Manuscript accepted on :11-02-2023
Published online on: 24-10-2023
Plagiarism Check: Yes
Reviewed by: Dr. Rand R. Hafidh
Second Review by: Dr. Amit Gupta
Final Approval by: Dr. Ayush Dogra
Jannatul Firdous Siddique1 , Sajitha Lulu S2
, Sajitha Lulu S2 and Mohanasrinivasan Vaithilingam3*
and Mohanasrinivasan Vaithilingam3*
School of Bio Sciences and Technology, Vellore Institute of Technology, Vellore, Tamil Nadu, India.
Corresponding Author E-mail: v.mohan@vit.ac.in
DOI : https://dx.doi.org/10.13005/bpj/2798
Abstract
As there is no cure for cancer, hence it becomes one of the top causes of mortality worldwide. Bacterial peptide obtained from probiotic species possesses anti-cancer and anti-bacterial activity and shows specificity towards the prominent target of cancer cells. To find such peptides with anti-cancer activity, a thorough literature review was conducted. Major 9 peptides have been selected as a promising candidate, and their interactions with 9840 possible target genes were retrieved using the STITCH database and the STRING 11.0b database, respectively. In the Cytoscape software version 3.8.2 network analysis, 2465 nodes and 10871 edges were found. Cytoscape version 3.8.1 was used to build, visualize and analyze target-disease and target-pathway networks for anti-cancer peptides to advance current understanding. There were 3127 nodes identified as disease-associated. In total, 271 nodes and 318 edges were discovered, along with 27 distinct genes linked to 243 distinct pathways. The hub genes were found by centrality analysis to comprehend how peptides and highly significant genes interact. Thus, six key hub genes for the peptides—GRB2, APP, COPS5, CDK1, RPS3, and XRCC6—were identified based on topological criteria such as MCC, degree, closeness, betweenness, and radiality. Major genes were discovered to be linked to Neoplasm Metastasis, Neoplasms, Non-Small Cell Lung Carcinoma, and Primary Malignant Neoplasm among the 57 genes. The expression of the disease-related genes and their regulation of the molecular mechanisms causing the disease, together with their transcription factors, have been identified.
Keywords
Anti-cancer; Bacteriocin; GO analysis; Hub Genes; Peptide; Transcription factors
Download this article as:| Copy the following to cite this article: Siddique J. F, Lulu S. S, Vaithilingam M. Investigating the Hub Genes of Lactic Acid Bacterial Peptides and their association with Anti-Cancer Role. Biomed Pharmacol J 2023;16(4). |
| Copy the following to cite this URL: Siddique J. F, Lulu S. S, Vaithilingam M. Investigating the Hub Genes of Lactic Acid Bacterial Peptides and their association with Anti-Cancer Role. Biomed Pharmacol J 2023;16(4). Available from: https://bit.ly/3SdoYO1 |
Introduction
In general, Lactic acid bacteria (LAB) are used as starters for the production of fermented food traditionally. Recently, it showed promising food preservatives and therapeutic effects1.
Since LAB is Generally Recognized as Safe (GRAS), its simple metabolism and production of multi-valuable biochemicals such as food ingredients and high-value pharma precursors emerged as a promising cell factory. Cancer is still one of the deadliest diseases and the conventional treatment strategies ((e.g., chemotherapy, surgery, radiotherapy, etc.) used to combat, has numerous shortcomings due to associated side effect and significant challenges to patient health. A bacterial protein produced by a group of prokaryotic microorganisms is of great potential for cancer therapy2.
Bacteria like Clostridium spp. and Bifidobacterium longum strains have been found to endure and progress in hypoxic conditions surrounding the tumor and hence inhibiting its proliferation and causing cell death3. The lactic acid bacterium Lactococcus lactis produces nisin, a lantibiotic of class I bacteriocins possessing broad-spectrum antibacterial activity against Gram-positive bacteria4. It is injected intramuscularly into the babies to prevent the deadly disease and showed 80% effectiveness primarily in the laboratory. Moreover, certain bacteria showed tumoricidal effects such as Clostridia, Shigella, Lactococcus, Bifidobacteria, Listeria, Salmonella, Vibrio and Escherichia coli. They have been reported to have an extraordinary ability to treat cancer5. The immune system’s ability to fight is worth noting irrespective of any infection or disease, still, some bacteria were found to enhance its ability positively. The cross-talk between bacteria and the immune system can improve the response against malignant cells, such as CD8 + T-cells was found to be the most influential component to inhibit tumor cell growth6, 7.Actinomycin D, bleomycin, doxorubicin, mitomycin C) and diphtheria toxin are some of the reported Anticancer antibiotics8. Bacteriocins discovered almost 100 years ago are proteinaceous in nature 9 and are produced by various species of bacteria and archaea. Some of the well-known bacteriocins produced by Gram-negative are microcin E492, colicins, (A, D, E1, E2, E3) and Gram-positive bacteria-produced bacteriocin-like nisin. Bacterial proteins and peptides are strong anti-proliferative agents in cancer treatment. Likely, in the nuclei of Lactobacillus sp., an enormous amount of unmethylated dinucleotide repeat sequences are present which bind to a specific receptor on the surface of human cells and activate the innate immune response. Findings also suggested the combined activity of the bacteriocin enterocin A with colicin E1 showed anti-cancer activity against AGS human cancer cells which further lead to caspase activation and cell death10. Bifidobacterium Derived Bacteriocins Bifidin B1 and Bifidin B2, showed potential antioxidant and Antitumor Activity against MCF-7 and Skov-3 with IC50 values of 28.9± 8.76 µg/ml, and 29.87± 9.13 µg/ml, respectively11.
Cancer is a genetic disease; hence it alters the way how a cell functions, divides and grows. Recently therapeutic approach has been explored using bacteriocin; a cationic antimicrobial and anti-cancer peptide. The current research deals with a similar type of peptide of interest possessing antibacterial and anti-cancer activity. The gene-gene interaction network has organized the complex data and provided a comprehensive structure hence the peptide-gene network reveals a visual understanding of the interconnection while the target disease and target-pathway network gave a deeper understanding of the connection and the major genes involved. This study was conducted to identify highly influential genes and the transcription factors associated with cancer, and it may help to discover drug-able targets in the future.
Material and methods
Construction of Dataset
In this era, identification of the molecular targets of active small-molecule or peptides is crucial and presently an unachieved challenge. Hence, network pharmacology and metabolomics play an important role to bring light to the possible targets of the peptides, which is solely possible via the system biology approach. The peptides responsible for the defense mechanism of the bacterial cell were identified from an extensive literature survey using the keyword “bacteriocin”. The proteins interacting with these kinds of peptides were determined and the UniProt ID of target proteins was obtained from UniProtKB (https://www.uniprot.org/) database by limiting the species to “Homo sapiens”. The potential target genes of the proteins were collected from the STITCH database.
 |
Chart 1: Flowchart the Construction of Dataset |
Target gene- Disease network construction
DisGeNET is a search tool that contains integrated human diseases, genes and experimental research12 combines information on human gene-disease associations (GDAs) and variant-disease associations (VDAs). The keyword “Cancer” was used to search throughout the databases to screen the disease concerning the target genes. The target-disease network was constructed, visualized and analyzed. The associated disease of the target genes was obtained from the DisGeNET database. Target proteins and their associated disease interaction were constructed, visualized and analyzed using Cytoscape 3.5.1.
