The flow rate of the solvent was set at 1ml/ min with a 5ml injection volume. Sample AMP ran for optimizing the gradient elution program at initially 5% solvent B with a 6-minute duration. Followed by the gradient uptake of 10% (7mins), 15% (7 mins) and 20% (10 mins) of solvent B. Electron spray Ionization (ESI) Interference was performed to analyze the AMP’s Mass by Mass Spectrometer. Fragmented MS spectra obtained from the UPLC-MS are compared to the literature records to characterize the positive and negative modes12.
Receptor and Ligand Preparation for In Silico studies
Identified tentative Bioactive compounds from AME & AMP from the analyzed GC-MS and LC-MS chromatogram was subjected to the ligand preparation for computational molecular interaction with Phosphoinositide 3-kinase (PI3K) receptors. All the respective ligands (53 Åmolecules) were fetched from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) in 3D SDF format. Copansilib, a standard drug treated against the PI3K receptor was also downloaded in SDF format. The receptor responsible for the PI3K /Akt/mTOR pathway is PI3K was downloaded from the Protein Data Bank as PDB Files in the name of PDB ID:4JPS (https://www.rcsb.org/). This also checked for the binding site in the UniProt database to confer the amino acid residues 13. The process of dehydration and elimination of external ligands is done with Discovery Studio Client Software 4.5 and saved as a PDB file.
Molecular Docking
SeeSAR 9.2 commercial molecular docking software is used for structural interaction between the Phytomolecules from AME & AMP and PI3K receptor. The resolution of the receptor 4JPS (PDB DOI: 10.2210/pdb4JPS/pdb) is found to be 2.20 Å with a sequence length of 1074. The systematic interaction between the ligands and receptor along with the standard was subjected to the identification of Hydrogen Dehydration Calculation (Hyde), Ligand Efficiency (LE) and Estimated Binding affinity in the Docking mode. The binding poses of the interaction are confirmed for the top 5 poses. A binding score of best ligand-receptor hits is determined using the command prompt in the build software FlexX 4.1. Hydrogen bond (H Bonds) interaction promotes cellular function and increases the binding energy of bonded receptors and Ligands as the actual binding site of the receptor was highly preferred. The final selected Receptor-Ligand complex was analysed using DS Visualizer 4.5 software 14.
Results
Extraction of AME and AMP
50g of coarse powdered A. mexicana Leaf undergoing hot Soxhlet extraction yielded 9.8 ± 0.20 mg of AME extract taken for GC-MS analysis. 2g of coarsely powdered A. mexicana subjected to sonication and degassing yielded 0.85 ± 0.18μl of AMP extracts taken for LC-MS analysis.
Analysis of Volatile phytocompounds from AME
Qualitative estimation of AME in the determination of volatile medicinally important bioactive compounds was carried out with GC-MS analysis. Every phytocompounds has been identified using the highest similarity search hits with NIST Library having patterns of more than 2,00,000 by the spectrum interpretation. They were focused to attain the Molecular Weight (MW), Molecular Formula (MF) and percentage of peak area (Concentration) according to the Retention time. About 34 Bioactive tentative phytocompounds were identified from GC-MS analysis of AME shown in Table 1. The highest abundant peaks identified from the GC-MS analysis are Cryptopine (17.5), beta-sitosterol (9.57), Protopine (8.6), Butane 2,2,–thiobis (5.52) beta-amyrin (4.88), Phytol(4.72), triethylphosphine (4.69), 1,2-Benzenedicarboxylic acid diisooctyl ester (4.43) and 2,4,6-Cycloheptatrien-1-one, 3,5-b is-trimethylsilyl- (3.9). The least compounds found in AME are 1-Ethyl1-Ethyl-2-pyrrolidinecarboxylic acid, methyl ester (0.49), 3H-Benz[e]indene, 2-methyl- (0.52), Benzofuran, 2, 3-dihydro-4-tert-Butoxystyrene (0.54). Many of the phytocompounds were reported in previous studies but the study of GC-MS analysis demonstrated 40 bioactive phytocompounds. The total TIC chromatogram is shown in Figure 1.
