Mohamed Thoufic Ali A M  and Vino Sundararajan*
and Vino Sundararajan*
Integrative Multiomics Laboratory, Department of Bio Sciences, School of Bio Sciences and Technology, Vellore Institute of Technology, Vellore, Tamil Nadu, India.
Corresponding Author E-mail: svino@vit.ac.in
DOI : https://dx.doi.org/10.13005/bpj/2930
Abstract
With the advances in the field of medicine there is an increase in the geriatric population and rheumatoid arthritis is one of the common diseases that affect this cohort. The modern medicines that are used for the treatment of rheumatoid arthritis provide a symptom-based treatment and there are studies showing severe side effects for some of the medicines being used. But there are shreds of evidence in traditional medical texts for the treatment of rheumatoid arthritis which gives an increased therapeutic coverage with less to no side effects. Bhadradarvadi kashayam (concoction) is one of the most commonly preferred and prescribed Ayurvedic medicine for managing the disease. In this study, we are investigating the mode of action of this kashayam by employing a network pharmacology-based framework which included the analysis of the cross-talks between the active ingredients of the kashayam and major molecules involved in the disease, the transcription factors and various pathways in which they are involved. Based on the systems pharmacology approach, 57 active compounds and a total of 377 potential targets with their interacting partners, and the targets associated with comorbidities were identified. The PPI network was analyzed to understand the topological index for screening the hub proteins such as MAPK1, MAPK14, FYN and CXCL8, which were found to be enriched in various signaling pathways. Furthermore, molecular docking analysis validated the strong physical interaction between the hub proteins and the corresponding active compounds from BDK. Overall, the study sheds light on the pharmacological mechanism of Bhadradarvadi kashayam against Rheumatoid Arthritis and also highlights that there are traditional herbal remedies imparted by the Ayurveda system of medicine which has the least side effects compared to modern medicines.
Keywords
Auto-immune diseases; Bhadradarvadi; Inflammation; Rheumatoid arthritis; Signaling pathways; Systems pharmacology
Download this article as:| Copy the following to cite this article: Ali A. M. M. T, Sundararajan V. Omics-based Analysis of Bhadradarvadi Kashayam in Managing Rheumatoid Arthritis via CXCL8-CXCR1/2 axis, MAPK and NF-κB Signaling Pathways - A Network Pharmacology Approach. Biomed Pharmacol J 2024;17(2). |
| Copy the following to cite this URL: Ali A. M. M. T, Sundararajan V. Omics-based Analysis of Bhadradarvadi Kashayam in Managing Rheumatoid Arthritis via CXCL8-CXCR1/2 axis, MAPK and NF-κB Signaling Pathways - A Network Pharmacology Approach. Biomed Pharmacol J 2024;17(2). Available from: https://bit.ly/3y9xJkq |
Introduction
Rheumatoid arthritis (RA) is a chronic autoimmune disease designated with symmetrical inflammation, progressive disability, bone erosion and cartilage destruction1–3. As an increasingly common inflammatory disease, the average prevalence of RA is about 0.5%-1.0% worldwide with the male-to-female ratio nearly 1:4, and the incident rate nearing approximately 0.6% in India4. However, the pathogenesis and/or etiology of RA remains convoluted and unclear5. Several studies have reported that a variety of immune cells and inflammatory mediators are critically implicated in the disease development and progression6,7. The processes are driven through the secretion of various pro-inflammatory cytokines and inflammatory enzymes by monocytes/macrophages in synovial fluid. These inflammatory mediators play a critical role in regulating the growth and proliferation of immune T cells, which further stimulates the release of tissue-degrading enzymes, all of which enhance the inflammatory reaction and joint damage8. Hence, aberrant activation of various important regulatory signaling cascades including mitogen-activated protein kinase (MAPK), toll-like receptor, CXCL8-CXCR1/2 axis, and nuclear factor kappa B (NFκB) pathways are recognized as significant contributors to accelerating the progression of RA9,10.
Currently, non-steroidal anti-inflammatory medications (NSAIDs), disease-modifying anti-rheumatic medicines (DMARDs), biologics, and immunosuppressants have been prescribed for the treatment of RA11. Even though they can mitigate the symptoms including inflammation and pain, they are found to come up with noteworthy side effects such as hepatic and pulmonary toxicity, peptic ulcers, infections, gastro-intestinal damage, renal failure and so forth12–14. Therefore, an exploration for efficient alternative and additional therapeutic options that aim to mitigate the symptoms concomitant with RA instead of affording a complete cure to the patients still endures.
In the current situation, complementary and alternative medicine (CAM) are getting greater prominence by providing a variety of treatment choices that outperforms traditional medications. The usage of CAM is on an increasing trend in all continents and is not restricted only to Asia15. As reported by National Health Interview Survey (NHIS) from the Centers for Disease Control (CDC) in 2007, more than 200K US adults selected to use Ayurvedic medications in the past year to treat their health issues. Seventy percent of the Indian population relies on the system of medicine that was traditionally considered like Ayurveda, homeopathy, yoga and Siddha16–18. Of the known traditional medical practices, Ayurveda is among the holistic medicinal approach and is in practice since olden times and emerged from a verbal tradition that was subsequently chronicled in the ancient Indian texts called Vedas, which were originally written in Sanskrit. It always emphasizes the significance of endorsing health by taking a holistic outlook of mind, body, and spirit as well as using natural remedies acquired from medicinal plants and minerals. Ayurvedic system of medicine is very less studied and documented due to the difficulty in understanding strict logical descriptions and less conceptual development. Ayurvedic pharmacology (Dravyaguna) encircles with three concepts i) taste (rasa), ii) properties (guna), iii) active principles (virya), iv) biotransformation (vipak) and v) specific actions (prabhav)19 and it affords to bring a favorable cure to an array of ailments which include RA20,21. One of the highly cited oral medications for RA is Bhadradarvadi decoction (BDK), a polyherbal concoction of fifteen herbs with influential active ingredients.
A blend of system biology with the study of levels of biological information along with the clinical pharmacology profile, brings in the module of system pharmacology, that investigates the relationship between molecular properties and their pharmacological effect. The arena integrates the omics (proteomics, genomics, metabolomics, and transcriptomics) outlines and metabolites to construct networks to ingress the underlying drug mechanisms22–24. In addition to finding unique drug targets that are novel, the network medicine strategy can be efficiently used to establish combinations of drugs in conditions like neurovascular diseases, CVD and cancer25–28. Consequently, the strategy could be extended to explore the involvement of plant-derived medicines in holistically treating a variety of diseases.
The herbal formulations encircle perplexed chemical systems incorporating a mixture of a wide array of compounds. The formulations derived from medicinal plants were deemed to work on a web of targets specific to the phenotype of diseases. The mode of action and efficacy of the herbal components could be predicted by network-based methods. The investigation of the network uncovers the effective therapeutic mechanism of the herbal compositions and the adeptness to comprehend the unexposed pharmacological properties. The network medicine strategy identifies the prospective on and off-targets that can be used to create a sensible design of drug interactions. The present study employs a network pharmacology strategy to probe into the mode of action of the polyherbal formulation of BDK in treating RA. The models of pharmacokinetic properties such as oral bioavailability (OB), drug-likeliness (DL), and ADMET parameters were evaluated to explore the therapeutic effects of the formulation. In this study, we attempted to utilize systematic pharmacology and network analysis to interpret the connections at the molecular level generated by several bioactive components of the herbal formulation to comprehend its mechanism of action.