KEGG pathway enrichment analysis
Analysis of the pathways associated with the target genes was done using the Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway database. Screening of the genes associated with non-cancer and cancer-related pathways was determined. The KEGG pathway enrichment analysis was performed to discover the pathways associated with the potential targets of the peptides with a p-value < 0.05. Target proteins and their associated pathway interaction were constructed and visualized using Cytoscape 3.5.1. Venn Diagram gives a pictorial representation of the two sets of data in an overlapping circle sharing a common pathway. It illustrates the pathways related to cancer and non-cancer diseases using Venny 2.0 software.
Peptide-target gene network construction
In this era of big data, inspecting the interaction of the peptides with their respective targets is unfeasible and time-consuming. System biology provides a platform, to establish the network via network analysis tools. Moreover, in an unbiased fashion, the network analysis assists us to identify the unknown potent targets of the active component. The target genes of the peptides were obtained using the STITCH database. The peptide-target gene network was constructed, visualized and analyzed using Cytoscape 3.4.013.
Identification of IP and network construction
The potent peptide possessing anti-bacterial and anti-cancer roles and its respective target genes were screened. The interacting partners of the target genes were obtained using the “Search tool for retrieval of interacting genes” (STRING v10.5) database. A confidence score cut‐off >0.9 reported through experiments and databases were only considered for the gene-protein network construction. The target-gene-IPs network was constructed, visualized and analyzed using Cytoscape 3.4.013.
Identification of highly influential genes
The closely related or highly interconnected hub genes were analyzed through cytohubba plugin20 of Cytoscape 3.5.1. CytoHubba plugin14 of Cytoscape was utilized for assessment based on centrality to identify the modules and the top-ranked nodes. Centrality analysis helps to identify the key genes involved in the biological process. Genes with a degree ≥9 were defined as hub genes in the present study. The highly influential genes were identified from the gene-protein interaction network based on topological properties like MCC, Degree, Closeness, Betweenness, Radiality and EPC. Moreover, the measure depends upon the network based on Degree. Topological properties of the networks refer to how the individual genes are interrelated or arranged. The cellular state was maintained indirectly by Hub genes, for it interacts with a large number of partners and offers control over it. The gene list enrichment analysis15 was performed to retrieve molecular function and biological processes using ToppGene Suite (http://toppgene.cchmc.org)16. The p-value cut-off was set as 0.05, gene limits 1 ≤ n ≤ 2000 and correction as FDR to extract the functional gene ontology for the set of hub genes.
Relationship between expression levels of hub genes and survival
The hub genes were found to regulate varied biological processes which in terms, are associated with the disease17. The biological processes were searched using GenCLiP 2.0:a web server18, tabulated and represented in graphical form.
Identification of the transcription factors concerning the gene regulation
The activation and repression activity of the associated TFs respective to the hub genes are studied using the transcriptional regulatory relationships unraveled by sentence-based text-mining (TRRUST), v2: an expanded reference database of human and mouse transcriptional regulatory interactions19. (TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Research 26 Oct 2017)
Results
Extraction and dataset construction of peptides from Lactic acid bacteria
 |
Table 1: The antimicrobial peptides and their role as an antibacterial &/or anticancer agent |
 |
Figure 1: Peptide – Target Network – Interaction between peptides and target are constructed and presented as a circular image Total of 9 peptides interacted with 9840 target proteins. |
The interaction between the peptides and the potent target was stabilized, visualized and analyzed by the construction of the network using Cytoscape software 3.8.2. [Figure 1]. The peptide with their respective target genes was attached as Supplementary File 1.
The Gene- disease associations have revealed, that several diseases share common genes. Specific gene expressions are involved in controlling varied biological functions, regulations and the advancement of diseases. The network was constructed and the disease gene association was extracted from the DisGeNET database. Further, the study revealed 3126 unique nodes were associated with 17112 diseases as shown [Figure 2].
 |
Figure 3: Genes-pathway network construction – Using KEGG software, the Circular network has been constructed for Target – Associated Pathway network. |
A total number of 3127 genes was found to be associated with various kinds of diseases. Among them, ANG, APP, AR, ATIC, BRE, CKB, CS, DES, GAN, GLA, IDE, IDS, IK, LAT, MET, MICA, MOS, MYOC, NSF, PIP, RAN, REL, RELA, RET, SET, SMS, SON and TRO, the 28 unique genes were associated with 243 unique pathways. For the pathways associated with the genes, a total of 271 nodes and 318 edges were observed in the constructed network. The network was constructed, visualized and analyzed using Cytoscape software 3.8.2 as shown in [Figure 3].
 |
Figure 4: Target–associated related and Target associated- other pathways have been illustrated using the Venn diagram |
Among the 242 pathways, 141 pathways were found to be associated with cancer and 100 pathways are related to other diseases. Only one pathway is commonly found in both cancer and non-cancer-related pathways as shown [Figure 4].
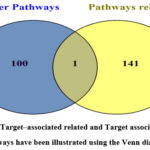 |
Figure 5: Using string database IPs were collected and the network was constructed using Cytoscape software v.3.8.2. |
The network shows 2465 nodes and 10871 edges concerning the target genes of the active peptide as shown in [Figure 5] in which darker color and larger size represented bigger degree values. The Target genes and their respective IP are listed and attached as Supplementary File 2 and File 3.
Identification of Hub genes based on centrality analysis
Based on the core network, these genes were identified as the crucial genes in the gene-gene core network of 10871 nodes. The sub-network was derived from the gene-gene network and described using topological features degree, MCC, Closeness, Betweenness, Radiality and EPC using the cytohubba plugin of Cytoscape software 3.8.2. The study showed 57 genes with varied scores for each of the features [Table 2a]. Among them, two genes CDK1 and RPS3 were found to be common in all the six hub categories and genes like GRB2, APP, XRCC6 and COPS5 are found common for four hub categories [Table 2b].
Table 2a: Cytohubba analysis of Target- IP network (Topological features). Hub genes are selected based on centrality, among 10871 nodes (gene-gene).