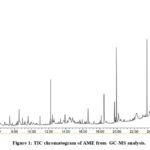 |
Figure 1: TIC chromatogram of AME from GC-MS analysis. |
Table 1: GC-MS analysed volatile phytocompounds from AME
|
S.NO |
Identified compound |
Retention Time |
Area% |
Molecular Weight |
Molecular Formula |
Doc score interacted to 4FA6 |
|
1. |
2-Cyclopentene-1,4-dione |
4.505 |
0.66 |
96.084 |
C5H4O2 |
-6.170 |
|
2. |
Triethylphosphine |
4.676 |
4.69 |
118.13 |
C6H15P |
3.070 |
|
3. |
2-Propenoic acid |
5.123 |
1.35 |
128.13 |
C6H8O3 |
–5.560 |
|
4. |
Butane,2,2–thiobis |
6.107 |
5.52 |
146.30 |
C8H18S |
2.560 |
|
5. |
Propionic acid, mercapto |
6.324 |
0.63 |
241.3 |
C10H11NO2S2 |
–10.560 |
|
6. |
6-methyluracil |
7.434 |
0.85 |
126.112 |
C5H6N2O2 |
– |
|
7. |
4H-Pyran-4-one, 2,3-dihydro-3,5-dihydroxy-6-methyl- |
8.533 |
2.99 |
144.12 |
C6H8O4 |
–10.380 |
|
8. |
Pyrrolidine, N-(4-methyl-3-pentenyl)- |
8.922 |
0.76 |
153.26 |
C10H19N |
–4.010 |
|
9. |
2,3- Dihydrobenzofuran |
9.603 |
0.62 |
120.15 |
C8H8O |
–11.080 |
|
10. |
Propane,1,1,3,3-tetraethoxy |
9.941 |
0.8 |
220.31 |
C11H24O4 |
2.230 |
|
11. |
2-Methoxy-4-Vinylphenol |
10.982 |
0.77 |
150.17 |
C9H10O2 |
2.230 |
|
12. |
Thiazolidine-2,5-dione |
12.229 |
1.53 |
117.13 |
C3H3NO2S |
-8.240 |
|
13. |
2-(4-hydroxyphenyl)ethanol |
12.447 |
0.7 |
138.16 |
C8H10O2 |
-8.610 |
|
14. |
Benzene, 1-hexyl-4-nitro |
12.624 |
0.69 |
275.27 |
C13H16F3NO2 |
-6.920 |
|
15. |
1-Ethyl1-Ethyl-2-pyrrolidinecarboxylic acid, methyl ester |
13.374 |
0.49 |
171.24 |
C9H17NO2 |
– |
|
16. |
Galactitol |
13.746 |
1.29 |
182.17 |
C8H14O6 |
–18.800 |
|
17. |
Benzofuran, 2, 3-dihydro-4-tert-Butoxystyrene |
13.912 |
0.54 |
134.13 |
C8H6O2 |
–11.080 |
|
18. |
3H-Benz[e]indene, 2-methyl- |
14.181 |
0.52 |
180.24 |
C14H12 |
-13.520 |
|
19. |
3-Deoxy-d-mannoic lactone |
14.432 |
1.189 |
162.14 |
C6H10O5 |
-10.380 |
|
20. |
3-Hydroxy-beta-damascone |
14.804 |
1.04 |
208.30 |
C13H20O2 |
-0.340 |
|
21. |
5,5,8a-Trimethyldecalin-1-one |
16.613 |
1.05 |
194.31 |
C13H22O |
2.500 |
|
22. |
n-Hexadecanoic acid |
18.4732 |
3.43 |
256.42 |
C16H32O2 |
4.090 |
|
23. |
1-octadecene |
19.697 |
1.151 |
252.5 |
C18H36 |
13.00 |
|
24. |
Phytol |
19.931 |
4.72 |
296.5 |
C20H40O |
2.180 |
|
25. |
9,12,15-Octadecatrienoic acid, (Z,Z,Z)- |
20.166 |
1.85 |
278.42 |
C18H30O2 |
– |
|
26. |
Benzo[h]quinoline, 2,4-dimethyl- |
22.294 |
3.52 |
207.27 |
C15H13N |
-18.280 |
|
27. |
1,2-Benzenedicarboxylic acid diisooctyl ester |
23.508 |
4.43 |
823.7 |
C38H56O8S2Sn |
-2.810 |
|
28. |
Acetamide,N-[4-(trimethyl)-phenyl]- |
24.56 |
0.89 |
204.34 |
C11H17NO |
-7.600 |
|
29. |
beta,-sitosterol |
25.098 |
9.57 |
414.7 |
C29H50O |
– |
|
30. |
4a,7,7,10a-Tetramethyl-dodecahydro -benzo[f]chromen-3-one |
25.453 |
0.64 |
264.4 |
C17H28O2 |
– |
|
31. |
.beta.-Amyrin |
25.762 |
4.88 |
426.7 |
C30H50O |
– |
|
32. |
Cryptopine |
27.193 |
17.5 |
369.4 |
C21H23NO5 |
– |
|
33. |
Protopine |
27.267 |
8.16 |
353.4 |