Materials and Methods
Profiling of compound
BDK is prepared from the combination of fifteen herbs Lignum of Cedrus deodara (CD), Radix of Abutilon Indicum (AI), Radix of Valeriana jatamansi (Vj), Radix of Desmodium gangeticum (Dg), Radix of Saussurea costus (Sc), Radix of Gmelina arborea (Ga), Radix of Aegle marmelos (Am), Radix of Stereospermum colors (Sc), Radix of Oroxylum indicum (Oi), Radix of Premna corymbosa (Pc), Radix of Solanum anguivi (Sa), Radix of Solanum surattense (Ss), Fructus of Tribulus terrestris (Tt), Radix of Pseudarthria viscida (Pv) and Radix of Sida cordifolia (Scf)29. The bioactive compounds from these fifteen plants were retrieved from the databases such as Traditional Chinese Medicine Systems Pharmacology (TCMSP)30, Universal Natural Products Database (UNPD)22, and Dr. Duke’s Phytochemical and Ethnobotanical Databases31. The compound structures were fetched from the Chemical book, PubChem and drawn using the Chemsketch tool.
Oral bioavailability (OB) and drug-likeness (DL) assessment
Traditional medicine formula decoctions are mostly orally administered, which have a greater number of compounds, but only a part of them create a therapeutic effect. OB signifies the capability of a compound to disseminate in the body after oral ingestion. OB can determine the ability of active compounds in a formula to be transported throughout the body and cause a physiochemical effect. Analysis of several molecular descriptors incited a series of rules connecting them with OB including Lipinski’s rule of 532. In this study, the QikProp filter from Schrödinger software (Schrodinger, LLC, New York, USA) was used with a set of descriptors (% human oral absorption, and rule of 5) were selected to delineate the activity, permeability, and metabolism. Together with Lipinski’s parameters, the other molecular descriptors supporting OB as recommended by Veber were also calculated33. Pharmacological descriptors including molecular weight (MW), Ghose-Crippen-Viswanadhan octanol-water partition coefficient (ALogP), number of donor atoms for hydrogen bond (nHDon), number of acceptor atoms for hydrogen bond (nHAcc), and total polar surface area (TPSA) were predicted using PaDEL descriptor tool (version 2.18)34.
DL is a pointer for identifying the likeness of a compound, which possess functional groups and physicochemical properties similar to the conventional drugs and could have a possible desired therapeutic effect. Evaluating DL according to the concept of desirability termed as a quantitative estimate of drug-likeness (QED), aids to investigate the chemical homology of the compounds with known drugs. The individual desirability functions were calculated for the molecular descriptors MW, RotB, ALogP, nHDon, nHAcc, and TPSA. The QED score is calculated by obtaining the geometric mean of all individual desirability functions using the following equation:

Where di is calculated with the desirability score of the nth molecular descriptor. The QED score ranges from 0 (complete unfavorable properties) to 0.49 (all favorable properties). DruLiTo tool was utilized to calculate the score.
Toxicity prediction
Furthermore, to the above assessments, the compounds chosen were evaluated for the probable effect of toxicity. The compound’s toxicity was determined by utilizing the ProTox-II (prediction of toxicity of chemicals) server35. The server predicts rodent oral toxicity using a globally harmonized system of classification of labeling of chemicals (GHS), hepatotoxicity using the Random Forest (RF) algorithm and immunotoxicity using the Bernoulli-Naïve Bayes algorithm.
Target fishing for potential active compounds
The archetype of “one drug – one target – one disease” has been transfigured to “one drug – multi targets – multi diseases” owing to the progression in the studies and approaches of polypharmacology. To discern the multi-target hypothesis, the bioactive compounds were further assessed and the protein targets were recognized. The protein targets for the identified compounds were gathered from several resources including extensive literature hunts and database searches from TCMSP, Therapeutic Target Database (TTD)36, and Traditional Chinese Medicine Integrated Database (TCMID)37. The compounds with yet unknown targets were imposed to reverse drug targeting strategy using STITCH (search tool for interacting chemicals) and BindingDB38. Based on the similarity index (>0.85) and other parameters like the Tanimoto index (>0.9) from Binding DB and STITCH respectively, the targets for the compounds were obtained.
Enrichment and comorbidities analysis
The Gene Ontology (GO) functional and pathway enrichment analysis provides a standardized depiction and annotation for genes and gene products that can possess the response to a biological query. Over GO analysis, researchers can retrieve complete insights into various aspects including cellular component (CC), biological process (BP), and molecular function (MF). GO and pathway analysis was performed using ToppGene Suite (a one-stop portal for functional and pathway enrichment analysis)39. The portal identifies the functional and pathway enrichment based on various databases including the Kyoto Encyclopedia of Genes and Genomes (KEGG), Reactome, and so forth. GO and KEGG pathway enrichment analysis for the predicted target proteins from the compounds were performed using the ToppFun tool in the Toppgene Suite. The default parameters like the probability density function for the p-value scheme and FDR correction were chosen. Gene count >2 and FDR B&H (q value) as 0.01 were selected as the significant cut-off limit for enrichment analysis. Further, the targets were explored to identify their concomitant diseases using DisGeNET (v7.0), which integrates human disease-gene correlations and their variants. Comorbidities associated with RA were filtered out to spotlight the intricacies of the target proteins by involving manifold diseases.
Protein-protein interaction (PPI) network construction
Upholding cellular homeostasis needs a blend of proteins with other molecules including genes, small particles, and other proteins. A PPI network was ascertained to explicate the connection between the human proteins and their predicted targets, as well as it has loomed as an ideal method for drug discovery40. Interacting partners for the target proteins of related ingredients were obtained using the “Search tool for retrieval of interacting proteins” (STRING v11.5), which is a database for constructing a protein-protein interaction network. The species were set as “Homo sapiens” and the interacting proteins retrieved with a confidence score cut-off as >0.8, which ensures to be reported through experiments and databases were only considered for the analysis. The networks were then constructed, visualized, and further analyzed using Cytoscape v3.8.241.
Hub protein identification and cluster analysis
In the PPI network, each node specifies a target protein and an edge specifies the interaction between the target proteins. Hub proteins are highly influential proteins having high interaction with a large number of partners that could play a crucial role in disease progression. The highly influential proteins from the constructed network were analyzed through the cytoHubba42 plugin of Cytoscape for computing the topological metrics such as degree centrality (t), betweenness centrality (CB), closeness centrality (CC) and maximal clique centrality (CMC) measures.
Degree centrality (t), indicates the number of interactions upon by a node or the edges connected with other nodes in the network and assists in calculating the node impact in regulating the network.

where Tu represents the node-set comprising all the neighbors of node u, and x(u, v) denotes the edge weight connecting node u along with node v.