|
MCC |
Degree |
Closeness |
Betweenness |
Radiality |
EPC |
||||||
|
Name |
Score |
Name |
Score |
Name |
Score |
Name |
Score |
Name |
Score |
Name |
Score |
|
RPL4 |
1.55E |
RPS3 |
64 |
RPS3 |
702.2 |
XRCC6 |
1003 |
RPS3 |
23.17 |
CDK1 |
803.943 |
|
RPL27 |
1.55E |
HSPA8 |
56 |
CDK1 |
678.8 |
COPS5 |
8170 |
APP |
23.14 |
TP53 |
803.18 |
|
RPL23A |
1.55E |
RPL8 |
54 |
APP |
675.0 |
APP |
7635 |
COPS5 |
23.11 |
CTN |
803.171 |
|
RPL27A |
1.55E |
CDK1 |
53 |
COPS5 |
673.2 |
RPS3 |
7359 |
GSK3B |
23.10 |
RPS3 |
802.677 |
|
RPS8 |
1.55E |
RPL4 |
51 |
HSPA5 |
669.9 |
YBX1 |
6223 |
HSPA5 |
23.10 |
CCT5 |
802.677 |
|
RPS3A |
1.55E |
EEF |
50 |
CSN |
669.4 |
VAV1 |
6186 |
CDK1 |
23.10 |
HSP |
802.677 |
|
RPS16 |
1.55E |
UQC |
50 |
ACTB |
667.3 |
MRPS6 |
6033 |
XRCC6 |
23.10 |
CCT8 |
802.677 |
|
RPS28 |
1.55E |
RPLP0 |
50 |
DHX9 |
661.7 |
PEBP1 |
5893 |
ACTB |
23.07 |
RPL4 |
802.677 |
|
RPS25 |
1.55E |
RPL7 |
50 |
GSK3B |
659.9 |
CDK1 |
5753 |
CSN |
23.07 |
RPS3A |
802.677 |
|
RPS15A |
1.55E |
RPSA |
49 |
CTN |
653.9 |
CDK7 |
5689 |
HNR |
23.05 |
RPL32 |
802.677 |
|
RPL12 |
1.55E |
RPL11 |
49 |
XRCC6 |
653.6 |
NUP |
5294 |
DHX9 |
23.0 |
GNB2L1 |
802.677 |
|
RPL18A |
1.55E |
RPL |
48 |
YBX1 |
651.2 |
NEK9 |
506 |
TBK1 |
23.02 |
UBC |
802.677 |
|
RPL21 |
1.55E |
RPS13 |
48 |
HNR |
649.5 |
MBP |
485 |
GRB2 |
23.00 |
RPL18 |
802.677 |
|
RPL30 |
1.55E |
RPS20 |
47 |
GRB2 |
643.1 |
MRPS9 |
4607 |
ALB |
22.98 |
RPL27 |
|
|
RPL7 |
1.55E |
GNB |
47 |
ALB |
637.4 |
GRB2 |
4546 |
CTN |
22.97 |
HNRN |
802.677 |
Table 2b: The genes possessing the common factors of at least 4 categories among, MCC, Degree, Closeness, Betweenness, Radiality and EPC were selected
|
Genes |
3 factors |
4 factors |
|
GRB2 |
Yes |
– |
|
APP |
Yes |
– |
|
XRCC6 |
Yes |
– |
|
COPS5 |
Yes |
– |
|
CDK1 |
Yes |
|
|
RPS3 |
Yes |
Gene ontology
Table 3: Gene Ontology analysis: Molecular functions and biological processes were identified for the highly influential genes
|
Genes |
Molecular function |
p Value |
Biological processes |
pValue |
|
GRB2 |
Ephrin receptor binding |
3.93E-05 |
regulation of MAPK cascade |
6.00E-07 |
|
– |
cellular response to DNA damage stimulus |
1.01E-06 |
||
|
– |
MAPK cascade |
1.86E-06 |
||
|
APP |
Ephrin receptor binding |
3.93E-05 |
regulation of MAPK cascade |
6.00E-07 |
|
– |
MAPK cascade |
1.86E-06 |
||
|
– |
positive regulation of protein kinase activity |
1.20E-05 |
||
|
XRCC6 |
Cyclin binding |
1.22E-04 |
cellular response to DNA damage stimulus |
1.01E-06 |
|
Carbon-oxygen lyase activity |
2.61E-04 |
DNA repair |
9.88E-06 |
|
|
damaged DNA binding |
1.74E-04 |
positive regulation of protein kinase activity |
1.20E-05 |
|
|
COPS5 |
– |
regulation of MAPK cascade |
6.00E-07 |
|
|
– |
cellular response to DNA damage stimulus |
1.01E-06 |
||
|
– |
MAPK cascade |
1.86E-06 |
||
|
– |
DNA repair |
9.88E-06 |
||
|
CDK1 |
Hsp70 protein binding |
9.69E-05 |
regulation of MAPK cascade |
6.00E-07 |
|
cyclin binding |
1.22E-04 |
cellular response to DNA damage stimulus |
1.01E-06 |
|
|
– |
MAPK cascade |
1.86E-06 |
||
|
– |
DNA repair |
9.88E-06 |
||
|
– |
positive regulation of protein kinase activity |
1.20E-05 |
||
|
RPS3 |
Hsp70 protein binding |
9.69E-05 |
||
|
damaged DNA binding |
1.74E-04 |
regulation of MAPK cascade |
6.00E-07 |
|
|
carbon-oxygen lyase activity |
2.61E-04 |
cellular response to DNA damage stimulus |
1.01E-06 |
|
|
– |
MAPK cascade |
1.86E-06 |
||
|
– |
DNA repair |
9.88E-06 |
||
|
– |
positive regulation of protein kinase activity |
1.20E-05 |
The functional annotation-based method was adopted for the gene enrichment analysis using ToppGene as represented in Table 3. The molecular functions and biological processes were retrieved and the association of the gene and the respective cellular role was identified. This study focused on the peptides and their immediate gene targets and their interacting partners. It is the first report to identify the hub genes (GRB2, APP, XRCC6, COPS5, CDK1 and RPS3) associated with the anti-cancer peptides and their gene ontology analysis was performed. The highly influential genes were prioritized based on the molecular functions and biological processes which may play a vital role in anti-cancer activity.
Expression levels of hub genes and their survival
The six hub genes showed a significant correlation with the disease and showed to be involved with cell growth and cell death. As per the analysis, APP and CDK1 were positively correlated with overall survival (OS), and RPS3 was negatively correlated with OS. Moreover, the expression of GRB2, COPS5, RPS3 and XRCC6 were found to vary in different biological processes. The genes and their respective biological processes were tabulated in Table 4.
Table 4: The gene expression was determined for the highly influential genes based on the biological processes they regulate.
|
S. No. |
Biological Processes |
APP |
COPS5 |
RPS3 |
XRCC6 |
GRB2 |
CDK1 |
|
1. |
DNA repair |
√ | √ | √ | √ |
x |
√ |
|
2. |
DNA binding |
√ | √ | √ | √ | √ | √ |
|
3. |
Immune response |
√ | √ | √ | √ | √ | √ |
|
4. |
Cell death |
√ | √ | √ | √ | √ | √ |
|
5. |
Cell growth |
√ | √ | √ | √ | √ | √ |
|
6. |
S phase |
√ | √ |
x |
√ | √ | √ |
|
7. |
Squamous cell carcinoma |
√ | √ |
x |
√ | √ | √ |
|
8. |
Protein complex |
√ | √ |
x |
√ | √ | √ |
|
9. |
Kinase inhibitor |
√ | √ |
x |
√ | √ | √ |
|
10. |
Kinase activity |
√ | √ |
x |
√ | √ | √ |
|
11. |
Protein-protein interaction |
√ | √ |
x |
√ | √ | √ |
|
12. |
Breast cancer cell |
√ | √ |
x |
√ | √ | √ |
|
13. |
RNA interference |
√ | √ |
x |
√ | √ | √ |
|
14. |
DNA helicase |
√ |
x |
x |
√ |
x |
√ |
|
15. |
Histone deacetylase inhibitor |
√ |
x |
x |
√ |
x |
√ |
|
16. |
Poly ADP ribose |
√ |
x |
x |
√ |
x |
√ |
|
17. |
Ubiquitin proteasome pathway |
√ | √ |
x |
x |
x |
√ |
|
18. |
Cyclin-dependent kinase |
√ | √ |
x |
x |
√ | √ |
|
19. |
Translation initiation factor |
√ |
x |
√ |
x |
x |
√ |
|
20. |
RNA binding |
√ |
x |
√ |
x |
√ | √ |
 |
Figure 6: Using GenCLiP 2.0: a web server, the graph was plotted and has been presented. |
Moreover, the genes COPS5, XRCC6 and GRB2 are involved in processes like DNA binding, Immune response. Cell death, Cell growth, S phase, Squamous cell carcinoma, Protein complex, Kinase inhibitor, Kinase activity, Protein-protein interaction, Breast cancer cell, and RNA interference. In addition, COPS5 and XRCC6 are involved in the DNA repair mechanism solely. Alone, COPS5 regulates the process like the Ubiquitin proteasome pathway and Cyclin-dependent kinase. The gene XRCC6 is solely involved with DNA helicase, Histone deacetylase inhibitor, and Poly ADP ribose. Moreover, the gene GRB2 was found to be expressed in the processes like Cyclin-dependent kinase and RNA binding [Figure 6].
