Manuscript accepted on :13-10-2023
Published online on: 06-02-2024
Plagiarism Check: Yes
Reviewed by: Dr. Omage, Joel and Dr. Bhavana Gundavarapu
Second Review by: Dr. Md. Sarwar Hossain
Final Approval by: Dr. Anton R Keslav
Prashant Nayak and R Narayan Charyulu*
and R Narayan Charyulu*
Nitte (Deemed to be University), NGSM Institute of Pharmaceutical Sciences, Department of Pharmaceutics, Deralakatte, Mangaluru-575018, India.
Corresponding Author E-mail: narayana@nitte.edu.in
DOI : https://dx.doi.org/10.13005/bpj/2847
Abstract
siRNAs(Small interfering RNA) have emerged as new nucleic acid drugs to treat life-threatening diseases such as malignant tumors as our understanding of the molecular mechanisms of endogenous RNA interference has increased. Synthetic small interfering RNAs (siRNA) or short hairpin RNAs (shRNA) have been shown to have clinical potential in dental illnesses, eye infections, cancer, metabolic syndromes, neurological disorders, and other illnesses in subsequent RNAi investigations. Although various siRNA are used as a medication for respiratory and ophthalmic illnesses in clinical trials, there are problems in developing siRNA for malignancy treatments because systemic delivery would be required in the treatment of the majority of patients. Aside from nonspecific off-target effects and immunological stimulation issues, proper administration remains a significant challenge. The technologies that have been created for the formulation of siRNA therapeutics, including antisense oligonucleotides and plasmid DNA, have prepared the path for rapid advancement in in-vivo siRNA delivery. This review focuses on the Potential uses of siRNA in different diseases and its challenges in usage.
Keywords
Cancer; Clinical trials; Diseases; Formulation; illnesses; RNA interference
Download this article as:| Copy the following to cite this article: Nayak P, Charyulu R. N. Small Interfering RNA Drug Delivery System in Cancer. Biomed Pharmacol J 2024;17(1). |
| Copy the following to cite this URL: Nayak P, Charyulu R. N. Small Interfering RNA Drug Delivery System in Cancer. Biomed Pharmacol J 2024;17(1). Available from: https://bit.ly/3HOXbgz |
Introduction
Small interfering RNAs (siRNAs) are becoming increasingly used as sequence-specific transcription inhibitors. When siRNAs, or short RNAs (double-stranded), are targeted on the cells, they mediate gene silencing of protein post-transcriptional phase with a definite target by destroying messenger RNAs (mRNAs) with matching sequences1,2. It is possible to target any disease-causing gene, as well as any cell type or tissue. RNA interference (RNAi), a naturally occurring mechanism that regulates gene expression, has emerged as a powerful tool for modifying gene expression in a variety of domains, including functional genomics, drug validation, and transgenic design3,4
The RNAi mechanism was primarily discovered in plants and then proven in the Caenorhabditis elegans roundworm using a microinjection approach to deliver dsRNA. In C. elegans, the introduction of dsRNA molecules can cause intrusive activity and very accurately inhibit complementary gene expression. Inhibition of complementary gene expression5.Recent research has shed light on the molecular approach of RNAi, In which dsRNA causes homologous mRNA to be silenced. Dicer, an enzyme found in the cytoplasm of mammalian cells, crafts RNA silencing which breaks down long dsRNA to produce small interfering RNA (siRNA) with a length of 21–23 nucleotides. The siRNAs are then integrated into an RNA-induced silencing complex (RISC) and unraveled into single-stranded RNA (ssRNA), with the sense strand ssRNA then degraded6.
The antisense strand of the RISC then binds to compatible mRNA molecules. The Argonaute 2 protein, which is a protein of the Argonaute family and is the reason for mRNA degradation and ssRNA synthesis, is one of the primary components of the RISC complex.7
Argonaute 2 (formerly known as eIF2C2), a catalytic engine inside RISC, allows the anti-sense strand of RNA to complement mRNA sequences and damages target mRNAs via the PIWI domain of an Ago protein, which is a structural homolog of RNase H. Although Adenosine 5′-triphosphate boosts endonuclease activity, this reaction is not essential for the RISC aimed breakdown. The hydrolysis mechanism that releases the 5′-PO4 and 3′-OH groups from the target mRNA phosphodiester backbone requires a divalent metal ion (Mg2+)8-12.
Mechanisms and Potential Applications of RNAi
siRNAs are dsRNAs with a short stretch (19–30 nucleotides) that can degrade complementary mRNA in the cytoplasm. Long dsRNAs are cleaved into short dsRNA duplexes or siRNA in the cytoplasm by the endoribonuclease Dicer. RNA-induced silencing complexes are loaded with siRNA (RISC). Argonaute 2 (Ago-2) is a protein that cuts and discharges one strand of double-strand RNA, resulting be an initiated form of RISC with an RNA which is single-strand acts as guide siRNA that guides the selectivity of target mRNA recognition by corresponding base pairing13.The target mRNA is then cleaved between bases 10 and 11 relative to the 5′ end of the siRNA antisense strand, resulting in mRNA degradation and gene silence14..
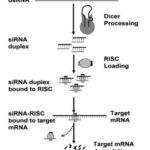 |
Figure 1: Shows the mechanism of gene silencing by siRNA14 |
RNAi Therapeutics a Comparison
siRNA, short hairpin RNA (shRNA), and micro-RNA (miRNA) are the three kinds of RNAi14. MiRNAs are single-stranded noncoding RNAs that are transcribed by RNA polymerase II from their genes or introns. After transcription, the main miRNA is first made into siRNA, which has advantages over shRNA in terms of transfection efficiency and distribution. In the transfection process the DNA in the chromosome need not be dependent on siRNA’s action, but shRNA-articulating pDNA needs a properly structured promoter.RNAi functions on the cytoplasm and during its delivery need not be specific in the nucleus, but shRNA will act on the nucleus. The quiescent cells which have limited nuclear envelope permeability possess extra tasks later in the mechanism. In cells with modest proliferative activity, shRNA has less transfection activity as compared to siRNA15.
so comparison to shRNA, siRNA has a 100-fold smaller molecular weight (19–30 bp), which makes it easier to distribute and modify. The rest of this article will be about siRNA.
Assessment of siRNA with other RNAi therapeutic classes
The ability to quickly create sequences for very selective inhibition of the object of concern is one of the key advantages of siRNA over small molecule medicines. In addition, siRNA manufacturing is quite simple and does not require a cellular manifestation scheme, refolding procedure, or sophisticated protein purification method16.
Anti-mRNA approaches can be allocated into four classes: Single-stranded antisense oligonucleotides (ODNs) are chemically produced, short single-stranded oligonucleotides that block the translation of a definite gene by hybridizing to the suitable mRNA by Watson-Crick binding17. Ribozymes, which are catalytically active RNAs that use transesterification or hydrolysis processes to break single-stranded sections of the RNA18, 19, are the second and third anti-mRNA techniques, correspondingly17.
In cell culture and in vivo, the knockdown effects of antisense siRNA and ODNs concluded the effectiveness of siRNA. The proteins are targeted by siRNA with high specificity at lower doses than the antisense ODNs. Gene expression is less by 100-1000 times as compared to ODNs20.
In human cells, there was a second comparative research. The potency, maximal potency, duration of action, and sequence specificity of optimized RNase H-dependent ODNs and siRNA-ODN duplexes were examined. With notable exceptions, the activity of RNase H-dependent ODNs targeted to the same locus was frequently correlated with the activity of 80 siRNA-ODN duplexes designed to bind to RNA from four different human genes. Only RNase H-dependent ODNs were found to be active when directed against pre-mRNA targets, whereas siRNAs were not.