C20H19NO5 |
-14.940 |
|
34. |
1H-Indole,1-methyl-2-phenyl- |
27.673 |
0.66 |
207.27 |
C15H13N |
-16.150 |
Non-volatile phytoconstituents from AMP
Quadrupole Time Of Flight (Q-TOF) plays important role in the finding of the mass by spectral data with a limited absorbance range directly proportionate to the retention time in LC-MS analysis. According to the literature, phenolic compounds are most responsible for the anticancer potential and lie in the mass range of 200-500 m/z 15. Non-volatile phytoconstituents of AMP have been screened by utilizing UPLC-QTOF-ESI-MS which was done with the positive and negative modes. The Electron Spray Ionization (ESI) denotes the Charge of the mass ESI (M+H+/M-H–). Totally 13 tentative compounds have been identified concerning the indexed literature 16-21. All the mass fragments showed at the highest 100 % were taken into the count and denoted in Table 2. Six tentative phytocompounds identified from the negative and seven from the positive mode were identified. The Total Ion Chromatogram obtained from AMP is shown in Figure 2a and 2b.
Table 2: LC-MS analysed phenolic phytoconstituents against AMP
|
S.NO |
Tentative |
Molecular |
Molecular |
Retention |
Observed |
Observed |
Referred |
Doc |
Reference |
|
|
1. |
Succinic |
11 |
C4H6O4 |
26.39 |
117.49 |
– |
117 |
-10. |
[15] | |
|
2. |
N-feruloylt |
31 |
C18H19O4 |
3.75 |
– |
314.19 |
314 |
-17. |
[15] |
|
|
3. |
3-Hydroxy |
45 |
C21H24O11 |
5.45 |
451.29 |
– |
451.1246 |
-13. |
[16] | |
|
4. |
Iso |
44 |
C21H20O11 |
6.65 |
447.16 |
– |
447.0936 |
– |
[17] |
|
|
5. |
Leachianol |
47 |
C28H24O7 |
17.09 |
471.65 |
– |
471.1450 |
-12. |
[17] | |
|
6. |
Catechin |
29 |
C15H14O6 |
13.45 |
– |
291.18 |
291.058 |
-18. |
[17] |
|
|
7. |
Genkwanin |
28 |
C16H12O5 |
23.94 |
283.77 |
– |
283.06 |
-19. |
[18] | |
|
8. |
Liquiritigenin |
25 |
C15H12O4 |
22.13 |
255.59 |
– |
255.068 |
-18. |
[19] |
|
|
9. |
Caffeic acid |
18 |
C9H8O4 |
11.66 |
– |
181.43 |
181 |
-45. |
[20] | |
|
10. |
Berberrubine |
35 |
C19H16CINO4 |
7.93 |
– |
336.22 |
336.12 |
-18. |
[21] |
|
|
11. |
Curcumin |
36 |
C21H20O6 |
7.04 |
– |
369.86 |
369.13 |
-15. |
[21] | |
|
12. |
Protopine |
35 |
C20H19NO5 |
7.50 |
– |
354.12 |
354.13 |
-14. |
[21] |
|
|
13. |
Linarin |
59 |
C28H32O14 |
21.10 |
– |
593.21 |
593.18 |
– |
[21] |
|
 |
Figure 2a: ES negative mode TIC chromatogram from AMP
|
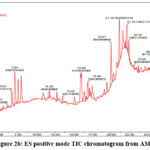 |
Figure 2b: ES positive mode TIC chromatogram from AMP
|
Molecular interaction of phytocompounds from AME and AMP with PI3K receptor