 |
Figure 7: Identification of the transcription factors concerning the gene regulation along with expression levels of hub genes and their survival are schematically presented |
The peptides influence the role of the hub genes in the mammalian cell. Moreover, the associated TFs activate or repress the specific hub genes by specific interaction which ultimately leads to varied biological processes. The role of a transcription factor in association with the activation and repression of the hub genes is delineated attached as Supplementary File 4 and illustrated [Figure 7].
Discussion
The peptides precisely hold anti-cancer and anti-microbial activities that are produced by probiotic bacteria including Lactobacillus, Bifidobacterium, and Pediococcus, etc. were retrieved using extensive literature search with the search keyword used “Bacteriocin, Antimicrobial peptides, Antibacterial, Anti-cancerous agent, Cytotoxic, Apoptosis”. Further, using the Uniprot kB function Uniprot (http://www.uniprot.org/)database, species were restricted to humans, target genes of the peptides were retrieved. The redundant duplicates were removed and the target genes of the respective peptides were obtained and converted into gene symbols. The major peptides with a critical role in cancer identified are, Laterosporulin10, M2163, Nisin Q, Weissellicin M, BaCf3, Bovine lactoferrin whereas peptides like BacF1, BacF2 and Small Bacteriocin (7E)-3-hydroxy-N-(2-oxooxolan-3-yl) tetradec-7-enamide (C18H31NO4) exhibit antibacterial role. A total of nine major peptides are associated with 9840 target proteins found to be critically involved in an antibacterial and or anti-cancer activity and were used to construct the dataset as listed in Table 1.
Recently, experimental data revealed the cytotoxic role of bacteriocin against, selective cancer cell lines such as breast cancer, prostate cancer, colon cancer, bone cancer etc. It has also been reported, the underlying mechanism mainly involved are blockage of angiogenesis, apoptosis, depolarization of cell membrane, inhibition of cell proliferation and inhibition of tumorigenesis in vivo28. Hence, in this research well known bacteriocin with a potential role was collected using an extensive literature survey. Further concerning the peptides, the target genes collected from the STITCH database restricted the search items to homo sapiens.
Through the DisGeNET database, the associated diseases of the target genes were retrieved. Cancers like Acute lymphocytic leukemia, Childhood Acute Lymphoblastic Leukemia, Non-Small Cell Lung Carcinoma, Breast Carcinoma, Childhood Leukaemia, Secondary Neoplasm, CNS metastases, Malignant mesothelioma, Precursor Cell Lymphoblastic Leukemia Lymphoma and many such more cancer are found to be associated with the genes. Whereas non-cancerous diseases like Gout, Arthritis, Obesity, Schizophrenia, Major Depressive Disorder, Human immunodeficiency virus (HIV) II infection category B1, Malaria, Myocardial Infarction, Coronary heart disease, Premature aging syndrome, ankylosing spondylitis, Seizures, Pancreatitis and Alzheimer’s Disease like other diseases are found to share the same genetic origin. The number of nodes was found to be 862 and edges 1669 [Figure 1]. In [Figure 2] the blue bubbles indicate genes are interconnected and are associated with multiple diseases. The Target Gene-Disease network was constructed, visualized and analysedusing Cytoscape software 3.8.2. The Gene- disease associations have revealed, that several diseases share common genes. Specific gene expressions are involved in controlling varied biological functions, regulations and the advancement of diseases. The network was constructed and the disease gene association was extracted from the DisGeNET database. Further, the study revealed 3126 unique nodes were associated with 17112 diseases.
Major pathways directly associated with cancer are identified as, Basal cell carcinoma, Breast cancer, Endometrial cancer, Pancreatic cancer, and Viral carcinogenesis. Next, the pathways associated with non-cancer diseases like Acarbose and validamycin biosynthesis, Biosynthesis of 12-, 14- and 16-membered macrolides, Maturity onset diabetes of the young, Renin-angiotensin system, etc. The common pathway associated with both cancer and non-cancer disease was found to be 2-Oxocarboxylic acid metabolism. Graphical representation of genes and their associative diseases is presented in Venn diagrams for cancer related and non- cancer related pathways [Figure 4].
Genes identified were CDK1 (cyclin-dependent kinase 1), RPS3 (Ribosomal Protein S3), GRB2 (Growth factor receptor-bound protein 2), APP (Amyloid Beta Precursor Protein), XRCC6 (X-ray repair cross-complementing 6) and COPS5 (COP9 Signalosome Subunit 5). GRB2, APP and XRCC6 are associated with Ataxia, Bladder Neoplasm, Breast Carcinoma, Carcinoma, Diabetes Mellitus, Non-Insulin-Dependent, Malignant neoplasm of the urinary bladder, Mammary Neoplasms, Neoplasm Metastasis, Neoplasms, Neuroblastoma, Non-Small Cell Lung Carcinoma, Primary malignant neoplasm, Renal Cell Carcinoma and Tumor Progression. In case of genes COPS5, CDK1 and RPS3 they are associated with Childhood Osteosarcoma, Hyperactive behavior, Liver carcinoma, Malignant Neoplasms, Neoplasm Metastasis, Neoplasms, Non-Small Cell Lung Carcinoma, Osteosarcoma, Osteosarcoma of bone, Primary malignant neoplasm and Tumor Cell Invasion. Alternative to this work, an experiment has been performed28, that has investigated the role of four miRNAs and their target genes in the cross-resistance of RR sub-line using Real-time PCR analysis.
Based on the topological features, centrality was measured. A set of 6 genes were identified in this study. The findings suggested that the 6 hub genes were found to interact mainly with 4894 genes. A similar report has been published29 , and their findings stated that they have screened the top five hub nodes, such as PHLPP1, UBC, ACACB, TGFB1, and ACTB. In that case, they performed GSEA which resulted in the identification of the five hub genes. Their study revealed that the hug genes were essentially enriched in the ribosomal pathway. Whereas, as reported in 201830, they performed the assay experimentally and checked for the expression of AKT1 and TGFB2, upregulation of CASP3 HCT116 cells, using quantitative real-time PCR analysis. A similar report31 , also investigated the levels of Bax, Bcl2, Trp53, and Casp3 mRNA expression by RT-PCR analysis to check the therapy efficacy of the venom. In this current research, we have identified the hub genes like GRB2, APP, XRCC6, COPS5, CDK1 and RPS3. As reported in 202132 , 7 key hub genes like RAB3IP, CENPN, NEDD1, CENPU, SPC25, POLR2D and CDC25A were identified and among them, CDC25A was found to be negatively correlated with CRC the survival of a patient.