Finally, microRNAs are endogenous, tiny, double-stranded, noncoding RNA molecules that have been discovered in a variety of organisms and viruses. This family of “new” molecules influences gene expression and development by being transcribed primarily from introns, exons, and intergenic regions. MicroRNAs are typically 20–24 nucleotides long and interact with partially mismatched sequences in the messengers’ 3′ untranslated regions to alter target mRNAs post-transcriptionally. As a result of these interactions, target mRNAs are either repressed or destroyed21.
Hurdles of siRNA Usage
Clinical trials and siRNA
SiRNAs are emerging as new-generation drugs due to their precise and strong RNAi-triggering potential. Several research have backed up siRNA’s medicinal potential. The effectiveness of viral mRNA-targeted siRNA in blocking different stages of the HIV lifetime phase has been demonstrated22.
There was autoimmune hepatitis seen in mice after injecting Fas-specific siRNA liver failure was reduced 23. Several prospective siRNA candidates are currently being tested in clinical trials for macular degeneration, respiratory disorders, and cancer treatment.
In biopharmaceutical therapy, siRNA treatment has immense potential.
Because siRNA affects translation rather than transcription in DNA, RNAi might not intermingle with DNA in the chromosome. Because there is no DNA contact, there are fewer concerns about undesirable gene changes that could occur as a result of gene therapy which is DNA-based. Because siRNA interacts with mRNA rather than protein molecules, it can limit the creation of dangerous proteins before they are synthesized.
Another advantage of siRNA as a therapeutic medication is that it can be used to silence a wide spectrum of target proteins to cure illnesses 24. Traditional chemical drug targets have been restricted to specific types of ion channels, enzymes, and receptors. Monoclonal antibodies and cytokines, for example, are currently used to target moieties that are mostly found in the blood or on the cell surface. An RNAi-built drug, on the other hand, can aim at any mRNA of interest, independent of translated protein location on the cell. Furthermore, a few siRNA strands are required per cell in effective gene silencing25, 26.
Hurdles of siRNA Remedy
The siRNAs have significant benefits as potential novel medications, but further research will have to overcome some obstacles. The possibility of an ‘off-target’ impact, which is the inhibition of a gene whose expression should not be targeted because the gene shares partial homology with the siRNA, is one such difficulty. Inadvertently silencing nontarget genes can cause challenges with data interpretation and even harm. To avoid this problem, great consideration should be given to the choice of effective siRNAs. In selecting siRNA it has to be taken care of internal repeating sequences, GC content, secondary structure, and base preference in sense strand, the length of siRNA should be 19-22 bps ideally. Several siRNA firms offer online design algorithms that take into account secondary structure, siRNA duplex end-stabilities, and mRNA target sequence, at the same time reducing the sequence-dependent off-target effects26.
The latest reading found that substituting a 2′-O-methyl ribosyl group at position 2 in the guide strand with complementarity to the siRNA guide could minimize the silence of most off-target transcripts Several computer techniques have been developed for forecasting the role by sequence of siRNA.
Immune stimulation, or the innate immune system’s detection of a siRNA duplex, is another obstacle to siRNA therapy23, 24.
Due to the stimulus that activates innate immune reactions, too much siRNA has been shown to cause nonspecific effects. Protein kinase R, a dsRNA sensor, is thought to trigger the immune system. siRNA is identified by toll-like receptors such as (TLR7), TLR8, and TLR927, and activation of NF-kB and interferon regulatory factors was discovered to generate inflammatory cytokines and interferons.TLR3, a viral RNA that is double-stranded and acts as a sensor, was recently found to be activated by 21-nucleotide or longer siRNAs, which suppressed neovascularization in a sequence and target-nonspecific way28.
Nevertheless, additional research has found that not all siRNAs can stimulate the immune system29, 30.
As per the discussion by MacLachlan et al siRNA stimulates the immune response which is found to be significant in nucleotide sequence. It was discovered that induction by siRNA by TLR7-mediated interferon alpha is always sequence-specific. As a result, the sequencing issue of siRNA-mediated immune activation must be studied further.
In-vivo Application of siRNA Delivery Systems
Several RNAi vectors for distribution into the cell’s cytoplasm have been reported by researchers and biotechnology companies, and while these are adequate for in vitro applications, this delivery method is frequently ineffectual in vivo.In clinical trials, siRNAs are currently delivered locally to particular target locations such as the lungs and eye, eluding the complexities of systemic distribution. To cure most tumors and other disorders, however, siRNA must be administered systemically.
The following characteristics should be included in the best siRNA in vivo systemic delivery. First and foremost, delivery systems must be biodegradable, biocompatible, and safe for the immune system. Next, the formulation must reach the target site or tissues, whereas it should be an intact strand formulation thereby avoiding serum nuclease degradation. Following that, after systemic administration, the delivery systems must provide target tissue-specific distribution to avoid rapid hepatic or renal clearance. To end with, the endocytosis has delivered siRNA to target cells; the systems must encourage the endosomal discharge of RNAi into the cytoplasm, permitting siRNA to interact with endogenous RNA31.
Advantages and Drawbacks
Most creatures, from plants to vertebrates, have already been shown to be affected by RNA interference. It’s also had a big impact on biomedical research, and it’ll lead to some interesting medical applications. Most creatures, from plants to vertebrates, have already been shown to be affected by RNA interference. This will light up biomedical research with news on medical applications. Infections, cancer, antiviral diseases (e.g., human immunodeficiency virus 1, HIV-1and viral hepatitis), neurodegenerative illnesses, and antiviral diseases (e.g., human immunodeficiency virus 1, HIV-1 and viral hepatitis) could all benefit from RNAi in the future. RNAi could be a game-changing new treatment option for infections, neurodegenerative illnesses, cancer, and antiviral diseases (e.g., human immunodeficiency virus 1, HIV-1, and viral hepatitis), unfocused exceptionally profound cell lines study then relatively high doses, siRNAs longer than 30 nucleotides activate the immune system32-35.
In vivo–administered siRNAs also have poor tissue penetration, low transfection efficiency, and nonspecific immune activation, which have hindered their beneficialefficacy. However, the absence of an effective delivery technique to aim and inject siRNA into the required cells is a significant constraint for this approach’s full therapeutic perspective.
Scientists are trying for an effective delivery system that can injected and should be compatible with minimal side effects and comfirmly achieve the region or tissue of interest.
Cancer Therapy of siRNA
The siRNA and RNAi phenomena ensure innovative possibilities for the improvement of inventive treatments to cure earlier incurable ailments, like as cancer. Because it uses the endogenous RNAi system, allowing for the careful lessening of disease-associated genetic factors, and can be applied to any gene with a corresponding arrangement, siRNA has intrinsic efficacy36. Many essential genes involved in many malignancies have been learned, the mutations carefully defined, and the trials via the act are categorized since cancer is a genetic illness37.
The argument for siRNA-mediated gene therapy is supported by the genetic nature of cancer. Several siRNAs have been engineered to target dominant oncogenes, dysregulated oncogenes, or viral oncogenes involved in carcinogenesis. Therapeutic siRNAs have also been studied for their ability to silence target molecules important for tumor-host interactions and tumor resistance to chemotherapy. Anti proliferative and/or apoptotic effects have been shown when siRNAs are used to turn off key cancer-associated target proteins.