Each identified tentative phytoconstituent from GC-MS and LC-MS analysis was allowed to interact in In silico with the 4FA6 (PI3K) receptor of human origin. Table 1 & 2 shows the Doc score value between the ligand and receptor. The highest negative score shows their stronger interaction bonding which has been highlighted in the table. Thus, the selected high Doc score ligands have been narrowed down for analyzing their interaction sites. Total 47 ligands (34 from GC-MS, 13 from LC-MS and a standard Copansilib). Ligands such as 6-methyluracil, 1-Ethyl1-Ethyl-2-pyrrolidinecarboxylic acid, methyl ester, 9,12,15-Octadecatrienoic acid, (Z,Z,Z)-cyclotrisiloxane, hexamethyl-, beta,-sitosterol, 4a,7,7,10a-Tetramethyl-dodecahydro-benzo[f]chromen-3-one, beta.-Amyrin, Methyltris(trimethylsiloxy)silane and Cryptopine from the GC- MS analysis doesn’t provide the Doc score by SeeSAR 9.2 software that may be due to the ligands weight, length or ungenerated poses respectively. From the LC-MS analysis, Iso-orientin and Linarin didn’t show the docked poses and binding score. Table 3 shows the estimated affinity range within the Picomolar (pM) to millimolar(mM), Log. P value, Number of Hydrogen Bonds, Binding residues and distance of interaction in Angstroms. The highlighted three residues, SER A: 806, LYS A: 890 and ASP A: 950 are the unmarked binding site that interacted with the amino acid residue of the PI3K receptor. 29 selected ligands from both GC-MS and LC-MS are denoted in Table 3. Most of the ligands bonded in the exact binding site (residue) of the receptor. Copansilib revealed the binding site value as 6.780 within the log. P value of -0.73 in μM affinity range with binding site residue as given in figure 3.
Table 3: Selected Ligands with best hits.
|
S.NO |
Name of the Ligand |
Source |
Estimated affinity |
Log P. |
No. H Bonds |
Residue interacted |
Distance in Å |
|
1. |
2-Cyclopentene-1,4-dione |
GC-MS |
mM |
0.08 |
1 |
TYR A: 867 |
2.76 |
|
2. |
2-Propenoic acid |
GC-MS |
>mM |
-1.08 |
1 |
TYR A: 867 |
2.60 |
|
3. |
Propionic acid, mercapto |
GC-MS |
μM |
1.26 |
3 |
TYR A: 867 ASP A:964 |
2.88,3.06 2.82 |
|
4. |
4H-Pyran-4-one, 2,3-dihydro-3,5-dihydroxy-6-methyl- |
GC-MS |
μM |
-0.26 |
2 |
GLU A:880 VAL A:882 |
2.70 2.78 |
|
5. |
Pyrrolidine, N-(4-methyl-3-pentenyl)- |
GC-MS |
μM |
1.02 |
– |
– |
– |
|
6. |
2,3- Dihydrobenzofuran |
GC-MS |
μM |
1.62 |
1 |
VAL A:882 |
2.85 |
|
7. |
Thiazolidine-2,5-dione |
GC-MS |
μM |
-0.03 |
1 |
VAL A:882 |
2.84 |
|
8. |
2-(4-hydroxyphenyl) ethanol |
GC-MS |
μM |
0.93 |
3 |
ASP A:964 TYR A:867 VAL A:882 |
2.81 2.96 2.77 |
|
9. |
Benzene, 1-hexyl-4-nitro |
GC-MS |
nM |
4.74 |
1 |
VAL A:882 |
2.80 |
|
10. |
Galactitol |
GC-MS |
μM |
-3.59 |
7 |
ALA A:885 VAL A:882 GLU A:880 |
2.70,2.86,2.64 2.78,2.89 3.09,2.66 |
|
11. |
Benzofuran, 2, 3-dihydro-4-tert-Butoxystyrene |
GC-MS |
μM |
2.14 |
1 |
VAL A:882
|
2.85 |
|
12. |
3H-Benz[e]indene, 2-methyl- |
GC-MS |
μM |
3.80 |
– |
– |
– |
|
13. |
Benzo[h]quinoline, 2,4-dimethyl- |
GC-MS |
μM |
4.00 |
– |
– |
– |
|
14. |
1,2-Benzenedicarboxylic acid diisooctyl ester |
GC-MS |
μM |
-0.38 |
1 |
VAL A:882
|
2.90 |
|
15. |
Acetamide,N-[4-(trimethyl)-phenyl]- |
GC-MS |
μM |
2.19 |
2 |
ASP A: 964 TYR A: 867
|
2.80 2.70 |