Betweenness centrality (CB) calculates the number of times a node tumbles on the shortest path with other neighboring nodes. It depicts a node’s competency to regulate the signal processing and data drift in the network.

where σtg (u) represents the number of interactions from t to g that passes through u, and σtg is the sum of all the interactions between node t and g.
Closeness centrality (CC) measures the distances of all the shortest paths from one node to all the other nodes in a network. Therefore, that highest centrality is nearer to the other nodes. It is measured as
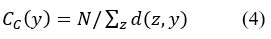
where d(z,y) denotes the distance between y and z nodes and N is the number of nodes in the network.
Maximal clique centrality (CMC) measures the level of the highly modular nodes in the complex network by identifying a large number of vertices (clique in a graph) within a network and a larger number of links between these vertices. It is represented as

where S(u) represents a collection of maximal cliques which consist of u, and (|C|-1)! is the multiplication of all positive integers <|C|. In case, there is no edge between the neighbors of node u, then CMC(u)=degree.
Further, clusters or topology modules of the PPI network signify the highly interconnected regions, which render to reflect molecular biological functions and essential protein progressions. In our network pharmacology study, highly interconnected regions in the PPI network were filtered by the MCODE (Molecular Complex Detection) plugin to identify the densely interconnected regions. MCODE with the node cut-off = 0.2, fluff-density cut-off = 0.2, K-core = 2, and a max-depth = 100 was initialized as advanced options.
Pathway mapping
We have explored all the RA pathways associated with the highly influential proteins; they were impelled by the multifaceted disease via modulating certain pathways. The obtained hub proteins along with their correlated bioactive compounds were investigated by mapping onto the pathways that are closely related and regulating the disease progression with the help of extensive literature studies manually.
Validation of compound-target interaction
Molecular docking technology toils based on the “Lock-Key principle” to predict the small molecule ligands-protein receptors association by calculating the repulsion, spatial effect, hydrogen bonds, molecular flexibility, hydrophobic interaction, and affinity score. The structure of identified hub proteins and the active compounds were retrieved from the Protein Data Bank database (PDB, https://www.rcsb.org/) and PubChem database (https://pubchem.ncbi.nlm.nih.gov/), respectively. The protein structures were prepared by the addition of hydrogen molecules as well as the removal of water molecules in the receptor. Molecular docking was performed with the selected hub proteins and the ligand using the AutoDock Vina 4.2.643. Ligand-binding affinities or binding energies between the receptor and ligand molecule were attained as negative Gibbs free energy (ΔG) scores (kcal/mol). The interactions of the post-docking/docked complex were visualized by Discovery Studio Visualizer. Ultimately, the potent inhibitor has been prompted from the docked complex according to the hydrophobic interaction, hydrogen bond and binding score for RA treatment.
Results
Screening of active compounds in BDK
A total of 205 BDK ingredients were obtained after structural confirmation and removal of duplicates. Further, screening was performed based on OB, DL and ADMET-related parameters, which obtained 57 compounds and the same is listed in Table.1, including 9 AMs, 1 AI, 6 CDs, 2 DGs, 2 GAs, 3 OIs, 1 PC, 7 PVs, 1 SA, 10 SCs, 6 SCFs, 1 SCO, 2 SSs, 2 TTs, and 6 VJs. The IDs of the molecules that were distributed by 2 or 3 medicinal herbs were indicated accordingly. The average QED value of the 57 compounds was 0.604. 57 compounds had a QED value = or > 0.49 and < 0.6; only biochanin A (OI) had a higher QED value than 0.8, which was 0.88.
Construction of compound-target (C-T) network
The construction of a compound-target network (C-T) helped us to highlight the inter-relation of the BDK active ingredient and the respective candidate target. After idleness elimination, the C-T network was obtained by connecting 57 compounds and 377 protein targets. The network consisted of 434 nodes and 814 edges with an average degree of 3.75 nodes per target and 18.27 edges per compound (Fig. 1), suggesting that a target could be common to multiple compounds, including synergistic action of BDK active ingredients. As exposed in this network, the degree value of cryptomeridol, 1,1-diethoxy-3-methyl butane, deodarone, nerolidol and coumaran were 37, 30, 30, 26 and 25, respectively, and they interact with many protein targets. This could justify the pleiotropic effects revealed by the active ingredients of BDK according to the prime positioning in the network.
Functional and pathway enrichment transcripts of targets
GO enrichment analysis was performed on the 377 targets to understand the biological functions involved in it, using three functional classes such as biological process (BP), cellular component (CC) and molecular function (MF). Ultimately, 2416 BPs, 230 CCs, and 367 MFs were enriched among the potential targets. The top 20 significantly enriched GO terms within BP, CC and MF were displayed in Tables 2, 3, and 4. BP was predominantly related to response to oxygen-containing compounds, response to nitrogen compounds, regulation of cell death, regulation of cell communication, and so forth. CC was mainly intricate in membrane stack, membrane switch, and membrane region. MF was associated with neurotransmitter receptor activity, signaling receptor activity, amino acid binding, oxidoreductase activity and so forth.
Furthermore, KEGG enrichment analysis was performed to identify the pathways impacted substantially by BDK and the top ones were TNF signaling pathway, MAPK signaling pathway, IL-17 signaling pathway, PI3K-Akt signaling pathway, rheumatoid arthritis, apoptosis and so forth. Among the top 20 highly enriched pathways, seven were directly related to the inflammatory condition in RA including the HIF-1 signaling pathway, TNF signaling pathway, IL-17 signaling pathway, PI3K-Akt signaling pathway, MAPK signaling pathway, NFκB signaling pathway, and JAK-STAT signaling pathway. BDK may probably endeavor an anti-arthritic effect mainly over modulating inflammation, which could be validated by in vitro and in vivo experiments.
Targets diseaseome – concerning RA with its comorbidities
Rheumatoid arthritis is recurrent manifold-syndromic and therefore influences the chances of having associated diseases coupled with its manifestations. Our dataset of BDK target proteins was traced for its association and was delivered with 198 different diseases, which were further categorized into four groups rheumatoid arthritis, RA-associated diseases (heart diseases, diabetes, joint pain, and irritable bowel disease (IBD)), autoimmune diseases (Lupus erythematosus, celiac disease, Sjogren’s syndrome, multiple sclerosis and Addison disease) and other diseases (hepatitis, cancer, thyroid diseases, leprosy, Alzheimer disease, atherosclerosis and Gaucher disease). The categorization identified 136 targets possibly responsible for rheumatoid arthritis, 71 targets for RA-associated diseases, 172 targets for autoimmune diseases, and 292 targets for other diseases, respectively.