Nonetheless, the majority of RNAi-mediated gene silencing for cancer therapy has been done in cell cultures in the lab, and there are still major roadblocks in the move to the bedside due to delivery issues. It is necessary to design delivery strategies that can improve siRNA stability and cancer cell selectivity while minimizing off-target and nonspecific immune stimulatory effects. The formulation methods need to be tailored for definite tumors because the method of management may vary based on the form of the malignancy.
Breast Tumour
Breast sarcoma is characterized by uncontrolled cell growth in breast tissues and ranks as the second most common cancer worldwide, following lung sarcoma. Among females in the United States, breast cancer is the most frequently diagnosed cancer and a leading cause of cancer-related deaths in this population. Surgical intervention is the primary treatment for localized breast cancer, with additional options including hormone therapy, chemotherapy, immunotherapy, and/or radiotherapy. Researchers have explored the use of lipid or polymer-based delivery systems for anti-cancer siRNA in breast cancer cells and mouse models with human breast tumor xenografts38.
To transport siRNAs to breast cancer cell lines, cationic liposomes containing dicta decyl amido glycyl spermidine (DOGS) and DOPE were employed. These liposomes showed successful delivery of siRNAs and specific localization in cytoplasmic compartments near the nucleus. They exhibited low cytotoxicity and high uptake of cyclin D1-specific siRNA in MCF-7 breast cancer cells, as well as efficient delivery of plasminogen activator inhibitor type I-specific siRNA to MDA MB 231 breast cancer cells39.
In another study, a hybrid molecule consisting of DNA and RNA, targeting HER-2, was encapsulated in immune cationic liposomes modified with a single-chain anti-transferrin receptor antibody fragment. Intravenous administration of these liposomes in mice with MDAMB-435 human breast cancer tumors resulted in suppression of HER-2 expression.
PEI-g-PEG copolymers have been utilized as safe carriers for siRNA targeting the clusterin secretory signal peptide. These complexes effectively inhibited clusterin secretion and enhanced the lethality of ionizing radiation in MCF-7 human breast cancer cells40.
Chitosan nanoparticles containing quantum dots were employed to deliver HER2/neu siRNA. The nanoparticles efficiently entered SKBR3 breast cancer cells, thanks to the entrapped fluorescent quantum dots, and achieved precise delivery of siRNA to HER2-overexpressing cells, resulting in gene silencing41-43.
Furthermore, self-assembling cationic core-shell nanoparticles composed of biodegradable amphiphilic copolymers were developed for the co-delivery of small-molecule anticancer drugs and siRNA. This approach showed enhanced sensitivity to paclitaxel in MDA-MB-231 human breast cancer cells when combined with siRNA targeting Bcl-244.
In summary, various delivery systems, including liposomes, nanoparticles, and copolymers, have been investigated for the targeted delivery of siRNA in breast cancer treatment. These approaches offer promising strategies for improving therapeutic outcomes in breast cancer patients.
Ovarian Cancer
Ovarian cancer ranks as the eighth most commonly diagnosed cancer in women. It is a particularly lethal gynecologic cancer due to challenges in early detection and the limited effectiveness of chemotherapy. Although several theories exist, the exact causes of ovarian cancer remain unknown. Treatment options for ovarian cancer have traditionally included surgery, chemotherapy, and radiation therapy. However, the toxic nature of the tumor has prompted extensive research into in vivo siRNA treatments using various synthetic techniques in animal models45.
In one study, low-molecular-weight linear PEI was utilized to deliver HER-2 receptor-targeting siRNA via intraperitoneal administration in mice with subcutaneously xenografted SKOV-3 ovarian cancer cells. The complexation of siRNA with PEI resulted in a significant reduction in tumor growth by downregulating HER-2 expression in the animal model. Notably, this effect was observed with the PEI-complexed siRNA, but not with free siRNA46.
Sood et al. conducted a series of ovarian cancer treatment trials involving siRNA-based approaches, including the use of neutral liposomes. They successfully encapsulated EphA2-targeting siRNA within neutral liposomes based on 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC) with an encapsulation efficiency of 65 percent. In xenografted mouse models of ovarian carcinoma, intravenous administration of liposomal EphA2-targeting siRNA at a dosage of 0.15 mg/kg inhibited tumor growth. Furthermore, combining siRNA with paclitaxel resulted in a significant reduction in ovarian tumor growth in a mouse model. The efficacy of intraperitoneal treatment with liposomal EphA2-targeting siRNA in reducing tumor growth in ovarian cancer mouse models was comparable to that achieved through intravenous delivery of siRNA liposomes. Other studies using neutral DOPC-based liposomes demonstrated the delivery of siRNAs targeting -2 adrenergic receptor, interleukin-8, and focal-adhesion-kinase, resulting in the suppression of ovarian tumor growth47-50.
In summary, ovarian cancer poses significant challenges, but siRNA-based therapies using various delivery methods such as PEI complexes and liposomes have shown promise in inhibiting tumor growth in animal models. These approaches hold the potential for advancing ovarian cancer treatment strategies.
Lung Cancer
Lung cancer is a leading cause of cancer-related mortality in men and the second leading cause in women. It can be categorized into small-cell lung carcinoma and non-small-cell lung carcinoma, requiring distinct treatment approaches. Recent advancements in lung cancer therapy have identified molecular targets such as gefitinib (Iressa), erlotinib (Tarceva), and bevacizumab, which selectively inhibit specific proteins involved in tumor growth and angiogenesis51-52.
In the context of lung cancer, liposomes loaded with human double-minute gene 2-specific siRNA were developed with arginine octamers on their surface. These siRNA-loaded liposomes exhibited good stability in the bloodstream and efficiently transfected multiple lung cancer cell lines after 24 hours of incubation53.
LPD (liposome-polycation-DNA) nanoparticles were utilized for RNA interference in lung carcinoma. PEGylated LPD nanoparticles containing RNAi targeting survivin demonstrated antitumor effects by promoting apoptosis, suppressing tumor cell proliferation, and enhancing the sensitivity of tumor cells to anticancer drugs. In an in vivo lung cancer xenograft mouse model, LPD nanoparticle formulations exhibited substantial tumor growth inhibition. Intravenous administration of epidermal growth factor receptor-specific siRNAs using LPD nanoparticles, in combination with cisplatin, synergistically suppressed lung cancer tumor activity54.
To enable systemic delivery, cationic immunoliposomes carrying siRNA were employed in an animal model of lung cancer metastasis. These immunoliposomes, coupled with a single-chain antibody fragment targeting the transferrin receptor, were administered intravenously to mice. The fluorescently labeled siRNA delivered by cationic immunoliposomes specifically accumulated in lung tissues with tumor metastases, while sparing the liver55.
In summary, siRNA-based therapies using liposomes, LPD nanoparticles, and cationic immunoliposomes have shown promising results in targeting lung cancer. These delivery systems offer efficient transfection, tumor growth inhibition, and specific siRNA distribution, providing potential avenues for the development of effective lung cancer treatments.
Liver Carcinoma
While various factors contribute to the development of liver cancer, infection with hepatitis B virus (HBV) or hepatitis C virus (HCV) is considered significant. Surgery, including liver transplantation, is the primary treatment option for hepatocellular carcinoma (HCC) and large liver tumors. Chemotherapy and radiotherapy can be used as adjuvant therapies56.
Chronic HBV infection is known to increase the risk of cirrhosis and HCC, and RNA interference (RNAi)-based therapeutics have shown potential in treating HBV infection. In animal models with HBV replication, chemically modified siRNAs targeting HBV RNA were administered intravenously. High-dose siRNA treatment at 30 mg/kg resulted in a significant decrease in serum HBV DNA levels, highlighting the importance of chemically altered siRNAs for HBV treatment57.