|
16. |
2,4,6-Cycloheptatrien-1-one, 3,5-b is-trimethylsilyl- |
GC-MS |
μM |
2.14 |
2 |
ASP A: 964 TYR A: 867
|
3.18 2.92 |
|
17. |
Protopine |
GC-MS |
μM |
1.14 |
2 |
VAL A:882 SER A:806
|
3.29 3.42 |
|
18. |
1H-Indole,1-methyl-2-phenyl- |
GC-MS |
μM |
3.85 |
– |
– |
– |
|
19. |
Succinic acid |
LCMS |
μM |
-2.73 |
5 |
CYS A: 869 ASP A: 841 TYR A: 867 ASP A: 964 |
3.48 2.91 2.82, 2.83 2.91 |
|
20. |
3-Hydroxyphloretin 2’-O-glucoside |
LCMS |
μM |
-0.50 |
3 |
GLU A: 880 ASP A: 964 LYS A: 890
|
2.72 2.73 3.22 |
|
21. |
Leachianol F |
LCMS |
μM |
4.67 |
2 |
LYS A: 890 ASP A: 950 |
2.62 2.41 |
|
22. |
Genkwanin |
LCMS |
nM |
2.88 |
4 |
VAL A: 882 ASP A:964 |
2.94, 2.54, 3.05 3.28 |
|
23. |
Liquiritigenin |
LCMS |
μM |
2.80 |
4 |
VAL A: 882 ASP A: 964 TYR A: 867 |
2.65, 3.01 2.86, 2.93 |
|
24. |
Caffeic acid |
LCMS |
μM |
1.20 |
3 |
TYR A: 867 VAL A: 882 MET A: 953 |
2.73 2.90, 2.96 |
|
25. |
N-feruloyltryamine |
LCMS |
nM |
2.48 |
6 |
TYR A: 867 ASP A: 964 VAL A: 882 GLU A: 880 |
3.14 2.86 2.84, 3.30, 3.17 2.65 |
|
26. |
Berberrubine |
LCMS |
μM |
2.96 |
1 |
TYR A: 867
|
2.70 |
|
27. |
Curcumin |
LCMS |
pM |
3.37 |
4 |
TYR A: 867 VAL A: 882 GLU A: 880
|
2.71 2.83, 3.20 2.69 |
|
28. |
Protopine |
LCMS |
μM |
1.14 |
2 |
SER A: 806 VAL A: 882 |
2.90 3.39 |
|
29. |
Catechin |
LCMS |
μM |
1.55 |
3 |
TYR A: 867 VAL A: 882
|
2.71 3.20, 2.83 |
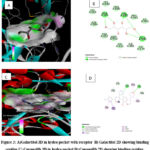 |
Figure 3: A)Galactitol 3D in hydro pocket with receptor B) Galactitol 2D showing binding residue C) Copansilib 3D in hydro pocket D) Copansilib 2D showing binding residue. |
Discussion
Overactive PI 3-kinase (PI3K) in cancer and immunological dysregulation has prompted substantial research into PI3K inhibitors for therapeutic use having several categories. Early investigations have shown that the pan-PI3K inhibitors (LY294002) might reverse cancer cell resistance to a wide range of treatments, including chemotherapy, radiation, and targeted therapies 22. Epigenetic modulators have been found to manage the epigenome by participating in the PI3K/AKT pathway contributing to oncogenicity in certain malignancies, especially in Non-small Cell Lung Cancer which has a higher projectile prognosis23. Induction of apoptosis in lung cancer and prevention of metastasis could be healthily induced with the help of bioactive phytoconstituents from medicinal plants24. A. mexicana has been proven for anticancer activity against several cell lines 25. The study of In silico anticancer modeling from the bioactive volatile and phenolic constituents against the PI3K receptor, responsible for lung cancer metastasis has been researched less. Regarding previous studies, we attempted the identification of phytoconstituents of the extracts AME and AMP have been done through GC-MS and LC-MS analysis. GC-MS analysis of AME revealed the presence of cryptopine with the highest peak percentile