Gene GRB2 Growth factor receptor-bound protein 2 also known as Grb2 is an adaptor protein involved in signal transduction/cell communication and hence also play an important role in the process like tumor growth, invasiveness and metastasis. Reports have revealed, it is called a high priority target protein for anti-cancer drug development purposes and its structural knowledge may aid in the development of anti-cancer drugs. The GRB2 protein consists of Src homology domains and its respective structure-function relation also contribute to drug development. As the drug can easily bind with the protein domain after it invade the cell and counteract GRB2 signalling33.
APP gene provides instructions for making a protein called amyloid precursor protein. This protein is found in many tissues and organs named an Amyloid beta precursor protein (APP).
APP or amyloid precursor-like protein 2 (APLP2) contributes to the migration, invasion, or proliferation of tumor cells. Reports also exposed its role in the causation of Alzheimer’s disease, breast cancer progression and are linked with neurological disorders34.It has also been delineated, its increased overexpression contributes to rapid cell division and invasion in advanced breast cancer. Hence targeting this specific protein may aid to control the progression and act as an effective therapy35.X-ray repair cross-complementing 6 (XRCC6) exhibit the role of an Activator, DNA-binding, Helicase, Hydrolase, Lyase and as a multifunctional enzyme. In the case of cancer, it is involved in tumor growth and development. Its overexpression is directly related to the size and progression of the disease36.Hence it may serve as a potential target for therapy. Moreover, it has also been disclosed, in the repair of double–strand breaks (DSBs), this DNA non-homologous end-joining repair gene XRCC6/Ku70 contributes widely. It has also been reported there is an association of genetic polymorphisms in XRCC4, XRCC5, XRCC6 and XRCC7 with cervical cancer and rural population is more prone37. Hence targeting this gene may also contribute to the therapeutic approach.
COPS5 (COP9 Signalosome Subunit 5) is a Protein Coding gene. It is a highly conserved protein complex that functions as an important regulator in multiple signaling pathways. COPS5 is also reported to be one of the frequently altered genes in ERα+ breast cancer samples from TCGA. The study suggested, its deregulation contributes to the activation of oncogenes, the deactivation of tumor Suppressor genes and hence leads to oncogenesis38. In addition, it also confirmed its role as a biomarker for prognosis, diagnosis and as a therapeutic tool for the development of drugs. Cyclin dependent kinase 1(CDK1) or cell division cycle protein 2 homolog is a highly conserved protein that functions to regulate progression of the cell cycle progression, transcription and other biological processes. In human cells, any kind of mutation, deregulation or increase in the expression of this gene leads to hyperactivity of CDK. These changes further lead to progression, cell proliferation and metastasis. Though it acts as a very attractive target for drug development, its highly conserved structure and ATP-binding pocket make it less accessible. Other approaches like targeting the protein-protein interface by the peptides and targeting allosteric sites by a smaller group of compounds had also been studied39. The activation of Cdk1 also contributes to HIV-1-induced apoptosis and upstream of the p53-dependent mitochondrial permeabilization step, however, unscheduled activation led to neurological disorders due to neuronal apoptosis40.Well-established research has confirmed the link between overexpression of CDK with the progression of cancer, and CDK1 gene silencing induces apoptosis and cell cycle arrest which led to inhibition of retinal angiogenesis41. Cytoplasmic ribosomal protein S3 (rpS3) is a Protein Coding gene of 243 amino acid components of the 40S ribosomal small subunit. It greatly contributes to apoptosis, DNA repair and the translation process. It is generally secreted by all types of cancer cells and confirmed in case of melanoma growth, over-expressed in colon adenocarcinoma, colorectal cancers, hepatocarcinogenesis etc. Hence this gene will serve as a potent therapeutic target in drug development research [Table 2].
The gene ontology study revealed the molecular functions of the genes GRB2 and APP were found to be linked with ephrin receptor binding. Genes XRCC6 and RPS3 were responsible for damaged DNA binding and carbon-oxygen lyase activity. Molecular functions like cyclin binding were found to be associated with Gene XRCC6 and CDK1. Moreover, CDK1 and RPS3 were associated with the Hsp70 protein binding role. In addition, COPS5 was not found to be associated with any molecular functions. Based on the biological processes, genes responsible for the cellular functions were majorly MAPK cascade, regulation of MAPK cascade, positive regulation of protein kinase activity, cellular response to DNA damage stimulus and DNA repair. All of this information was retrieved from ToppGene and the data collected are tabulated as shown in [Table 3]. A similar kind of study has also been executed42 , where they have performed the gene ontology (GO), pathway enrichment analysis using g: Profiler, Protein-protein interaction network (PPIN) constructed by Search Tool for the Retrieval of Interacting Genes (STRING) and analyzed using Cytoscape successfully.
In conclusion, our findings suggest that APP and CDK1 may play an essential role in the development of Cancer and may serve as a potential biomarker for future diagnosis and treatment. The image depicted the expression of genes in percentages associated with diseases like cancer. In addition, it has been shown in graphical illustration [Figure 6] 43,44.
APP assists with high levels of transcription in a specific type of cells. CCCTC-Binding Factor (CTCF) is a zinc finger protein, a multifunctional transcription factor that binds to the APBbeta domain of APP to initiate transcription from the APP promoter. In this regard, CTCF is cap of both activating and repressing the gene APP45. ETS2, ETS Proto-Oncogene 2, Transcription Factor which chromosome 21 transcription factor ETS2 transactivates the beta-APP gene. APP is up-regulated in the prostate, colon, pancreatic tumor, and oral squamous cell carcinoma, hence controlling the growth in the case of pancreatic and colon cancer46. NFIL3, Nuclear Factor, Interleukin 3 Regulated is an immune system modulator transcription factor that regulates the expression of the human gene APP as well as APPB gene in zebrafish47. The increase in activity of the SP1, Sp1 Transcription Factor, leads to the upregulation of the expression of the APP gene48. Moreover, transcription factors like AP1, SP1 and TFIID interact with the -203 to +55 bp of the human APP promoter sequence. Whereas STAT1, Signal Transducer and Activator of Transcription 1 involve in suppressing the tumor growth49. TBP, TATA-Box Binding Protein regulates the activity of most of the genes throughout the system, and are specifically found to be involved in DNA repair mechanism while associated factor TBPL1 is involved in processes like cell cycle arrest and apoptosis50. In the case of squamous cell carcinoma, it has been especially noted of the transcription role of the TFAP2A, Transcription Factor AP-2 Alpha in regulating the gene Amyloid precursor protein and chromatin accessibility study of lung cancer cells demonstrates that TFAP2A increases angiogenesis in the development of anlotinib resistance51.
In the case of gene CDK1, ARID3A, AT-Rich Interaction Domain 3A isolated as protein binds to E2F1, which stimulates transcription via transcription factor like E2F that regulates the progression of the cell cycle. Though the role of ARID3A has not yet been fully revealed. E2F1, E2F Transcription Factor 1 along with DP-1 controls the various cellular promoters and the respective gene products involved in the cell cycle. Moreover, the level of E2F1 is correlated with transcriptional targets like (TYMS, DHFR, PCNA, RRM1, CCNE1, CDC2, and MYBL2), known to target cyclin-dependent kinase 1 (Cdk1), which is involved in G1 checkpoint control52.