To improve the delivery of siRNA targeted to HBV RNA, a lipid-based SNALP (stable nucleic acid-lipid particle) technology was utilized. SNALP-encapsulated siRNA, administered through three daily intravenous injections at a dose of 3 mg/kg, exhibited an extended half-life in the liver. This led to a substantial 95 percent reduction in HBV serum titers, with the reduction being dose-dependent and sustained for up to 7 days after the final dose. Importantly, SNALP did not induce immune responses or the production of interferon’s or inflammatory cytokines58.
In a liver metastasis model, a lipid-based system was employed to deliver siRNA in vivo. Liver tumors were induced in nude mice through intrasplenic injection of A549 cell lines. Anti-human bcl-2 siRNA was combined with a cationic liposome LIC-101 composed of specific lipid components. Intravenous administration of the siRNA and LIC-101 complex for two 5-day cycles resulted in significant shrinkage of liver tumor nodules. Additionally, the use of LIC-101 facilitated siRNA transport to the liver, unlike bare siRNA59.
In summary, RNAi-based therapies using chemically modified siRNAs and lipid-based delivery systems show promise in the treatment of HBV infection and liver cancer, offering potential advancements in therapeutic approaches.
Prostate Cancer
Prostate cancer ranks among the most prevalent malignancies in men in the United States, and it is the third leading cause of cancer-related death among males. Due to its high incidence, there is a critical need for the development of new therapeutic approaches. Clinical studies are currently underway to evaluate the efficacy of novel treatments such as kinase inhibitors, antisense oligonucleotides, and heat shock protein inhibitors for prostate cancer60.
In animal models, cationic liposomes have been utilized to deliver various RNAi therapies for prostate tumor treatment. One study conducted by Pal et al. employed cardiolipin liposomes as a delivery system for siRNA targeting Raf-1. The siRNA complexed with cardiolipin liposomes was administered intravenously in a mouse xenograft model of human prostate cancer. This treatment effectively inhibited tumor progression by targeting Raf-1 expression within the tumor tissue61.
Another study by Bisanz et al. demonstrated the therapeutic effect of cationic liposome’s containing dipalmitoyl ethyl phosphocholine, dioleoyl phosphoethanolamine, dipalmitoyl phosphoethanolamine, and polyethylene glycol, which were used to deliver integrin alphaV-specific siRNA. Intratumoral administration of anti-integrin alpha V siRNA and liposomes effectively suppressed tumor growth in xenograft models of human PC3 prostate cancer cells grown in the flank and tibia62.
Furthermore, cationic liposome’s comprising AtuFECT01, 1,2-diphytanoyl-sn-glycero-3-phosphoethanolamine, and DSPE-PEG were used to complex CD31 siRNA, which selectively targets endothelial cells. In rats with prostate tumors, intravenous administration of anti-CD31 siRNA lipoplexes resulted in reduced tumor growth and metastases63.
In a different approach, subcutaneous injection of LIC-101 cationic liposomal anti-bcl-2 siRNA effectively reduced tumor size in mice with subcutaneously xenografted prostate cancer (PC-3) cells.
Overall, these studies highlight the potential of cationic liposomes as delivery systems for siRNA-based therapies in prostate cancer treatment, demonstrating promising results in animal models64.
Others
Numerous alternative delivery techniques have been explored for targeted delivery to cancer tissues, including cationic liposomes, polymers, inorganic nanoparticles, and antibody-based systems. In vivo, assessments of siRNA-mediated RNA interference effects have frequently been conducted in mice using subcutaneous xenograft models. SiRNAs have been administered intratumorally or intravenously using various delivery systems to target diseases such as brain cancer, glioblastoma, nasopharyngeal cancer, gastric cancer, and prostate cancer. Efficient delivery of siRNA to the brain has posed challenges due to the limited transport of siRNA across the blood-brain barrier. However, a recent advancement utilized a receptor-specific monoclonal antibody delivery technique coupled with avidin-biotin interaction to successfully deliver siRNA to brain cancer models in vivo65.
In this approach, mono-botinylated siRNA specific to luciferase was linked to a combination of streptavidin and an anti-transferrin receptor monoclonal antibody. Intravenous administration of the biotinylated siRNA at a dose of 0.27 mg/kg in animals with intracranial luciferase-producing glial cell tumors resulted in a substantial reduction (69-81%) in luciferase gene expression within the intracranial brain tumors. Another study employed siRNA complexed with PEI specifically targeting the secreted growth factor pleiotrophin (PTN) for intracerebral therapy. This approach demonstrated reduced tumor growth and cell proliferation in an orthotopic glioma mouse model without any reported toxicity or abnormal behavior in the animals66.
Different delivery systems of siRNA for cancer treatment are shown in Table 1
Table 1: SiRNA delivery system for treatment of cancer67-92.
|
Delivery system |
Property |
Target gene |
Animal model |
Route |
|
liposomes |
SNALP |
HBV |
HRV vector Based mouse |
i.v |
|
|
Cationic liposome |
Bcl-2 |
Liver metastasis mouse model |
i.v |
|
|
Cationic liposome |
Integrin |
Prostate cancer xenograft |
i.v |
|
|
Cationic liposome |
CD31 |
Prostate cancer xenograft |
i.v |
|
|
Cationic liposome |
Bcl-2 |
Prostate cancer xenograft |
i.v |
|
|
Cationic cardiolipin liposome |
Raf-1 |
Prostate cancer xenograft |
i.v |
|
|
Cationic cardiolipin analogue-based liposomes |
c-raf |
Breast cancer xenograft |
i.v |
|
|
Neutral liposomes (DOPC) |
EphA2 |
Ovarian cancer xenograft |
i.v/i.p |
|
|
Neutral liposomes (DOPC) |
FAK |
Ovarian cancer xenograft |
i.p |
|
|
Neutral liposomes (DOPC) |
ADRB2 |
Ovarian cancer xenograft |
i.p |
|
|
Neutral liposomes (DOPC) |
IL-8 |
Ovarian cancer xenograft |
i.p |
|
|
Liposome-polycation-DNA |
EGFR |
Lung cancer xenograft |
i.v |
|
|
Cationic immunoliposome |
|
Lung metastasis |
i.v |
|
|
Immunoliposome |
Her-2 |
Breast cancer xenograft |
i.v |
|
Nanoparticles |
Caco3 nanoparticle |
VEGF |
Gastric cancer xenograft |
i.t |
|
|
Chitosan-coated nanoparticles |
RhoA |
Breast cancer xenograft |
i.v |
|
|
Folated lipid nanoparticle |
Her-2 |
Nasopharyngeal cancer xenograft |
i.t |
|
Polymers |
PEI |
Her-2 |
Ovarian cancer xenograft |
i.p |
|
|
PEI |
PTN |
Orthotopic gliobastoma |
i.c |
|
|
Poly (ester amine) |
AKT1 |
Urethane induced lung cancer |
Inhalation |
|
Others |
Atelocollagen |
HPV |
cervical cancer xenograft |
i.t |
|
|
Chemical Modification |
HBV |
HRV vector.based mouse |
i.v |
|
|
Carbon nanotube |
TERT |
Lewis lung Tumor |
i.t |
|
|
Cyclodextrin containing polycation |
EWS-FLII |
Metastatic ewings sarcoma |
i.v |
|
|
Fusion protein |
c-myc, MDM2, VEGF |
Subcutaneous B16 Melanoma Tumor |
i.t/i.v |
|
|
Electroporation |
EGFP |
Subcutaneous B16F10 expressing ECFP |
i.t |
i.v= intravenous injection, i.p = intraperitoneal injection, i.t intratumoral injection, i.c = intracerebral injection
Design and Synthesis of Target-Specific siRNAs
The selection of siRNAs specific to mRNA is a crucial step in designing effective RNA interference (RNAi) strategies for targeting specific genes. Despite the existence of numerous algorithms for siRNA selection, many of them are inefficient, lack transparency, or have commercial restrictions93.To address this, a study introduced an open-source JAVA tool that accurately predicts active siRNAs, with a Pearson correlation coefficient of 0.52 based on a dataset of 526 siRNAs. The release of version 1.0 of this tool also allows for community contributions to further improve the open-source code94.