of 17.5, which is found mostly in the species of Indian Opium and Protopine responsible for latex formation 10. The presence of phytol shows that A. mexicana has potential antioxidant and anticancer activity.Beta-sitosterol and Beta-amyrin are reported to be non-toxic to normal cells (Table 1) 26. UPLC-Q-TOF-ESI-MS analysis of AMP (Table 2) revealed 13 major phenolic components reported to possess anti-cancer activity in several studies. Genkwanin is the Flavonoid found in AMP at the retention time of 23.959 in the negative mode also found in antioxidant-rich rosemary oil 18,4. Virtual screening in silico is a good choice before the preliminary research of in vitro or in vivo to inhibit cancer proliferation 15. Out of 47 ligands found from analysing the GC-MS and LC-MS spectrum peaks, 29 were filtered out from the ligand-receptor complex analysis with good binding affinity parameters. 4FA6 receptors possess 20 different binding sites analysed from the Uniprot database compared with binding sites of ligand residues. All ligands showed the exact binding nature except Leachinol F. Galactitol has the highest binding affinity range (μM) having 7 hydrogen bonds with a log. P value of -3.59 in the binding site of ALA A:885 (2.70, 2.86, 2.64), VAL A:882 (2.78,2.89), GLU A:880 (3.09,2.66). Next lies N-feruloyltryamine with 6 H Bonds and 4 H bonds shown in Genkwanin, Liquiritigenin, and Curcumin with the highest log p-value with exact binding residue. Ligands such as Propionic acid, mercapto, 2-(4-hydroxyphenyl) ethanol, 3-Hydroxyphloretin 2’-O-glucoside, Caffeic acid, and Catechin have 3 H Bonds with the PI3K receptor. This may inhibit the capacity of metastatic proliferation in PI3K/Akt/mTOR pathways by controlling the 4FA6 (PI3K) protein found in phytocompounds found from A. mexicana Leaves.
Conclusion
Different extracts such as AME and AMP from the leaves of A. mexicana were checked for bioactive phytoconstituent screening and carried through molecular docking to reveal the anticancer potential at In silico level. We have screened the impact of phytochemicals through GC-MS analysis with 34 different phytoconstituents. LCMS revealed 13 phenolic and flavonoid phytocompounds which is none other reported in any of the research studies in the A. mexicana plant. Totally 47 compounds including the standard drug were targeted against the PI3K receptor and narrowed down to 29 ligands by analyzing the binding score parameter. Phytoconstituents such as Galactitol (GC-MS), Succinic acid (LC-MS) and N-feruloyltryamine (LC-MS) resulted in good binding affinity and best hydrogen bonding with the exact binding site of the 4FA6 (PI3K) receptor paving to analyze the anticancer target against the Phosphoinositide kinase pathway by inducing apoptosis and controlling the cell growth. This would lead to greater alternatives in the drug discovery from A. mexicana.