PPI analysis and revelation of highly influential proteins
To meticulously comprehend the mechanism of action of BDK on rheumatoid arthritis, the interplay and crosstalk between proteins must be elucidated. The PPI (T-IP) network aids in the discernment of the biological activities of the cell and its interior and exterior response to the environment. Therefore, the PPI network of 377 putative therapeutic targets and the interacting partners (PPI data) which were obtained from the STRING database was constructed. The PPI network was constructed using Cytoscape and composed of 377 nodes and 3992 edges with a clustering coefficient estimated to be 0.302. Subsequently, the topological parameters of the network were analyzed using NetworkAnalyzer and illustrated in Table 1. Based on different algorithms such as high centrality measures like MCC, degree, closeness and betweenness in the cytoHubba plugin of Cytoscape, the top 20 hub proteins were identified in each category as illustrated in Table 2. A total of 4 hub proteins commonly occurred almost in three categories implying their prominence in sustaining the highly interconnected structure and interaction. The commonly appeared hub proteins were MAPK1, MAPK14, FYN, and CXCL8 which are highlighted in Table 2. Moreover, the mainstream of the hub proteins was found to be actively engaged in the regulation of cell proliferation, response to cytokine and chemokine, immune and inflammatory response, kinase activity, cell-cell signaling, stress-activated protein kinase signaling cascade, and transcription factor binding.
The clustering modules were ascertained from the PPI network consisting of 377 proteins (nodes). Using the MCODE plugin for cluster analysis, the top three highly interconnected sub-networks (modules) were obtained from the main network based on the clustering score, each one was autonomous of the others and exerted a distinctive task. Notably, interacting partners of interlinked hub proteins (MAPK1 and MAPK14) were observed in the first module with a clustering score of 11.808 (Fig.2A), the second module with a cluster score of 7.576 containing the interconnected hub protein CXCL8 (Fig.2B), whereas the third module with a cluster score of 7.037 including the connected hub protein FYN (Fig.2C). These three highly interlinked modules again highlight the significance of hub proteins in the global network structure and are enriched in topmost ontologies including response to cytokine and chemokine, immune and inflammatory response, cellular response to endogenous stimuli, protein kinase activity, and cell-cell signaling.
Mapping the connectivity to design a molecular roadmap
The hub proteins identified from the dataset were mapped with RA-related pathways including MAPK signaling, CXCL8-CXCR1/2 axis, toll-like receptor signaling and NFκB signaling cascades along with their associated bioactive ingredients to target the molecules promoting disease progression. Among the highly influential proteins, CXCL8 plays a pleiotropic role in immune-inflammatory response related to disease pathogenesis through several pathways including MAPK and NF-κB signaling pathways. Involvement of other hub proteins like MAPK1, MAPK14, and FYN was found to play a crucial role in regulating the production of pro-inflammatory cytokines including TNF-α, IL-1β, IL-6 and IL-17, which ultimately promotes synovial hyperplasia, inflammation and bone erosion.
Validation of molecular docking
To validate the network pharmacology prediction results, we employed AutoDock Vina to assess the physical interaction between the hub proteins and prime active ingredients of BDK. The hub proteins along with their interlinked bioactive compounds were selected from the network analysis to identify the binding activity. The binding potency between the compound and the protein was indicated as the lower the binding energy (> negative value), the higher the binding ability. The calculated binding energies between the bioactive compounds and their corresponding proteins were displayed in Table 3. The binding energy of kanakonol with MAPK1, cedrol/cryptomeridol with MAPK14, deodarone with FYN, and hypaphorine with CXCL8 was the least and their score was -6.6 kcal/mol, -7.8 kcal/mol, -8.8 kcal/mol, and -6.5 kcal/mol, respectively (Fig. 3), which suggests that these compounds could have a strong binding affinity towards their target proteins.
Discussion
From the standpoint of traditional pharmacology, TIM patterns are just too intricate and extensive to comprehend. However, systems pharmacology-based strategy consents this issue to be tackled. Herbal decoctions also consist of several active ingredients that encounter a wide range of biological targets implicated in disease pathogenesis. First-ever, the present study executes a systems pharmacology approach to meticulously unveil the mechanism of action of BDK decoction (Ayurveda formulation) against RA. A total of 205 compounds were retrieved from various databases including TCMSP, UNPD, and Dr. Duke’s Phytochemical and Ethnobotanical Databases and we screened out 57 compounds that were found to possess OB, DL and ADMET properties. Among the 57 active compounds, six compounds including nerolidol44,45, alpha-phellandrene46, camphene, coumaran, myrcene and naphthalene47 exhibit diverse biological activity including anti-inflammatory, anti-oxidant, promoting immune responses, and induces mitochondrial apoptosis. Evidence also showed that some other compounds like camphene and myrcene are reported to have anti-inflammatory and antioxidant activities19. Interestingly, they also exhibited promising therapeutic impacts on rheumatoid arthritis. For instance, Zou revealed that β-elemene could be efficient in stimulating mitochondrial apoptosis of rheumatoid arthritis fibroblast-like synoviocytes (RA-FLS), which is regulated via the initiation of ROS production and activation of p38 MAPK signaling48. Hence, these compounds must be given high priority while exploring natural products, which can attenuate the rheumatic condition for further investigation.
To comprehend intuitively the potential mechanism of BDK, a PPI network was constructed for 377 targets, to represent the interplay link between the proteins. The top 20 hub proteins were screened out by investigating each topological attribute in the PPI network. The frequently occurring 4 highly influential proteins MAPK1, MAPK14, FYN, and CXCL8 were recognized, which could play a crucial role in BDK-mediated alleviation of the rheumatic condition. MAPK1 (ERK2) and MAPK14 (p38α) are the key members of the mitogen-activated protein kinase (MAPK) family49,50. These proteins are involved in various events including regulation of cell proliferation, cell differentiation and apoptosis, triggering growth factors and pro-inflammatory cytokines, and may initiate ERK signaling pathway, subsequently related to the chronic inflammatory response51,52. In the tissues of damaged joints, MAPKs are implicated in the downstream IL-1, IL-17, and TNF-α receptors signaling cascade as well as regulate the production of the pro-inflammatory cytokines53. ERK2 (MAPK1) controls the generation of TNF-α, IL-23, IL-12, and IL-6 in lipopolysaccharide (LPS) induced macrophages54. It also regulates the COX-2-dependent PGE2 production owing to the release of epidermal growth factor in RA-FLS of patients55. MAPK14 (p38α) are predominantly activated by several inflammatory cytokines (IL-1β, TNF-α, IL-8, IL-6, MMP1 and MMP3) and modulate their expression that consequently plays a critical role in immune-inflammatory response related to autoimmune diseases56,57. In the inflamed joint tissues, IL-1β and TNF-α arbitrate p38-MAPK-dependent production of IL-6, IL-8, and receptor activator of nuclear factor-κ B ligand (RANKL) in osteocytes and stroma cells of bone marrow, which eventually promote cartilage and bone degradation58.