In conventional siRNA design, the initial step involves identifying coding sequences that are devoid of translational or regulatory proteins. Regions approximately 115 bases downstream of the start codon are typically selected. Sequence motifs containing an AA (or NA) dinucleotide followed by about 20 bases with a G/C content ranging from 35% to 75% are then chosen. The choice of the dinucleotide leader determines the composition of the antisense 3′ overhangs, resulting in 20-base duplexes targeting AA (N20) having a 3′ termini of UU or dTdT.
The antisense strand of the siRNA is designed to perfectly match the target mRNA sequence. Using traditional methods, 65-75% of siRNA duplexes exhibit 50-65% gene silencing efficacy, although the effectiveness of gene knockdown may vary. In many cases, gene knockdown levels below 70% may not have significant physiological or therapeutic relevance. This necessitates the use of additional techniques to enhance the gene-silencing capacity of commonly produced siRNAs.Virus-mediated siRNA delivery for a variety of illnesses95.
Viral-mediated delivery of siRNA has emerged as a promising approach for in vitro and in vivo applications targeting various diseases. Lipid-based delivery reagents often face limitations in successfully transfecting desirable cell types like primary cells or those in the immune system. In such cases, viral delivery of RNAi cassette-containing vectors has become a favorable alternative. Viruses possess the ability to infect a wide range of mammalian cell types, including challenging-to-transfect cells, primary cells, and non-dividing cells, making them efficient carriers for gene delivery. Certain viral vectors, such as adenoviruses, can infect both dividing and non-dividing cells, and they offer advantages such as high stability of recombinant vectors, large insert capacity, and the ability to be produced at high titers96.
Viruses naturally possess the ability to efficiently deliver their genetic material into host cells, making them attractive candidates for constructing therapeutic gene delivery virus vector systems. Recent advancements have seen the utilization of viral vectors derived from RNA and DNA viruses with diverse genomic layouts and host preferences in both laboratory research and clinical practice.
Several viruses have been selected as gene delivery vehicles due to their capability to accommodate foreign genes and effectively transfer them, resulting in efficient gene expression97.
Retroviruses, adenoviruses, adeno-associated viruses, herpes viruses, and poxviruses are being extensively used in more than 60% of clinical gene therapy trials worldwide due to these reasons. For instance, the wild-type adenovirus genome has a length of approximately 35 kilobases (kb), with the potential to replace up to 30 kb of foreign DNA in the viral genome98.
Some viruses have been chosen as gene delivery vehicles because of their capacity to hold foreign genes and their ability to successfully transfer these genes, which correlates with efficient gene expression.
Retroviruses, adenoviruses, adeno-associated viruses, herpesviruses, and poxviruses are used in more than 60% of clinical gene therapy trials around the world for these reasons99.
Because recombinant adenovirus lacks key replication genes, infected cells can express the therapeutic gene without replicating the vector100.
An adenoviral vector that contains a tandem siRNA expression unit. This vector utilizes two human U6 promoters to transcribe the sense and antisense strands of the siRNA duplex separately101-103. The target of this siRNA is survivin, an antiapoptotic molecule that is typically over expressed in cancer cells but not detectable in terminally ill patients. This particular adenoviral vector is most suitable for adult tissues that have undergone differentiation. The introduction of Adv-siSurv into HeLa, U251, and MCF-7 cancer cells effectively triggered apoptosis and resulted in visible signs of infection. Both in vitro and in vivo experiments showed a significant reduction in the growth capacity of these cancer cells104. Intramuscular injections of Adv-siSurv also demonstrated substantial inhibition of tumor growth in a xenograft model, using U251 glioma cells. Another study focused on an adenoviral vector capable of expressing siRNA molecules targeting p53 or VprBP/KIAA0800, a cellular protein that interacts with the HIV auxiliary protein viral protein r (Vpr). In all cases, adenoviral infection led to a specific decrease in the target protein level, which correlated with a reduction in the corresponding mRNA level. Lentiviruses, similar to retroviruses, can infect both dividing and non-dividing cells. Lentiviral vectors derived from HIV are well-known in the field, and they can be produced at high concentrations of N10^9 virus particles per milliliter. Experimental data demonstrated that when lentiviral vectors were injected into mouse eyes, the transgenic expression persisted for at least 12 weeks without significant decline105.
Future Prospects
Traditional pharmaceutical medications have various advantages that siRNA therapies do not. Because siRNAi is an endogenous biological process, siRNA may effectively silence practically any gene. The development of highly selective and inhibitory sequences is much faster than the development of new pharmaceuticals, and synthesizing and manufacturing siRNA on a large scale is quite straightforward106.
Several oncogenes are related to excessively high expression in cancers. The use of endogenous RNAi machinery to interfere with specific oncogene expression could lead to the creation of a treatment method that is effective against a wide range of malignancies.
Because the RNAi system allows for the precise silence of pathogenic genes or gene targets involved in melanoma development, siRNA-based therapies are an appealing and potent option to treat numerous tumors.
Despite great progress, there are still challenges to overcome in the field of in vivo siRNA administration. Off-target effects and immunological activation must be avoided, which necessitates the development of solutions.
Synthetic systems based on lipids or polymers have recently been proven to produce powerful RNAi effects after systemic injection. After intravenous administration of siRNA utilizing adequate delivery mechanisms, nonhuman primates showed target-specific RNAi effects107, 108.
Interpretation of si RNA Formulation
Precision Medicine: The siRNA drug delivery system has the potential to revolutionize medicine by offering highly specific treatments tailored to individual patients. Since siRNAs can be designed to target a particular gene sequence, the therapy can be customized for various genetic mutations, providing more precise and effective treatments.
Gene Silencing: The core mechanism of the siRNA drug delivery system is gene silencing. By silencing specific genes involved in disease progression, it is possible to halt or mitigate the harmful effects of certain diseases without causing widespread collateral damage to healthy cells and tissues.
Therapeutic Potential: SiRNA-based therapies hold great promise for treating a wide range of diseases that were previously considered challenging to manage using traditional pharmaceuticals. These include genetic disorders, neurodegenerative diseases, viral infections, and certain types of cancers.
Hypothesis
Enhanced Drug Delivery: One of the critical challenges in siRNA therapy is delivering the siRNA molecules efficiently to the target cells. Hypotheses might explore novel drug delivery systems, such as lipid nanoparticles, polymer-based carriers, or viral vectors, that can protect and deliver siRNA to the desired tissues with minimal side effects.
Immune Response and Off-Target Effects: When introducing exogenous siRNA into the body, there is a possibility of triggering an immune response or causing unintended effects by targeting genes other than the intended ones. The hypothesis could focus on optimizing siRNA designs or exploring ways to minimize these off-target effects.
Stability and Half-Life: siRNAs are susceptible to degradation by cellular nucleases, which can limit their therapeutic efficacy. Researchers might hypothesize on methods to enhance siRNA stability and prolong their half-life within the body for sustained therapeutic effects.