Acknowledgment
All the authors contributed their sincere effort in designing, analyzing and reporting on this project.The authors acknowledge sincere gratitude to the Karpagam Academy of Higher Education for providing the network and molecular modeling facility for implementing this research manuscript.
Conflict of Interest
The authors declare no conflict of interest.
Funding Sources
The authors declare the funding sources from DST-FIST (SR/FST/LS-1/2018/187) for providing the infrastructure facilities and commercial software for this research work.
References
- Tan AC. Targeting the PI3K/AKT/mtor pathway in non‐small cell lung cancer (NSCLC). Thoracic Cancer. 2020;11(3):511-518. doi:10.1111/1759-7714.13328.
CrossRef - Govindammal M, Prasath M, Kamaraj S, Muthu S, Selvapandiyan M. Exploring the molecular structure, vibrational spectroscopic, Quantum Chemical Calculation and molecular docking studies of curcumin: A potential PI3K/AKT uptake inhibitor. Heliyon. 2021;7(4). doi:10.1016/j.heliyon.2021.e06646.
CrossRef - Le T, David J, Bryan LJ. Update on the role of Copanlisib in hematologic malignancies. Therapeutic Advances in Hematology. 2021;12:204062072110060. doi:10.1177/20406207211006027.
CrossRef - Ahmed B, Joseph A, Das S, et al. Structure-activity relationship insight of naturally occurring bioactivemolecules and their derivatives against non-small cell lungcancer: A comprehensive review. Current Medicinal Chemistry. 2022;29(39):6030-6062. doi:10.2174/0929867329666220509112423
CrossRef - Mallavarpu Ambrose J, Veeraraghavan VP, Kullappan M, et al. Molecular modeling studies of the effects of withaferin A and its derivatives against oncoproteins associated with breast cancer stem cell activity. Process Biochemistry. 2021;111:186-199. doi:10.1016/j.procbio.2021.09.007
CrossRef - Khan AM, Bhadauria S. Analysis of medicinally important phytocompounds from Argemone Mexicana. Journal of King Saud University – Science. 2019;31(4):1020-1026. doi:10.1016/j.jksus.2018.05.009
CrossRef - Dénou A, Ahmed A, Dafam DG, et al. Pharmacognostic, physicochemical and phytochemical investigations on aerial parts of Argemone mexicana L. Research Journal of Pharmacognosy . 2020;7(3):15-24. doi:10.22127/rjp.2020.220380.1559
- Paul D, Sanap G, Shenoy S, Kalyane D, Kalia K, Tekade RK. Artificial Intelligence in drug discovery and development. Drug Discovery Today. 2021;26(1):80-93. doi:10.1016/j.drudis.2020.10.010
CrossRef - Talevi A. Multi-target pharmacology: Possibilities and limitations of the “Skeleton key approach” from a medicinal chemist perspective. Frontiers in Pharmacology. 2015;6. doi:10.3389/fphar.2015.00205
CrossRef - Paul D, Sanap G, Shenoy S, Kalyane D, Kalia K, Tekade RK. Artificial Intelligence in drug discovery and development. Drug Discovery Today. 2021;26(1):80-93. doi:10.1016/j.drudis.2020.10.010
CrossRef - Kiran Chiliveryravi, Alagar Sangilimuthu, Manoharan Sowmyapriya. Volatile phytoconstituent profile of Argemone Mexicana L. Leaves and pharmacological importance. International Journal of Pharma and Bio Sciences. 2017;8(3). doi:10.22376/ijpbs.2017.8.3.b523-528
CrossRef - Sunisha K, Dhivya R. GC-MS analysis and antioxidant activity of organic extracts of Psidium cattleianum leaves. International Journal of Scientific Research in Biological Sciences . 2020;7(2):128-133. doi: https://doi.org/10.26438/ijsrbs/v7i2.128133
- Ananth DA, Rameshkumar A, Jeyadevi R, et al. Amelioratory effect of flavonoids rich Pergularia daemia extract against CFA induced arthritic rats. Biomedicine & Pharmacotherapy. 2016;80:244-252. doi:10.1016/j.biopha.2016.03.019