Among the highly influential proteins, CXCL8 (IL8) is one of the most extensively studied chemokines, released by various cell types such as macrophages, monocytes, fibroblasts and T lymphocytes26,59. CXCL8 is activated by several factors including cellular stress, lipopolysaccharide60, bacterial particles61, as well as by various cytokines (IL-1β, TNF-α, IL-6, interferon-γ, and so forth) (60). CXC chemokine receptors type 1 (CXCR1) and type 2 (CXCR2) are members of the G protein-coupled receptors (GPCR) family, which have a strong affinity for CXCL862. The CXCL8-CXCR1/CXCR2 axis has a pleiotropic role in the immune response including upregulation of granulocytes and neutrophils towards the inflammation site63, tending to pathogen elimination that results in high severity and chronic inflammatory immune response and tissue destruction and thus plays a vital role in all types of inflammatory disorders, for instance: arthritis64, bacterial infections65,66, sepsis67, ulcerative colitis68, and so forth. The CXCL8-CXCR1/CXCR2 axis also pertains to regulating angiogenesis, cell migration and immune cell infiltration during chronic inflammation46,69. The production of CXCL8 is intimately connected to the MAPK signaling pathway. ERK and JNK promote CXCL8 gene transcription via stimulating activating protein-1 (AP-1), a transcription activator primarily comprised of Jun and Fos proteins. The MAPK signaling pathway activates c-Jun and c-fos transcription factors, facilitating the translocation into the nucleus to create AP-1, which interacts with DNA target sequences and further promotes CXCL8 transcription. Subsequently, this leads to the regulation of cellular processes including migration, differentiation, proliferation, apoptosis, stress, and inflammation reaction70,71. Another hub protein Fyn regulates the various cellular functions including cell adhesion, survival, growth, motility, cytoskeletal remodeling and T-cell receptor signaling, which are immensely stretched to several pathological conditions. Fyn also upregulates the pro-inflammatory cytokines in macrophages, mast cells, and natural killer cells, which enhances the immune response that aggravates inflammatory disease conditions. Mkaddem revealed that downregulation and chronic activation of immunoreceptor tyrosine-based activation motif (ITAM)-containing immunoreceptor has been reported to be responsible for autoimmune and inflammatory disorders. ITAM present in the aggregated immunoreceptors can be phosphorylated by Fyn kinase72. Taken together, these recognized highly influential proteins, contribute to the explicit or implicit effect associated with MAPK signaling cascade on inflammation-induced arthritis, thus proving the rationale of the network pharmacology prediction outcomes.
We noted that the identified hub proteins are associated with rheumatoid arthritis and other inflammatory diseases, mediated by several pathways including TNF signaling pathway, IL-17 signaling pathway, toll-like receptor signaling pathway, IL-8 mediated signaling events and NOD-like receptor signaling pathway to name a few. The corresponding compounds of hub proteins or the synergistic effect of the compounds such as alpha-phellandrene, caffeic acid, cryptomeridol, kanokonol, cedrol, deodarone, hypaphorine and nerolidol, present in BDK decoction could facilitate the attenuation of rheumatic disease condition through the regulation of inflammatory-related pathways. Yangxinshi tablet was investigated to be efficient and reliable as a cardiovascular nourishing agent when administered at the time of cardiac arrest and enhances the immunological system [20]. Likewise, Zhang also established the protein interaction network of the potential targets of Wu-tou decoction as well as RA-associated targets by maintaining the coordination between immune and endocrine systems73. Therefore, multi-component herbal medicine is likely effective in managing the inflammatory disorders like rheumatoid arthritis primarily through two mechanisms: induction of immunomodulatory agents (like MAPK1, MAPK14, CXCL8, FYN) which inevitably assist in advancing the innate and adaptive immune system, and in the interim, regulation of pro-inflammatory mediators (like TNF-α, Il-1β, IL-6, IL-17, IL-8, COX-2) and inflammatory cytokines by herbal ingredients will possibly assist in reducing inflammation. In the future, the predicted targets and anti-inflammatory mechanism of BDK need to be validated along with their biological process and molecular function by a more comprehensive wet-lab experiment.
 |
Figure 1: Compound-Target (C-T) network connecting 57 compounds and 377 protein targets. |
 |
Figure 2: Clustering module analysis on the PPI network by MCODE. Nodes are displayed in turquoise green and the edges in pink colour. |
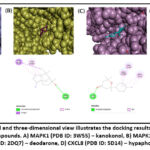 |
Figure 3: Two-dimensional and three-dimensional view illustrates the docking results of hub proteins and their corresponding active compounds. |
Table 1: Topological parameters of the T-IP network
|
Topological parameters |
Comprehended values |
|
Number of nodes |
377 |
|
Number of edges |
3992 |
|
Clustering co-efficient |
0.302 |
|
Network density |
0.002 |
|
Characteristic path length |
4.987 |
|
Average number of neighbors |
6.702 |
Table 2: The top 20 highly influential proteins (common between at least 3 – highlighted in bold) identified using distinct algorithms of Cytohubba plugin.
|
Degree |
Betweeness |
Closeness |
MCC |
||||
|
Terms |
Score |
Terms |
Score |
Terms |
Score |
Terms |
Score |
|
PPARA |
1187 |
PPARA |
1174870.293 |
MAPK3 |
1516.33333 |
CCR2 |
40901508 |
|
PTPN1 |
865 |
MAPK14 |
960585.1948 |
MAPK1 |
1491.96667 |
CXCR3 |
40857927 |
|
MAPK14 |
853 |
MAPK1 |
725003.9293 |
AKT1 |
1480.71905 |
CCR5 |
40712592 |
|
JAK2 |
765 |
HIF1A |
704480.7551 |
MAPK14 |
1476.13571 |
CCR3 |
39850964 |
|
JAK1 |
761 |
TP53 |
678181.2047 |
STAT3 |
1475.56667 |
CXCR2 |
39513387 |
|
PARP1 |
755 |
MDM2 |
601992.2102 |
PIK3CA |
1465.51667 |
CXCL8 |
38540127 |
|
HIF1A |
732 |
CXCL8 |
568776.8803 |
JUN |
1465.3 |
CXCR1 |
37922604 |
|
JAK3 |
636 |
CDK2 |
542526.8383 |
NFKB1 |
1461.7 |
CCR9 |
27105305 |
|
ESR1 |
538 |
GAPDH |
536164.1021 |
SRC |
1457.93571 |
CCR1 |
20743064 |
|
CDK2 |
524 |
PTGS2 |
520864.4916 |
TP53 |
1457.23333 |
CCR4 |
19162491 |
|
PTGS2 |
457 |
AKT1 |
513678.1772 |
CREBBP |
1452.16905 |
ADORA3 |
13848675 |
|
ALOX5 |
448 |
HSPD1 |
491127.8147 |
NR3C1 |
1445.51667 |
GRIN2A |
2993139 |
|
MAPK1 |
420 |
ESR1 |
491100.5705 |
FOS |
1433.08333 |
GRIN1 |
2993112 |
|
PTGS1 |
395 |
STAT3 |
490452.6036 |
MAPK11 |
1430.26905 |
GRIN2D |
2993064 |
|
MAPK8 |
390 |
CREBBP |
480520.6511 |
TNF |
1420.9381 |
GRIN2C |
2993064 |
|
EGFR |
381 |
SRC |
473425.1257 |
IL2 |
1420.88333 |
GRIA3 |
2989139 |
|
IL2 |
357 |
FYN |
459267.1117 |
FYN |
1419.76667 |
GRIA4 |
2989130 |
|
MMP9 |
342 |
NR3C1 |
456976.7236 |
NFKBIA |
1411.80238 |
GRIA2 |
2984050 |
|
FYN |
338 |
MAPK3 |
451730.9712 |
CXCL8 |
1409.63571 |
GRIN2B |
2912542 |
|
PTPN2 |
333 |
EPRS |
425404.4823 |
PDPK1 |
1406.65 |
NEFL |
2906643 |
Table 3: The binding affinity and interactions of selected top hit hub proteins docked with potential phytocompounds of Badradarvadi concoction.