Combination Therapy: To maximize therapeutic benefits, the hypothesis could explore the potential of combining siRNA therapy with other treatment modalities, such as chemotherapy or immunotherapy, to create synergistic effects and improve overall patient outcomes.
Target Identification: Identifying appropriate target genes for siRNA therapy is crucial. Hypotheses could center on advanced bioinformatics and screening techniques to identify and validate potential gene targets associated with specific diseases.
Conclusion
In conclusion, siRNA (small interfering RNA) drug delivery systems have emerged as a promising approach in cancer therapy. siRNAs offer a unique mechanism to silence specific genes involved in cancer progression, making them attractive targets for therapeutic intervention. However, the successful application of siRNA-based treatments relies on efficient and targeted delivery to cancer cells.
Various delivery systems have been developed to overcome the challenges associated with siRNA delivery, including nanoparticle-based carriers, lipid-based formulations, viral vectors, and conjugates. These systems aim to enhance stability, improve cellular uptake, and ensure specific delivery to cancer cells while minimizing off-target effects.
The development of siRNA drug delivery systems has shown promising results in preclinical studies, demonstrating effective gene silencing and tumour regression. Targeting specific oncogenes or pathways using siRNAs has shown potential for personalized medicine, allowing for tailored treatments based on the genetic profile of individual patients.
Despite these advancements, several hurdles remain to be addressed. Efficient systemic delivery, stability during circulation, avoidance of immune responses, and targeted delivery to tumors are ongoing challenges in siRNA delivery. Additionally, the optimization of dosing regimens and long-term safety evaluations are crucial for the successful translation of siRNA-based therapies from the laboratory to the clinic.
In conclusion, siRNA drug delivery systems have demonstrated significant potential in cancer therapy by selectively inhibiting the expression of disease-associated genes. Continued research and development efforts are needed to refine delivery strategies, optimize therapeutic efficacy, and address safety concerns. With further advancements, siRNA-based treatments have the potential to revolutionize cancer therapy, offering more precise and personalized approaches to combat this devastating disease.
Acknowledgment
The authors wish to express their gratitude to Nitte (Deemed to be University) and NGSM Institute of Pharmaceutical Sciences for generously providing the essential resources required for conducting this research.
Conflict of Interest
There is no conflict of interest.
Funding Sources
There are no funding sources.
References
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494-498. doi:10.1038/35078107
- de Fougerolles A, Manoharan M, Meyers R, Vornlocher HP. RNA interference in vivo: toward synthetic small inhibitory RNA-based therapeutics. Methods Enzymol. 2005;392:278-296. doi:10.1016/S0076-6879(04)92016-2.
- Caplen NJ. Gene therapy progress and prospects. Downregulating gene expression: the impact of RNA interference. Gene Ther. 2004;11(16):1241-1248. doi: 10.1038 /sj. gt.3302324.
- Napoli C, Lemieux C, Jorgensen R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell. 1990;2(4):279-289. doi:10.1105/tpc.2.4.279
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806-811. doi:10.1038/35888
- Doi N, Zenno S, Ueda R, Ohki-Hamazaki H, Ui-Tei K, Saigo K. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr Biol. 2003;13(1):41-46. doi:10.1016/s0960-9822 (02)01394-5
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404 (6775):293-296. doi:10.1038/35005107
- Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437-1441. doi:10.1126/science.1102513.
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185-197. doi:10.1016/j.molcel.2004.07.007
- Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23(24):4727-4737. doi:10.1038/sj.emboj.7600488
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434(7033):666-670. doi:10.1038/nature03514
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309(5740):1519-1524. doi:10.1126/science.1111444
- Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem Sci. 2005;30(2):106-114. doi:10.1016/j.tibs.2004.12.007
- Grimm D. Small silencing RNAs: state-of-the-art. Adv Drug Deliv Rev. 2009;61(9):672-703. doi:10.1016/j.addr.2009.05.002
- Takahashi Y, Yamaoka K, Nishikawa M, Takakura Y. Quantitative and temporal analysis of gene silencing in tumor cells induced by small interfering RNA or short hairpin RNA expressed from plasmid vectors. J Pharm Sci. 2009;98(1):74-80. doi:10.1002/jps.21398
- Sah DW. Therapeutic potential of RNA interference for neurological disorders. Life Sci. 2006;79(19):1773-1780. doi:10.1016/j.lfs.2006.06.011
- Stahel RA, Zangemeister-Wittke U. Antisense oligonucleotides for cancer therapy-an overview. Lung Cancer. 2003;41 Suppl 1:S81-S88. doi:10.1016/s0169-5002(03)00147-8
- Emilsson GM, Nakamura S, Roth A, Breaker RR. Ribozyme speed limits. RNA. 2003;9(8):907-918. doi:10.1261/rna.5680603
- Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418(6894):222-228. doi:10.1038/418222a
- Bertrand JR, Pottier M, Vekris A, Opolon P, Maksimenko A, Malvy C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem Biophys Res Commun. 2002;296(4):1000-1004. doi:10.1016/s0006-291x(02)02013-2
- Sall A, Liu Z, Zhang HM, et al. MicroRNAs-based therapeutic strategy for virally induced diseases. Curr Drug Discov Technol. 2008;5(1):49-58. doi:10.2174/157016308783769478
- Novina CD, Murray MF, Dykxhoorn DM, et al. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8(7):681-686. doi:10.1038/nm725
- Song E, Lee SK, Wang J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9(3):347-351. doi:10.1038/nm828
- de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6(6):443-453. doi:10.1038/nrd2310
- Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2(12):711-719. doi:10.1038/nchembio839
- Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8(3):173-184. doi:10.1038/nrg2006
- Marques JT, Williams BR. Activation of the mammalian immune system by siRNAs. Nat Biotechnol. 2005;23(11):1399-1405. doi:10.1038/nbt1161
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23(4):457-462. doi:10.1038/nbt1081
- Hornung V, Guenthner-Biller M, Bourquin C, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11(3):263-270. doi:10.1038/nm1191
- Juliano R, Alam MR, Dixit V, Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 2008;36(12):4158-4171. doi:10.1093/nar/gkn342
- Aigner A. Nonviral in vivo delivery of therapeutic small interfering RNAs. Curr Opin Mol Ther. 2007;9(4):345-352.
- Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov. 2004;3(4):318-329. doi:10.1038/nrd1345
- Persengiev SP, Zhu X, Green MR. Nonspecific, concentrationdependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). RNA., 2004; 10:12-18.
- Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5(9):834-839. doi:10.1038/ncb1038
- Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34(3):263-264. doi:10.1038/ng1173
- Leung RK, Whittaker PA. RNA interference: from gene silencing to gene-specific therapeutics. Pharmacol Ther. 2005;107(2):222-239. doi:10.1016/j.pharmthera .2005. 03.004
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789-799. doi:10.1038/nm1087
- Moulder S, Hortobagyi GN. Advances in the treatment of breast cancer. Clin Pharmacol Ther. 2008;83(1):26-36. doi:10.1038/sj.clpt.6100449
- Lavigne C, Thierry AR. Specific subcellular localization of siRNAs delivered by lipoplex in MCF-7 breast cancer cells. Biochimie. 2007;89(10):1245-1251. doi:10. 10 16/j.biochi.2007.05.002
- Meryet-Figuières M, Resina S, Lavigne C, Barlovatz-Meimon G, Lebleu B, Thierry AR. Inhibition of PAI-1 expression in breast cancer carcinoma cells by siRNA at nanomolar range. Biochimie. 2007;89(10):1228-1233. doi:10.1016/j. biochi.2007 .03. 017
- Hogrefe RI, Lebedev AV, Zon G, et al. Chemically modified short interfering hybrids (siHYBRIDS): nanoimmunoliposome delivery in vitro and in vivo for RNAi of HER-2. Nucleosides Nucleotides Nucleic Acids. 2006;25(8):889-907. doi:10.1080/ 1525777 0600793885
- Sutton D, Kim S, Shuai X, et al. Efficient suppression of secretory clusterin levels by polymer-siRNA nanocomplexes enhances ionizing radiation lethality in human MCF-7 breast cancer cells in vitro. Int J Nanomedicine. 2006;1(2):155-162. doi:10. 2147 / nano.2006.1.2.155
- Tan WB, Jiang S, Zhang Y. Quantum-dot based nanoparticles for targeted silencing of HER2/neu gene via RNA interference. Biomaterials. 2007; 28 (8):1565-1571. doi:10.1016/j.biomaterials.2006.11.018
- Wang Y, Gao S, Ye WH, Yoon HS, Yang YY. Co-delivery of drugs and DNA from cationic core-shell nanoparticles self-assembled from a biodegradable copolymer. Nat Mater. 2006;5(10):791-796. doi:10.1038/nmat1737
- Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12(5):461-466. doi:10.1038/sj.gt.3302425
- Landen CN Jr, Chavez-Reyes A, Bucana C, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65(15):6910-6918. doi:10.1158/0008-5472.CAN-05-0530
- Landen CN, Merritt WM, Mangala LS, et al. Intraperitoneal delivery of liposomal siRNA for therapy of advanced ovarian cancer. Cancer Biol Ther. 2006;5(12):1708-1713. doi:10.4161/cbt.5.12.3468
- Halder J, Kamat AA, Landen CN Jr, et al. Focal adhesion kinase targeting using in vivo short interfering RNA delivery in neutral liposomes for ovarian carcinoma therapy [published correction appears in Clin Cancer Res. 2019 May 15;25(10):3194]. Clin Cancer Res. 2006;12(16):4916-4924. doi:10.1158/1078-0432.CCR-06-0021
- Thaker PH, Han LY, Kamat AA, et al. Chronic stress promotes tumor growth and angiogenesis in a mouse model of ovarian carcinoma [published correction appears in Nat Med. 2021 Dec;27(12):2246]. Nat Med. 2006;12(8):939-944. doi:10. 1038 /nm 14 47
- Merritt WM, Lin YG, Spannuth WA, et al. Effect of interleukin-8 gene silencing with liposome-encapsulated small interfering RNA on ovarian cancer cell growth. J Natl Cancer Inst. 2008;100(5):359-372. doi:10.1093/jnci/djn024
- Zhang C, Tang N, Liu X, Liang W, Xu W, Torchilin VP. siRNA-containing liposomes modified with polyarginine effectively silence the targeted gene. J Control Release. 2006;112(2):229-239. doi:10.1016/j.jconrel.2006.01.022
- Li SD, Huang L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Mol Pharm. 2006;3(5):579-588. doi:10.1021/mp060039w
- Li SD, Chen YC, Hackett MJ, Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol Ther. 2008;16(1):163-169. doi:10.1038/sj.mt.6300323
- Li SD, Chono S, Huang L. Efficient gene silencing in metastatic tumor by siRNA formulated in surface-modified nanoparticles. J Control Release. 2008;126(1):77-84. doi:10.1016/j.jconrel.2007.11.002
- Pirollo KF, Zon G, Rait A, et al. Tumor-targeting nanoimmunoliposome complex for short interfering RNA delivery. Hum Gene Ther. 2006;17(1):117-124. doi:10.1089/hum.2006.17.117
- Arbuthnot P, Longshaw V, Naidoo T, Weinberg MS. Opportunities for treating chronic hepatitis B and C virus infection using RNA interference. J Viral Hepat. 2007;14(7):447-459. doi:10.1111/j.1365-2893.2006.00818.x
- Morrissey DV, Blanchard K, Shaw L, et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005;41(6):1349-1356. doi:10.1002/hep.20702
- Morrissey DV, Lockridge JA, Shaw L, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23(8):1002-1007. doi:10.1038/nbt1122
- Yano J, Hirabayashi K, Nakagawa S, et al. Antitumor activity of small interfering RNA/cationic liposome complex in mouse models of cancer. Clin Cancer Res. 2004;10(22):7721-7726. doi:10.1158/1078-0432.CCR-04-1049
- Rubin MA. Targeted therapy of cancer: new roles for pathologists–prostate cancer. Mod Pathol. 2008;21 Suppl 2:S44-S55. doi:10.1038/modpathol.2008.11
- Pal A, Ahmad A, Khan S, et al. Systemic delivery of RafsiRNA using cationic cardiolipin liposomes silences Raf-1 expression and inhibits tumor growth in xenograft model of human prostate cancer. Int J Oncol. 2005;26(4):1087-1091.
- Bisanz K, Yu J, Edlund M, et al. Targeting ECM-integrin interaction with liposome-encapsulated small interfering RNAs inhibits the growth of human prostate cancer in a bone xenograft imaging model. Mol Ther. 2005;12(4):634-643. doi:10.1016/j.ymthe.2005.05.012
- Santel A, Aleku M, Keil O, et al. RNA interference in the mouse vascular endothelium by systemic administration of siRNA-lipoplexes for cancer therapy. Gene Ther. 2006;13(18):1360-1370. doi:10.1038/sj.gt.3302778
- Yano J, Hirabayashi K, Nakagawa S, et al. Antitumor activity of small interfering RNA/cationic liposome complex in mouse models of cancer. Clin Cancer Res. 2004;10(22):7721-7726. doi:10.1158/1078-0432.CCR-04-1049
- Xia CF, Zhang Y, Zhang Y, Boado RJ, Pardridge WM. Intravenous siRNA of brain cancer with receptor targeting and avidin-biotin technology. Pharm Res. 2007;24(12):2309-2316. doi:10.1007/s11095-007-9460-8
- Grzelinski M, Urban-Klein B, Martens T, et al. RNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenografts. Hum Gene Ther. 2006;17(7):751-766. doi:10.1089/hum.2006.17.751
- Morrissey DV, Lockridge JA, Shaw L, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23(8):1002-1007. doi:10.1038/nbt1122
- Yano J, Hirabayashi K, Nakagawa S, et al. Antitumor activity of small interfering RNA/cationic liposome complex in mouse models of cancer. Clin Cancer Res. 2004;10(22):7721-7726. doi:10.1158/1078-0432.CCR-04-1049
- Bisanz K, Yu J, Edlund M, et al. Targeting ECM-integrin interaction with liposome-encapsulated small interfering RNAs inhibits the growth of human prostate cancer in a bone xenograft imaging model. Mol Ther. 2005;12(4):634-643. doi:10.1016/ j.ymthe.2005.05.012
- Santel A, Aleku M, Keil O, et al. RNA interference in the mouse vascular endothelium by systemic administration of siRNA-lipoplexes for cancer therapy. Gene Ther. 2006;13(18):1360-1370. doi:10.1038/sj.gt.3302778
- Chien PY, Wang J, Carbonaro D, et al. Novel cationic cardiolipin analogue-based liposome for efficient DNA and small interfering RNA delivery in vitro and in vivo. Cancer Gene Ther. 2005;12(3):321-328. doi:10.1038/sj.cgt.7700793
- Pal A, Ahmad A, Khan S, et al. Systemic delivery of RafsiRNA using cationic cardiolipin liposomes silences Raf-1 expression and inhibits tumor growth in xenograft model of human prostate cancer. Int J Oncol. 2005;26(4):1087-1091.