After the cell suffers DNA damage, E2F activity increases during the S/G2 phase where it fails to exit the cell cycle and goes through mitosis which further leads to genetic instability, encourage the spread of cancer and reduces therapeutic sensitivity53. According to a recent study, E2F3 promotes cellular proliferation during the G1/S transition. Several genes, including CDK1 and Survivin expression, are modulated by E2F354. In the case of cell proliferation, and cell differentiation ETS2, ETS Proto-Oncogene 2, Transcription Factor, regulates gene expression.
ETS2 is the substrate for CDK10/cyclin M and its phosphorylation leads to the inhibition of ETS2 transactivation55. IRF1, Interferon Regulatory Factor 1 regulates epithelial-mesenchymal plasticity in breast cancer cells. Plasticity is the ability to invade surrounding and distant tissues and to withstand chemotherapy56. IRF1, Interferon regulatory factor 1, a suppressor protein might bind to two HOTAIR promoter sites (53-64 and 136-148 bp) which contain related binding motifs. Moreover, the critical effect of HOTAIR activity has been described in the progression of the cell cycle and proliferation by the regulation of a variety of molecules. It has also been proposed, HOTAIR coordinates in a molecular pathway which leads to promoting proliferation through the activation of CDK1/CDK2/STAT3 signalling cascade57.
PTTG1, Pituitary tumor-transforming gene 1 or Securin is a PTTG1 Regulator of Sister Chromatid Separation. The main prognostic genes of the hepatitis B inflammation and cancer transformation are PTTG1, MAD2L1, CDK1, and NEK2 as reported58. RB1, RB Transcriptional Corepressor 1 is an RB. Tumor suppressor, which controls cell cycle progression, a key regulator of many biological processes, besides shows the downstream target for CDK4/6 inhibitors. In case the RB1 Is inactivated, it surely led to cancer, as it possesses a dual role to execute or resist apoptosis in the cell59. SMAD7, SMAD Family Member 7; SMAD7, a TGF-beta signalling inhibitor, showed a role inthe case of pancreatic adenocarcinoma. Though it has an inhibitory effect on TGF-β1 signalling, in a TGF-independent manner smad7 can also influence the expression and function of several molecules involved in the regulation of both fibrotic and cancerous processes60. To govern the cell cycle and the activity of tumor suppressor proteins, transcription factor Dp-1 (TFDP1) plays a major role in cell cycle regulation, binds to E2F, another essential transcription protein. It acts as a master candidate in the case of lung adenocarcinoma61. A malignant tumor eventually arises as a result of aberrant cell differentiation and cell cycle due to the abnormal regulation of CDK1 whereas TFDP1, Transcription Factor Dp-1 helps to activate the target CDK1.
TP53, Tumor Protein P53; represses the activity of CDK1 which in turn halts cell progression and proliferation. By preventing abnormally elevated CDK1 activity, the p53/p73-p21CIP1 tumor suppressor axis, linked to W-CIN in human cancer, protects against chromosome mis segregation and aneuploidy62.
This XRCC6 gene acts as a transcription factor and targets both CD40, CD40 Molecule and ERBB2, Erb-B2 Receptor Tyrosine Kinase 2. The mode of regulation concerning cancer is still unknown. The COPS5 gene itself acts as a transcription factor and targets both CDKN1B- Cyclin Dependent Kinase Inhibitor 1B and VEGFA-Vascular Endothelial Growth Factor A. Although the link between VEGFA and COPS5 still needs to be further investigated, COPS5 may control VEGFA to accelerate the advancement of breast cancer63. As a target gene of miRNA let-7d, COPS5 was also discovered to be linked to the metastasis of breast cancer64.COPS5, interacts functionally with several tumor-related genes, to inhibit the kinase activity in the case of CDKN1B and VEGFA. In addition, inhibition or control of the COPS5 can stop the cell growth and proliferation of the tumor cells. Transcription factors like GATA1, GATA Binding Protein 1 and STAT3, Signal Transducer and Activator of Transcription 3 regulate the gene COPS5 in case of breast carcinoma cells. Triple-negative breast cancer TNBC is generally defined by frequent visceral metastasis with no reliable clinical targets. In such a scenario GATA1 deSUMOylation promotes COPS5 transcription, which is further regulated by SENP1. This process encourages the invasion and metastasis of triple-negative breast cancer65. There is a relation between the Signal transducer and activator of transcription-3 (Stat3) expression with Jab1/Csn5 transcription. STAT3 knockdown in tumours consistently decreases Jab1/Csn5 expression, making NPC cells prone to cisplatin-induced apoptosis both in vitro and in vivo. In a mechanistic way, Jab1/Csn5 is transcriptionally regulated by STAT3. As per the R2 online database, colon cancer, breast cancer and glioblastoma rate of survival periods were found to be reduced when Stat3 or Jab1 mRNA expression levels were high66. The transcription factor for GRB23 and RPS3 is not reported.
Conclusion
The major gene which was upregulated and downregulated can be further explored as a targetable gene for therapeutic purposes. This study will help to understand the interaction of the genes associated with the peptides and their respective role in anti-cancer activity at the molecular level. This approach also may aid to give an in-depth insight into the effect of peptides for Homo sapiens as a potent anticancer peptide generally obtained from Lactic acid bacteria.
Acknowledgment
We are much indebted to the management of the Vellore Institute of Technology for the constant encouragement, help and support for extending necessary facilities.
Conflicts of Interest
The authors declare that there is no conflict of interest.
Funding Sources
No funding was obtained for this study
References
- Liu J, Chan SH, Chen J, Solem C, Jensen PR. Systems biology–A guide for understanding and developing improved strains of lactic acid bacteria. Front. Microbiol; 10:876 (2019).
CrossRef - Rommasi F. Bacterial-Based Methods for Cancer Treatment: What We Know and Where We Are. Oncol. Ther;10(1):23–54 (2021).
CrossRef - Janku F, Zhang HH, Pezeshki A, Goel S, Murthy R, Wang-Gillam A, Shepard DR, Helgason T, Masters T, Hong DS, Piha-Paul SA. Intratumoral Injection of Clostridium novyi-NT Spores in Patients with Treatment-refractory Advanced Solid TumorsPhase I Study of Intratumoral Clostridium novyi-NT. Clin. Cancer Res; 27(1):96-106 (2021).
CrossRef - Małaczewska J, Kaczorek-Łukowska E. Nisin—A lantibiotic with immunomodulatory properties: A review. Peptides;137:170479 (2021).
CrossRef - Deng W, Lira V, Hudson TE, Lemmens EE, Hanson WG, Flores R, Barajas G, Katibah GE, Desbien AL, Lauer P, Leong ML. Recombinant Listeria promotes tumor rejection by CD8+ T cell-dependent remodeling of the tumor microenvironment. Proc. Natl. Acad. Sci;115(32):8179-84 (2018).
CrossRef - Raskov H, Orhan A, Christensen JP, Gögenur I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br. J. Cancer; 124(2):359-67 (2021).
CrossRef - Stern C, Kasnitz N, Kocijancic D, Trittel S, Riese P, Guzman CA, Leschner S, Weiss S. Induction of CD 4+ and CD 8+ anti‐tumor effector T cell responses by bacteria mediated tumor therapy. Int. J. Cancer; 137(8):2019-28 (2015).
CrossRef - Karpiński TM, Adamczak A. Anticancer activity of bacterial proteins and peptides. Pharmaceutics;10(2):54 (2018).
CrossRef - Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LM. Functions and emerging applications of bacteriocins. Curr Opin Biotechnol; 49:23-8 (2018).