|
S. No |
Target |
Compound/Ligand |
PubChem Id |
Binding affinity (kcal/Mol) |
No. of Hydrogen bonds |
|
1 |
MAPK1 |
Alpha-Phellandrene |
7460 |
-6.1 |
0 |
|
Caffeic acid |
689043 |
-6.1 |
0 |
||
|
Cryptomeridol |
165258 |
-6 |
1 |
||
|
Ephedrine |
9294 |
-6 |
1 |
||
|
Kanokonol |
46173905 |
-6.6 |
1 |
||
|
Linalol |
6549 |
-5.7 |
0 |
||
|
2 |
MAPK14 |
Alpha-Phellandrene |
7460 |
-6.1 |
0 |
|
1,1-diethoxy-3-methylbutane |
19695 |
-5 |
1 |
||
|
Cedrol |
65575 |
-7.1 |
1 |
||
|
Cryptomeridol |
165258 |
-6.2 |
0 |
||
|
Deodarone |
14657303 |
-7 |
1 |
||
|
2,3-Dihydro-Benzofuran |
10329 |
-5.5 |
0 |
||
|
Kanokonol |
46173905 |
-6.7 |
1 |
||
|
Nerolidol |
5284507 |
-5.7 |
1 |
||
|
3 |
FYN |
Deodarone |
14657303 |
-7.5 |
1 |
|
Ephedrine |
9294 |
-5.9 |
1 |
||
|
Alpha-Phellandrene |
7460 |
-5.9 |
0 |
||
|
4 |
CXCL8 |
Hypaphorine |
442106 |
-6.5 |
1 |
|
2,3-Dihydro-Benzofuran |
10329 |
-5.5 |
0 |
Conclusion
This study for the first time elucidated the therapeutic mechanism of BDK against rheumatoid arthritis using network pharmacology approach employing multiple bioinformatic tools. We identified 57 active ingredients of BDK effective against RA and MAPK1, MAPK14, FYN and CXCL8 were identified as crucial proteins that are involved in the main signal transduction pathway of the disease like MAPK signaling cascade, CXCL8-CXCR1/CXCR2 axis and TNF signaling pathway which were targeted by BDK to employ its anti-inflammatory and anti-apoptotic effect on RA. Our study offers theoretical support for the pharmacological basis and clinical application for future research, wherein the results of which would support and validate our study-derived conclusion.
Acknowledgement
The authors are grateful to the management of VIT for providing the facilities to carry out this research work.
Funding support
The authors received no specific funding for this work.
Authors’ contributions
Mohamed Thoufic Ali A M: performed experiments and wrote the manuscript.
Vino S: conceptualized and designed the work, formal analysis, writing – review & editing.
Conflict of interest
The authors declare that there is NO conflict of interest.
References
- Ferguson LD, Siebert S, McInnes IB, Sattar N. Cardiometabolic comorbidities in RA and PsA: lessons learned and future directions. Nat Rev Rheumatol (2019) 15:461–474. doi: 10.1038/S41584-019-0256-0
- Koenders MI, Van Den Berg WB. Novel therapeutic targets in rheumatoid arthritis. Trends Pharmacol Sci (2015) 36:189–195. doi: 10.1016/J.TIPS.2015.02.001
- Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet (2016) doi: 10.1016/S0140-6736(16)30173-8
- Almutairi K, Nossent J, Preen D, Keen H, Inderjeeth C. The global prevalence of rheumatoid arthritis: a meta-analysis based on a systematic review. Rheumatol Int 2020 415 (2020) 41:863–877. doi: 10.1007/S00296-020-04731-0
- Ali AMMT, Vino S. Genetic markers as therapeutic target in rheumatoid arthritis: A game changer in clinical therapy? Rheumatol Int (2016) 36:1601–1607. doi: 10.1007/s00296-016-3563-7
- Alunno A, Carubbi F, Giacomelli R, Gerli R. Cytokines in the pathogenesis of rheumatoid arthritis: new players and therapeutic targets. BMC Rheumatol 2017 11 (2017) 1:1–13. doi: 10.1186/S41927-017-0001-8
- M Guo YSQGHDWZYWXHXJ. The protective mechanism of Ginkgolides and Ginkgo flavonoids on the TNF-α induced apoptosis of rat hippocampal neurons and its mechanisms in vitro. Heliyon (2015) 1:e00020. doi: 10.1016/j.heliyon.2015.e00020
- Szekanecz Z, Koch AE. Successes and failures of chemokine-pathway targeting in rheumatoid arthritis. Nat Rev Rheumatol (2016) 12:5–13. doi: 10.1038/NRRHEUM.2015.157
- Ibrahim SSA, Huttunen KM. Orchestrated modulation of rheumatoid arthritis via crosstalking intracellular signaling pathways. Inflammopharmacology (2021) 29:965–974. doi: 10.1007/S10787-021-00800-3/FIGURES/5
- Malemud CJ. Intracellular Signaling Pathways in Rheumatoid Arthritis. J Clin Cell Immunol (2013) 4: doi: 10.4172/2155-9899.1000160
- Smolen JS, Aletaha D, Barton A, Burmester GR, Emery P, Firestein GS, Kavanaugh A, McInnes IB, Solomon DH, Strand V, et al. Rheumatoid arthritis. Nat Rev Dis Prim (2018) 4: doi: 10.1038/NRDP.2018.1
- Abbasi M, Mousavi MJ, Jamalzehi S, Alimohammadi R, Bezvan MH, Mohammadi H, Aslani S. Strategies toward rheumatoid arthritis therapy; the old and the new. J Cell Physiol (2019) 234:10018–10031. doi: 10.1002/JCP.27860
- Mohamed Thoufic Ali AM, Agrawal A, Sajitha Lulu S, Mohana Priya A, Vino S. RAACFDb: Rheumatoid arthritis ayurvedic classical formulations database. J Ethnopharmacol (2017) 197:87–91. doi: 10.1016/J.JEP.2016.06.047
- Sang Q, Liu X, Wang L, Qi L, Sun W, Wang W, Sun Y, Zhang H. Curcumin protects an SH-SY5Y cell model of Parkinson’s disease against toxic injury by regulating HSP90. Cell Physiol Biochem (2018) 51:681–691. doi: 10.1159/000495326
- Xue CCL, Zhang AL, Lin V, Da Costa C, Story DF. Complementary and alternative medicine use in Australia: a national population-based survey. J Altern Complement Med (2007) 13:643–650. doi: 10.1089/ACM.2006.6355