CrossRef - Halder D, Das S, R. A, R. S. J. Molecular docking and dynamics based approach for the identification of kinase inhibitors targeting PI3Kα against non-small cell lung cancer: A computational study. RSC Advances. 2022;12(33):21452-21467. doi:10.1039/d2ra03451d
CrossRef - Bhardwaj VK, Singh R, Sharma J, Das P, Purohit R. Structural based study to identify new potential inhibitors for dual specificity tyrosine-phosphorylation- regulated kinase. Computer Methods and Programs in Biomedicine. 2020;194:105494. doi:10.1016/j.cmpb.2020.105494
CrossRef - Hao Y, Huo J, Wang T, Sun G, Wang W. Chemical profiling of Coptis Rootlet and screening of its bioactive compounds in inhibiting staphylococcus aureus by UPLC-Q-TOF/MS. Journal of Pharmaceutical and Biomedical Analysis. 2020;180:113089. doi:10.1016/j.jpba.2019.113089
CrossRef - Al-Yousef HM, Hassan WH, Abdelaziz S, Amina M, Adel R, El-Sayed MA. UPLC-ESI-MS/MS profile and antioxidant, cytotoxic, antidiabetic, and antiobesity activities of the aqueous extracts of three different hibiscus species. Journal of Chemistry. 2020;2020:1-17. doi:10.1155/2020/6749176
CrossRef - Tang J, Dunshea FR, Suleria HA. LC-ESI-QTOF/MS characterization of phenolic compounds from medicinal plants (hops and juniper berries) and their antioxidant activity. Foods. 2019;9(1):7. doi:10.3390/foods9010007
CrossRef - Avula B, Bae J-Y, Zhao J, et al. Quantitative determination and characterization of polyphenols from cissus quadrangularis L. and dietary supplements using UHPLC-PDA-MS, LC-QToF and HPTLC. Journal of Pharmaceutical and Biomedical Analysis. 2021;199:114036. doi:10.1016/j.jpba.2021.114036
CrossRef - Velamuri R, Sharma Y, Fagan J, Schaefer J. Application of UHPLC-ESI-QTOF-MS in phytochemical profiling of sage (salvia officinalis) and Rosemary (rosmarinus officinalis). Planta Medica International Open. 2020;07(04). doi:10.1055/a-1272-2903
CrossRef - Zhou J, Cai H, Tu S, et al. Identification and analysis of compound profiles of Sinisan based on ‘individual herb, herb-pair, herbal formula’ before and after processing using UHPLC-Q-TOF/MS coupled with multiple statistical strategy. Molecules. 2018;23(12):3128. doi:10.3390/molecules23123128
CrossRef - Kumar BR. Application of HPLC and ESI-MS techniques in the analysis of phenolic acids and flavonoids from green leafy vegetables (glvs). Journal of Pharmaceutical Analysis. 2017;7(6):349-364. doi:10.1016/j.jpha.2017.06.005
CrossRef - Vanhaesebroeck B, Perry MW, Brown JR, André F, Okkenhaug K. PI3K inhibitors are finally coming of age. Nature Reviews Drug Discovery. 2021;20(10):741-769. doi:10.1038/s41573-021-00209-1
CrossRef - Noorolyai, S. et al. (2019) “The relation between PI3K/akt signalling pathway and cancer,” Gene, 698, pp. 120–128. Available at: https://doi.org/10.1016/j.gene.2019.02.076.
CrossRef - Hosseini A, Ghorbani A. Cancer therapy with phytochemicals: evidence from clinical studies. Avicenna journal of phytomedicine, 2015; 5(2):84.
- Gali K, Ramakrishnan G, Kothai R, Jaykar B. In vitro anticancer activity of methanolic extract of leaves of Argemone Mexicana Linn. International Journal of Pharmtech Research, 2011; 3(3):1329-33.
- Aati HY, Anwar M, Al-Qahtani J, et al. Phytochemical profiling, in vitro biological activities, and in-silico studies of Ficus vasta forssk.: An unexplored plant. Antibiotics. 2022;11(9):1155. doi:10.3390/antibiotics11091155.
CrossRef