CrossRef - Fathizadeh H, Saffari M, Esmaeili D, Moniri R, Mahabadi JA. Anticancer effect of enterocin A-colicin E1 fusion peptide on the gastric cancer cell. Probiotics Antimicrob; 13(5):1443-51 (2021).
CrossRef - Alwendawi SA. In vitro Assessment the Potential Antioxidant and Antitumor Activities of Bifidobacterium Derived Bacteriocins. Int. J. Drug Deliv. Technol; 9(02):207-16 (2019).
CrossRef - Piñero J, Saüch J, Sanz F, Furlong LI. The DisGeNET cytoscape app: Exploring and visualizing disease genomics data. Comput. Struct. Biotechnol. J ; 19:2960-7(2021).
CrossRef - Ge H, Zhang B, Li T, Yang Q, Tang Y, Liu J, Zhang T. In vivo and in silico studies on the mechanisms of egg white peptides in relieving acute colitis symptoms. Food Funct; 12(24):12774-87(2021).
CrossRef - Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res; 13(11):2498-504 (2003).
CrossRef - Pavlidis P, Qin J, Arango V, Mann JJ, Sibille E. Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem. Res; 29(6):1213-22 (2004).
CrossRef - Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res; 37(suppl_2):W305-11 (2009).
CrossRef - Li CY, Cai JH, Tsai JJ, Wang CC. Identification of hub genes associated with development of head and neck squamous cell carcinoma by integrated bioinformatics analysis. Front. Oncol;10:681 (2020).
CrossRef - Wang J-H, Zhao L-F, Lin P, Su X-R, Chen S-J, Huang L-Q, et al. GenCLiP 2.0: a web server for functional clustering of genes and construction of molecular networks based on free terms. Bioinform; 30(17):2534–6 (2014).
CrossRef - Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res; 46(D1):D380-6 (2018).
CrossRef - Mohapatra AR, Jeevaratnam K. Inhibiting bacterial colonization on catheters: Antibacterial and antibiofilm activities of bacteriocins from Lactobacillus plantarum SJ33. J. Glob. Antimicrob. Resist; 19:85-92 (2019).
CrossRef - Baindara P, Gautam A, Raghava GP, Korpole S. Anticancer properties of a defensin like class IId bacteriocin Laterosporulin10. Sci. Rep; 7(1):1-9 (2017).
CrossRef - Tsai TL, Li AC, Chen YC, Liao YS, Lin TH. Antimicrobial peptide m2163 or m2386 identified from Lactobacillus casei ATCC 334 can trigger apoptosis in the human colorectal cancer cell line SW480. Tumor Biol; 36(5):3775-89 (2015).
CrossRef - Shin JM, Gwak JW, Kamarajan P, Fenno JC, Rickard AH, Kapila YL. Biomedical applications of nisin. J. Appl. Microbiol; 120(6):1449-65 (2016).
CrossRef - Kwak SH, Cho YM, Noh GM, Om AS. Cancer preventive potential of kimchi lactic acid bacteria (Weissella cibaria, Lactobacillus plantarum). J. Cancer Prev;19(4):253–8 (2014).
CrossRef - Bindiya, E. S., & Bhat, S. G. (2017). Bacteriocins BaCf3 and BpSl14 with anticancer and antibiofilm potential from probiotic Bacillus amyloliquefaciens BTSS3 and Bacillus pumilus SDG14 isolated from gut of marine fishes: Enhanced production, Purification and Characterization (Doctoral dissertation, Cochin University of Science and Technology).
- Zhang Y, Lima CF, Rodrigues LR. In vitro evaluation of bovine lactoferrin potential as an anticancer agent. Int. Dairy J; 40:6-15 (2015).
CrossRef - John J, Saranathan R, Adigopula LN, Thamodharan V, Singh SP, Lakshmi TP, CharanTej MA, Rao RS, Krishna R, Rao HS, Prashanth K. The quorum sensing molecule N-acyl homoserine lactone produced by Acinetobacter baumannii displays antibacterial and anticancer properties. Biofouling; 32(9):1029-47 (2016).
CrossRef - Afshar S, Pashaki AS, Najafi R, Nikzad S, Amini R, Shabab N, Khiabanchian O, Tanzadehpanah H, Saidijam M. Cross-resistance of acquired radioresistant colorectal cancer cell line to gefitinib and regorafenib. Iran. J. Med; 45(1):50 (2020).
- Lu X, Gao C, Liu C, Zhuang J, Su P, Li H, Wang X, Sun C. Identification of the key pathways and genes involved in HER2-positive breast cancer with brain metastasis. Pathol. Res. Pract; 215(8):152475 (2019).
CrossRef - Tanzadehpanah H, Mahaki H, Moradi M, Afshar S, Rajabi O, Najafi R, Amini R, Saidijam M. Human serum albumin binding and synergistic effects of gefitinib in combination with regorafenib on colorectal cancer cell lines. Colorectal Cancer; 7(2):CRC03(2018).
CrossRef - Moradi M, Najafi R, Amini R, Solgi R, Tanzadehpanah H, Esfahani AM, Saidijam M. Remarkable apoptotic pathway of hemiscorpius lepturus scorpion venom on CT26 cell line. Cell Biol. Toxicol; 35(4):373-85 (2019).
CrossRef - Manoochehri H, Jalali A, Tanzadehpanah H, Taherkhani A, Saidijam M. Identification of key gene targets for sensitizing colorectal cancer to chemoradiation: an integrative network analysis on multiple transcriptomics data. J. Gastrointest. Cancer; 25:1-20 (2021).
CrossRef - Kazemein Jasemi NS, Reza Ahmadian M. Allosteric regulation of GRB2 modulates RAS activation. Small GTPases;13(1):282-6 (2022).
CrossRef - Lee HN, Jeong MS, Jang SB. Molecular Characteristics of Amyloid Precursor Protein (APP) and Its Effects in Cancer. Int. J. Mol. Sci; 22(9):4999 (2021).
CrossRef - Zayas‐Santiago A, Martínez‐Montemayor MM, Colón‐Vázquez J, Ortiz‐Soto G, Cirino‐Simonet JG, Inyushin M. Accumulation of amyloid beta (Aβ) and amyloid precursor protein (APP) in tumors formed by a mouse xenograft model of inflammatory breast cancer. FEBS Open bio; 12(1):95-105 (2022).
CrossRef - Luo S, Wang W, Feng J, Li R. TEX10 Promotes the Tumorigenesis and Radiotherapy Resistance of Urinary Bladder Carcinoma by Stabilizing XRCC6. J. Immunol. Res; 1-11 (2021).
CrossRef - Datkhile KD, Patil MN, Durgawale PP, Korabu KS, Joshi SA, Kakade SV. Association of genetic polymorphisms in XRCC4, XRCC5, XRCC6 and XRCC7 in cervical cancer susceptibility from rural population: a hospital-based case-control study. Int. j. res. med. sci; 6(7):2453 (2018).
CrossRef - Liu G, Claret FX, Zhou F, Pan Y. Jab1/COPS5 as a novel biomarker for diagnosis, prognosis, therapy prediction and therapeutic tools for human cancer. Front. Pharmacol; 9:135 (2018).
CrossRef - Lu H, Zhou Q, He J, Jiang Z, Peng C, Tong R, Shi J. Recent advances in the development of protein–protein interactions modulators: mechanisms and clinical trials. Signal Transduct Target Ther; 5(1):1-23 (2020).