- Gandhi GR, Jothi G, Mohana T, Vasconcelos ABS, Montalvão MM, Hariharan G, Sridharan G, Kumar PM, Gurgel RQ, Li H Bin, et al. Anti-inflammatory natural products as potential therapeutic agents of rheumatoid arthritis: A systematic review. Phytomedicine (2021) 93: doi: 10.1016/J.PHYMED.2021.153766
- Lorenc A, Ilan-Clarke Y, Robinson N, Blair M. How parents choose to use CAM: A systematic review of theoretical models. BMC Complement Altern Med (2009) 9:1–12. doi: 10.1186/1472-6882-9-9/FIGURES/2
- Tandon M, Prabhakar S, Pandhi P. Pattern of use of complementary/alternative medicine (CAM) in epileptic patients in a tertiary care hospital in India. Pharmacoepidemiol Drug Saf (2002) 11:457–463. doi: 10.1002/PDS.731
- Choudhary M, Kumar V, Malhotra H, Singh S. Medicinal plants with potential anti-arthritic activity. J Intercult Ethnopharmacol (2015) 4:147. doi: 10.5455/JICE.20150313021918
- Anand U, Tudu CK, Nandy S, Sunita K, Tripathi V, Loake GJ, Dey A, Proćków J. Ethnodermatological use of medicinal plants in India: From ayurvedic formulations to clinical perspectives – A review. J Ethnopharmacol (2022) 284:114744. doi: 10.1016/J.JEP.2021.114744
- Basnyat S, Kolasinski SL. Ayurvedic medicine for rheumatoid arthritis. Curr Rheumatol Rep (2014) 16: doi: 10.1007/S11926-014-0435-6
- Gu J, Gui Y, Chen L, Yuan G, Lu HZ, Xu X. Use of Natural Products as Chemical Library for Drug Discovery and Network Pharmacology. PLoS One (2013) 8:e62839. doi: 10.1371/JOURNAL.PONE.0062839
- Geng J, Wang F, Huang Z, Chen X, Wang Y. Perspectives on anti-IL-1 inhibitors as potential therapeutic interventions for severe COVID-19. Cytokine (2021) 143: doi: 10.1016/J.CYTO.2021.155544
- Sivakumar TR, Surendhiran D, Chen K, Lv P, Vinothkanna A, Prathiviraj R, Sethupathy S, Sirajunnisa AR. Network pharmacology based analysis of Astragalus propinquus components for the treatment of rheumatoid arthritis and diabetes. South African J Bot (2021) 139:92–105. doi: 10.1016/J.SAJB.2021.01.034
- Dougados M, Soubrier M, Antunez A, Balint P, Balsa A, Buch MH, Casado G, Detert J, El-Zorkany B, Emery P, et al. Prevalence of comorbidities in rheumatoid arthritis and evaluation of their monitoring: Results of an international, cross-sectional study (COMORA). Ann Rheum Dis (2014) 73:62–68. doi: 10.1136/ANNRHEUMDIS-2013-204223
- Bie Y, Ge W, Yang Z, Cheng X, Zhao Z, Li S, Wang W, Wang Y, Zhao X, Yin Z, et al. The Crucial Role of CXCL8 and its receptors in colorectal liver metastasis. Dis Markers (2019) 2019: doi: 10.1155/2019/8023460
- Zhao S, Iyengar R. Systems Pharmacology: Network Analysis to Identify Multiscale Mechanisms of Drug Action. Annu Rev Pharmacol Toxicol (2012) 52:505. doi: 10.1146/ANNUREV-PHARMTOX-010611-134520
- Cao K, Zheng A, Xu J, Li H, Liu J, Peng Y, Long J, Zou X, Li Y, Chen C, et al. AMPK activation prevents prenatal stress-induced cognitive impairment: Modulation of mitochondrial content and oxidative stress. Free Radic Biol Med (2014) 75:156–166. doi: 10.1016/J.FREERADBIOMED.2014.07.029
- Astanga Hrdayam (Eng) : Dr. R. Vidyanath : Free Download, Borrow, and Streaming : Internet Archive.
- Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Li P, Guo Z, Tao W, Yang Y, et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform (2014) 6:1–6. doi: 10.1186/1758-2946-6-13/FIGURES/2
- Duke JA. Handbook of Medicinal Herbs. Handb Med Herbs (2002) doi: 10.1201/9781420040463/HANDBOOK-MEDICINAL-HERBS-JAMES-DUKE
- Lipinski C. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol (2004) 1:337–341. doi: 10.1016/j.ddtec.2004.11.007
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem (2002) 45:2615–2623. doi: 10.1021/JM020017N/SUPPL_FILE/JM020017N_S.PDF
- Yap CW. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J Comput Chem (2011) 32:1466–1474. doi: 10.1002/JCC.21707
- Banerjee P, Eckert AO, Schrey AK, Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res (2018) 46:W257. doi: 10.1093/NAR/GKY318
- Chen X, Ji ZL, Chen YZ. TTD: Therapeutic Target Database. Nucleic Acids Res (2002) 30:412–415. doi: 10.1093/NAR/30.1.412
- Xue R, Fang Z, Zhang M, Yi Z, Wen C, Shi T. TCMID: Traditional Chinese Medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res (2013) 41: doi: 10.1093/NAR/GKS1100
- Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res (2007) 35:D198. doi: 10.1093/NAR/GKL999
- Liang G, Zhou H, Wang Y, Gurley EC, Feng B, Chen L, Xiao J, Yang S, Li X. Inhibition of LPS-induced production of inflammatory factors in the macrophages by mono-carbonyl analogues of curcumin. J Cell Mol Med (2009) 13:3370–3379. doi: 10.1111/J.1582-4934.2009.00711.X
- Murakami Y, Tripathi LP, Prathipati P, Mizuguchi K. Network analysis and in silico prediction of protein-protein interactions with applications in drug discovery. Curr Opin Struct Biol (2017) 44:134–142. doi: 10.1016/J.SBI.2017.02.005
- Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J Proteome Res (2019) 18:623–632. doi: 10.1021/ACS.JPROTEOME.8B00702
- Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol (2014) 8:1–7. doi: 10.1186/1752-0509-8-S4-S11/TABLES/4
- O Trott AO. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem (2010) 31:455–461.