- Landen CN Jr, Chavez-Reyes A, Bucana C, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65(15):6910-6918. doi:10.1158/0008-5472.CAN-05-0530
- Halder J, Kamat AA, Landen CN Jr, et al. Focal adhesion kinase targeting using in vivo short interfering RNA delivery in neutral liposomes for ovarian carcinoma therapy [published correction appears in Clin Cancer Res. 2019 May 15;25(10):3194]. Clin Cancer Res. 2006;12(16):4916-4924. doi:10.1158/1078-0432.CCR-06-0021
- Landen CN, Merritt WM, Mangala LS, et al. Intraperitoneal delivery of liposomal siRNA for therapy of advanced ovarian cancer. Cancer Biol Ther. 2006;5(12):1708-1713. doi:10.4161/cbt.5.12.3468.
- Thaker PH, Han LY, Kamat AA, et al. Chronic stress promotes tumor growth and angiogenesis in a mouse model of ovarian carcinoma [published correction appears in Nat Med. 2021 Dec;27(12):2246]. Nat Med. 2006;12(8):939-944. doi:10.1038/nm1447
- Merritt WM, Lin YG, Spannuth WA, et al. Effect of interleukin-8 gene silencing with liposome-encapsulated small interfering RNA on ovarian cancer cell growth. J Natl Cancer Inst. 2008;100(5):359-372. doi:10.1093/jnci/djn024
- Li SD, Chen YC, Hackett MJ, Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol Ther. 2008;16(1):163-169. doi:10.1038/sj.mt.6300323
- Pirollo KF, Zon G, Rait A, et al. Tumor-targeting nanoimmunoliposome complex for short interfering RNA delivery. Hum Gene Ther. 2006;17(1):117-124. doi:10.1089/hum.2006.17.117
- Hogrefe RI, Lebedev AV, Zon G, et al. Chemically modified short interfering hybrids (siHYBRIDS): nanoimmunoliposome delivery in vitro and in vivo for RNAi of HER-2. Nucleosides Nucleotides Nucleic Acids. 2006;25(8):889-907. doi:10.1080/ 152 57770600793885
- He XW, Liu T, Chen YX, et al. Calcium carbonate nanoparticle delivering vascular endothelial growth factor-C siRNA effectively inhibits lymphangiogenesis and growth of gastric cancer in vivo. Cancer Gene Ther. 2008;15(3):193-202. doi:10.1038/sj.cgt.7701122
- Pillé JY, Li H, Blot E, et al. Intravenous delivery of anti-RhoA small interfering RNA loaded in nanoparticles of chitosan in mice: safety and efficacy in xenografted aggressive breast cancer. Hum Gene Ther. 2006;17(10):1019-1026. doi:10.1089/hum.2006.17.1019
- Yoshizawa T, Hattori Y, Hakoshima M, Koga K, Maitani Y. Folate-linked lipid-based nanoparticles for synthetic siRNA delivery in KB tumor xenografts. Eur J Pharm Biopharm. 2008;70(3):718-725. doi:10.1016/j.ejpb.2008.06.026
- Novina CD, Murray MF, Dykxhoorn DM, et al. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8(7):681-686. doi:10.1038/nm725
- Grzelinski M, Urban-Klein B, Martens T, et al. RNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenografts. Hum Gene Ther. 2006;17(7):751-766. doi:10.1089/hum.2006.17.751
- Xu CX, Jere D, Jin H, et al. Poly(ester amine)-mediated, aerosol-delivered Akt1 small interfering RNA suppresses lung tumorigenesis. Am J Respir Crit Care Med. 2008;178(1):60-73. doi:10.1164/rccm.200707-1022OC
- Fujii T, Saito M, Iwasaki E, et al. Intratumor injection of small interfering RNA-targeting human papillomavirus 18 E6 and E7 successfully inhibits the growth of cervical cancer. Int J Oncol. 2006;29(3):541-548.
- Morrissey DV, Blanchard K, Shaw L, et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005;41(6):1349-1356. doi:10.1002/hep.20702
- Zhang Z, Yang X, Zhang Y, et al. Delivery of telomerase reverse transcriptase small interfering RNA in complex with positively charged single-walled carbon nanotubes suppresses tumor growth. Clin Cancer Res. 2006;12(16):4933-4939. doi: 10. 1158 /10 78-0432.CCR-05-2831
- Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65(19):8984-8992. doi:10.1158/0008-5472.CAN-05-0565
- Song E, Zhu P, Lee SK, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23(6):709-717. doi:10. 1038 / nbt1101
- Golzio M, Mazzolini L, Ledoux A, et al. In vivo gene silencing in solid tumors by targeted electrically mediated siRNA delivery. Gene Ther. 2007;14(9):752-759. doi:10.1038/sj.gt.3302920
- Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26(2):199-213. doi:10.1016/S1046-2023(02)00023-3
- Holen T. Efficient prediction of siRNAs with siRNArules 1.0: an open-source JAVA approach to siRNA algorithms. RNA. 2006;12(9):1620-1625. doi:10.1261/rna.81006
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363-366. doi:10.1038/35053110
- Ghosh SS, Gopinath P, Ramesh A. Adenoviral vectors: a promising tool for gene therapy. Appl Biochem Biotechnol. 2006;133(1):9-29. doi:10.1385/abab:133:1:9
- Walther W, Stein U. Viral vectors for gene transfer: a review of their use in the treatment of human diseases. Drugs. 2000;60(2):249-271. doi:10.2165/00003495-20 0060020-00002
- Smith AE. Viral vectors in gene therapy. Annu Rev Microbiol. 1995;49:807-838. doi:10.1146/annurev.mi.49.100195.004111
- Yeh P, Perricaudet M. Advances in adenoviral vectors: from genetic engineering to their biology. FASEB J. 1997;11(8):615-623. doi:10.1096/fasebj.11.8.9240963
- Verma IM, Somia N. Gene therapy — promises, problems and prospects. Nature. 1997;389(6648):239-242. doi:10.1038/38410
- Uchida H, Tanaka T, Sasaki K, et al. Adenovirus-mediated transfer of siRNA against survivin induced apoptosis and attenuated tumor cell growth in vitro and in vivo. Mol Ther. 2004;10(1):162-171. doi:10.1016/j.ymthe.2004.05.006
- Zhao LJ, Jian H, Zhu H. Specific gene inhibition by adenovirus-mediated expression of small interfering RNA. Gene. 2003;316:137-141. doi:10.1016/s0378-1119(03)00750-9
- Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle [published correction appears in EMBO J 1992 Nov;11(11):4249]. EMBO J. 1992;11(8):3053-3058. doi:10.1002/j.1460-2075.1992.tb05376.x
- Naldini L, Blömer U, Gallay P, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263-267. doi:10.1126/science.272.5259.263
- Miyoshi H, Takahashi M, Gage FH, Verma IM. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc Natl Acad Sci U S A. 1997;94(19):10319-10323. doi:10.1073/pnas.94.19.10319
- Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2(12):711-719. doi:10.1038/nchembio839
- Zimmermann TS, Lee AC, Akinc A, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441(7089):111-114. doi:10.1038/nature04688
- Heidel JD, Yu Z, Liu JY, et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc Natl Acad Sci U S A. 2007; 104(14):5715-5721. doi:10.1073/pnas.0701458104