CrossRef - Galarreta A, Valledor P, Ubieto‐Capella P, Lafarga V, Zarzuela E, Muñoz J, Malumbres M, Lecona E, Fernandez‐Capetillo O. USP7 limits CDK1 activity throughout the cell cycle. EMBO Rep; 40(11):e99692 (2021).
CrossRef - Gao X, Zhang Y, Zhang R, Zhao Z, Zhang H, Wu J, Shen W, Zhong M. Cyclin‑dependent kinase 1 disruption inhibits angiogenesis by inducing cell cycle arrest and apoptosis. Exp. Ther. Med; 18(4):3062-70 (2019).
CrossRef - Manoochehri H, Asadi S, Tanzadehpanah H, Sheykhhasan M, Ghorbani M. CDC25A is strongly associated with colorectal cancer stem cells and poor clinical outcome of patients. Gene Rep; 25:101415 (2021).
CrossRef - Jin Y, Yang Y. Identification and analysis of genes associated with head and neck squamous cell carcinoma by integrated bioinformatics methods. Mol. Genet. Genomic Med; 7(8):e857 (2019).
CrossRef - Sunada S, Saito H, Zhang D, Xu Z, Miki Y. CDK1 inhibitor controls G2/M phase transition and reverses DNA damage sensitivity. Biochem. Biophys. Res. Commun; 550:56-61 (2021).
CrossRef - Liu F, Wu D, Wang X. Roles of CTCF in conformation and functions of chromosome. Seminars in Cell & Dev. Biol; 90:168–73 (2019).
CrossRef - Lee HN, Jeong MS, Jang SB. Molecular Characteristics of Amyloid Precursor Protein (APP) and Its Effects in Cancer. Int. J. Mol. Sci; 22(9):4999 (2021).
CrossRef - Jin W, Qazi TJ, Quan Z, Li N, Qing H. Dysregulation of transcription factors: A key culprit behind neurodegenerative disorders. Neuroscientist; 25(6):548-65 (2019).
CrossRef - Gao Q, Dai Z, Zhang S, Fang Y, Yung KK, Lo PK, Lai KW. Interaction of Sp1 and APP promoter elucidates a mechanism for Pb2+ caused neurodegeneration. Arch. Biochem. Biophys; 681:108265 (2020).
CrossRef - Loh CY, Arya A, Naema AF, Wong WF, Sethi G, Looi CY. Signal transducer and activator of transcription (STATs) proteins in cancer and inflammation: functions and therapeutic implication. Front. Oncol; 9:48 (2019).
CrossRef - Mishal R, Luna-Arias JP. Role of the TATA-box binding protein (TBP) and associated family members in transcription regulation. Gene; 833:146581 (2022).
CrossRef - Zhang LL, Lu J, Liu RQ, Hu MJ, Zhao YM, Tan S, Wang SY, Zhang B, Nie W, Dong Y, Zhong H. Chromatin accessibility analysis reveals that TFAP2A promotes angiogenesis in acquired resistance to anlotinib in lung cancer cells. Acta Pharmacol. Sin; 41(10):1357-65 (2020).
CrossRef - Qiao L, Zheng J, Tian Y, Zhang Q, Wang X, Chen JJ, Zhang W. Regulator of chromatin condensation 1 abrogates the G1 cell cycle checkpoint via Cdk1 in human papillomavirus E7-expressing epithelium and cervical cancer cells. Cell Death Dis; 9(6):1-2 (2018).
CrossRef - Segeren HA, van Rijnberk LM, Moreno E, Riemers FM, van Liere EA, Yuan R, Wubbolts R, de Bruin A, Westendorp B. Excessive E2F transcription in single cancer cells precludes transient cell-cycle exit after DNA damage. Cell reports; 33(9):108449 (2020).
CrossRef - Liu Z, Mo H, Liu R, Niu Y, Chen T, Xu Q, Tu K, Yang N. Matrix stiffness modulates hepatic stellate cell activation into tumor-promoting myofibroblasts via E2F3-dependent signaling and regulates malignant progression. Cell Death Dis; 12(12):1-9 (2021).
CrossRef - You Y, Bai F, Ye Z, Zhang N, Yao L, Tang Y, Li X. Downregulated CDK10 expression in gastric cancer: Association with tumor progression and poor prognosis. Mol. Med. Rep;17(5):6812-8 (2018).
CrossRef - Meyer-Schaller N, Tiede S, Ivanek R, Diepenbruck M, Christofori G. A dual role of Irf1 in maintaining epithelial identity but also enabling EMT and metastasis formation of breast cancer cells. Oncogene; 39(24):4728-40 (2020).
CrossRef - Mozdarani H, Ezzatizadeh V, Rahbar Parvaneh R. The emerging role of the long non-coding RNA HOTAIR in breast cancer development and treatment. J Transl Med;18(1):1-5 (2020).
CrossRef - Zhang J, Liu X, Zhou W, Lu S, Wu C, Wu Z, Liu R, Li X, Wu J, Liu Y, Guo S. Identification of key genes associated with the process of hepatitis B inflammation and cancer transformation by integrated bioinformatics analysis. Front. Genet; 2021;12.
CrossRef - Ou HL, Hoffmann R, González‐López C, Doherty GJ, Korkola JE, Muñoz‐Espín D. Cellular senescence in cancer: From mechanisms to detection. Mol. Oncol; 15(10):2634-71 (2021).
CrossRef - Stolfi C, Troncone E, Marafini I, Monteleone G. Role of TGF-beta and Smad7 in gut inflammation, fibrosis and cancer. Biomolecules; 11(1):17 (2020).
CrossRef - Wang Z, Embaye KS, Yang Q, Qin L, Zhang C, Liu L, Zhan X, Zhang F, Wang X, Qin S. Development and validation of a novel epigenetic-related prognostic signature and candidate drugs for patients with lung adenocarcinoma. Aging (Albany NY); 13(14):18701 (2021).
CrossRef - Schmidt AK, Pudelko K, Boekenkamp JE, Berger K, Kschischo M, Bastians H. The p53/p73-p21CIP1 tumor suppressor axis guards against chromosomal instability by restraining CDK1 in human cancer cells. Oncogene; 40(2):436-51 (2021).
CrossRef - Yin X, Liu J, Wang X, Yang T, Li G, Shang Y, Teng X, Yu H, Wang S, Huang W. Identification of Key Transcription Factors and Immune Infiltration Patterns Associated With Breast Cancer Prognosis Using WGCNA and Cox Regression Analysis. Front. Oncol; 11 (2021).
CrossRef - Wei Y, Liu G, Wu B, Yuan Y, Pan Y. Let-7d inhibits growth and metastasis in breast cancer by targeting Jab1/Cops5. Cell. Physiol. Biochem; 47(5):2126-35 (2018)
CrossRef - Gao Y, Wang R, Liu J, Zhao K, Qian X, He X, Liu H. SENP1 promotes triple-negative breast cancer invasion and metastasis via enhancing CSN5 transcription mediated by GATA1 deSUMOylation. Int. J. Biol. Sci;18(5):2186 (2022).
CrossRef - Pan, Y., Wang, S., Su, B., Zhou, F., Zhang, R., Xu, T., Zhang, R., Leventaki, V., Drakos, E., Liu, W., & Claret, F.X. (2017). Stat3 Contributes to Cancer Progression by Regulating Jab1/Csn5 Expression. Oncogene; 36(8):1069–79 (2016).
CrossRef