- Chan WK, Tan LTH, Chan KG, Lee LH, Goh BH. Nerolidol: A Sesquiterpene Alcohol with Multi-Faceted Pharmacological and Biological Activities. Molecules (2016) 21: doi: 10.3390/MOLECULES21050529
- Ni YL, Shen HT, Su CH, Chen WY, Huang-Liu R, Chen CJ, Chen SP, Kuan YH. Nerolidol suppresses the inflammatory response during lipopolysaccharide-induced acute lung injury via the modulation of antioxidant enzymes and the AMPK/NRf-2/HO-1 pathway. Oxid Med Cell Longev (2019) 2019: doi: 10.1155/2019/9605980
- Zhu Y, Yang S, Zhao N, Liu C, Zhang F, Guo Y, Liu H. CXCL8 chemokine in ulcerative colitis. Biomed Pharmacother (2021) 138:111427. doi: 10.1016/J.BIOPHA.2021.111427
- Tag HM, Khaled HE, Ismail HAA, El-Shenawy NS. Evaluation of anti-inflammatory potential of the ethanolic extract of the Saussurea lappa root (costus) on adjuvant-induced monoarthritis in rats. J Basic Clin Physiol Pharmacol (2016) 27:71–78. doi: 10.1515/JBCPP-2015-0044
- Zou S, Wang C, Cui Z, Guo P, Meng Q, Shi X, Gao Y, Yang G, Han Z. β-Elemene induces apoptosis of human rheumatoid arthritis fibroblast-like synoviocytes via reactive oxygen species-dependent activation of p38 mitogen-activated protein kinase. Pharmacol Rep (2016) 68:7–11. doi: 10.1016/J.PHAREP.2015.06.004
- Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J (2010) 429:403–417. doi: 10.1042/BJ20100323
- Morrison DK. MAP kinase pathways. Cold Spring Harb Perspect Biol (2012) 4: doi: 10.1101/CSHPERSPECT.A011254
- Clark AR, Dean J LE. Suppl 2: The p38 MAPK Pathway in Rheumatoid Arthritis: A Sideways Look. Open Rheumatol J (2012) 6:209. doi: 10.2174/1874312901206010209
- Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science (2002) 298:1911–1912. doi: 10.1126/SCIENCE.1072682
- McGeachy MJ, Cua DJ, Gaffen SL. The IL-17 Family of Cytokines in Health and Disease. Immunity (2019) 50:892–906. doi: 10.1016/J.IMMUNI.2019.03.021
- Lu N, Malemud CJ. Extracellular Signal-Regulated Kinase: A Regulator of Cell Growth, Inflammation, Chondrocyte and Bone Cell Receptor-Mediated Gene Expression. Int J Mol Sci (2019) 20: doi: 10.3390/IJMS20153792
- Nah SS, Won HJ, Ha E, Kang I, Cho HY, Hur SJ, Lee SH, Baik HH. Epidermal growth factor increases prostaglandin E2 production via ERK1/2 MAPK and NF-icB pathway in fibroblast like synoviocytes from patients with rheumatoid arthritis. Rheumatol Int (2010) 30:443–449. doi: 10.1007/S00296-009-0976-6
- Asih PR, Prikas E, Stefanoska K, Tan ARP, Ahel HI, Ittner A. Functions of p38 MAP Kinases in the Central Nervous System. Front Mol Neurosci (2020) 13:172. doi: 10.3389/FNMOL.2020.570586/BIBTEX
- Sakurai K, Dainichi T, Garcet S, Tsuchiya S, Yamamoto Y, Kitoh A, Honda T, Nomura T, Egawa G, Otsuka A, et al. Cutaneous p38 mitogen-activated protein kinase activation triggers psoriatic dermatitis. J Allergy Clin Immunol (2019) 144:1036–1049. doi: 10.1016/J.JACI.2019.06.019
- Koga Y, Tsurumaki H, Aoki-Saito H, Sato M, Yatomi M, Takehara K, Hisada T. Roles of Cyclic AMP Response Element Binding Activation in the ERK1/2 and p38 MAPK Signalling Pathway in Central Nervous System, Cardiovascular System, Osteoclast Differentiation and Mucin and Cytokine Production. Int J Mol Sci (2019) 20: doi: 10.3390/IJMS20061346
- Wanninger J, Bauer S, Eisinger K, Weiss TS, Walter R, Hellerbrand C, Schäffler A, Higuchi A, Walsh K, Buechler C. Adiponectin upregulates hepatocyte CMKLR1 which is reduced in human fatty liver. Mol Cell Endocrinol (2012) 349:248. doi: 10.1016/J.MCE.2011.10.032
- Brat DJ, Bellail AC, Van Meir EG. The role of interleukin-8 and its receptors in gliomagenesis and tumoral angiogenesis. Neuro Oncol (2005) 7:122–133. doi: 10.1215/S1152851704001061
- Protein Data Bank. RCSB PDB: Homepage. RCSB PDB (2019)
- Baggiolini M. Chemokines in pathology and medicine. J Intern Med (2001) 250:91–104. doi: 10.1046/J.1365-2796.2001.00867.X
- Waugh DJJ, Wilson C. The Interleukin-8 Pathway in Cancer. Clin Cancer Res (2008) 14:6735–6741. doi: 10.1158/1078-0432.CCR-07-4843
- Min SH, Wang Y, Gonsiorek W, Anilkumar G, Kozlowski J, Lundell D, Fine JS, Grant EP. Pharmacological targeting reveals distinct roles for CXCR2/CXCR1 and CCR2 in a mouse model of arthritis. Biochem Biophys Res Commun (2010) 391:1080–1086. doi: 10.1016/J.BBRC.2009.12.025
- Boonyanugomol W, Rukseree K, Kongkasame W, Palittapongarnpim P, Baik SC, Manwong M. Genetic Polymorphisms of CXCL8 (−251) Are Associated with the Susceptibility of Helicobacter pylori Infection Increased the Risk of Inflammation and Gastric Cancer in Thai Gastroduodenal Patients. Iran J Allergy, Asthma Immunol (2019) 18:393–401. doi: 10.18502/IJAAI.V18I4.1417
- Chen Z, Chen X, Li F, Cheng JW, Cheng HT, Yeh SC, Yu HY. A novel CXCL8-IP10 hybrid protein is effective in blocking pulmonary pathology in a mouse model of Klebsiella pneumoniae infection. Int Immunopharmacol (2018) 62:40–45. doi: 10.1016/J.INTIMP.2018.06.040
- Kaneider NC, Agarwal A, Leger AJ, Kuliopulos A. Reversing systemic inflammatory response syndrome with chemokine receptor pepducins. Nat Med (2005) 11:661–665. doi: 10.1038/NM1245
- Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res (2009) 37: doi: 10.1093/NAR/GKP427
- Hannelien V, Karel G, Jo VD, Sofie S. The role of CXC chemokines in the transition of chronic inflammation to esophageal and gastric cancer. Biochim Biophys Acta (2012) 1825:117–129. doi: 10.1016/J.BBCAN.2011.10.008
- Uddin MJ, Dorotea D, Pak ES, Ha H. Fyn Kinase: A Potential Therapeutic Target in Acute Kidney Injury. Biomol Ther (Seoul) (2020) 28:213. doi: 10.4062/BIOMOLTHER.2019.214
- Pearson G, Robinson F, Beers Gibson T, Xu B, Karandikar M, Berman K, Cobb MH. Mitogen-Activated Protein (MAP) Kinase Pathways:Regulation and Physiological Functions. Endocr Rev (2001) 22:153–183. doi: 10.1210/EDRV.22.2.0428
- Mkaddem S Ben, Benhamou M, Monteiro RC. Understanding Fc receptor involvement in inflammatory diseases: From mechanisms to new therapeutic tools. Front Immunol (2019) 10:811. doi: 10.3389/FIMMU.2019.00811/BIBTEX
- Zhang J, Fan W, Wang H, Bao L, Li G, Li T, Song S, Li H, Hao J, Sun J. Resveratrol Protects PC12 Cell against 6-OHDA Damage via CXCR4 Signaling Pathway. Evidence-based Complement Altern Med (2015) 2015: doi: 10.1155/2015/730121








