Manuscript accepted on :27-10-2023
Published online on: 01-12-2023
Plagiarism Check: Yes
Reviewed by: Dr. B. Kirthika
Second Review by: Dr. Cherry Bansal
Final Approval by: Dr. Patorn Piromchai
Sangeetha Narasimhan1* , Malathi Narasimhan2
, Malathi Narasimhan2 , Shishir Ram Shetty1
, Shishir Ram Shetty1 , Sharada T Rajan2
, Sharada T Rajan2 , Sausan Al Kawas1
, Sausan Al Kawas1  and Vijaya Nirmala Subramani2
and Vijaya Nirmala Subramani2
1Department of Craniofacial Health Sciences, College of Dental Medicine, University of Sharjah, Sharjah, United Arab Emirates.
2 Sri Ramachandra Dental College, Department of Oral Pathology and Microbiology, Sri Ramachandra Institute of Higher Education and Research, Chennai, Tamil Nadu, India.
Corresponding Author E-mail: snarasimhan@sharjah.ac.ae
DOI : https://dx.doi.org/10.13005/bpj/2781
Abstract
Objective: Oral mucosal cancers are the 11th most common human malignancies worldwide with a five-year survival rate of ≤50%. The lacunae of reliable diagnostic and prognostic markers pose an enormous challenge to the timely identification and prediction of disease progression in oral cancer. MicroRNAs (miRNAs) are emerging molecular markers associated with cancer initiation, progression, and therapy. The present study evaluated the microRNA -375(miR-375) expression and its target p53 gene in Oral squamous cell carcinoma (OSCC) to validate its utility as a diagnostic marker of the disease. Patients and Methods: This case-control study targeted histopathologically diagnosed cases of OSCC. miR-375 was quantified from 22 cases of OSCC and corresponding control tissues using qRT-PCR. Mutant p53 expression in cases and controls was determined by subjecting the tissues to immunohistochemical Results: Significant downregulation of miR-375 was noted in OSCC tissues (68.1%) compared to the control tissues with a mean fold change of 83.9 (p<0.05). Significant downregulation of miR-375 was noted in Paan and tobacco chewing patients (77.8%). Men exhibited considerable downregulation compared to women (p<0.05). The miR-375 expression levels did not correlate with the patient’s age, location, size, nodal status, and histopathological grade of the tumor. About 63.6 % of OSCC tissues showed mutant p53 positivity. Mutant p53 expression was noted in 73.3% of miR-375 downregulated tumors. Smokers exhibited higher expression of mutant p53 contrary to non-smokers(p<0.00). P53 immunopositivity showed a correlation with tumor size, histopathological grade, and nodal metastasis. Conclusion: The findings of the study indicate that miR-375 downregulation may have a crucial effect on oral carcinogenesis by targeting p53. miR-375 should be further evaluated as a potential marker for oral cancer diagnosis.
Keywords
immunohistochemistry; Oral cancer; MicroRNA; RT-PCR; p53; Tumor suppressor gene
Download this article as:| Copy the following to cite this article: Narasimhan S, Narasimhan M, Shetty S. R, Rajan S. T, Kawas S. A, Subramani V. N. Analyzing the Expression of MicroRNA-375 and its Target Gene p53 in Oral Squamous Cell Carcinoma and its Implication in Oral Carcinogenesis. Biomed Pharmacol J 2023;16(4). |
| Copy the following to cite this URL: Narasimhan S, Narasimhan M, Shetty S. R, Rajan S. T, Kawas S. A, Subramani V. N. Analyzing the Expression of MicroRNA-375 and its Target Gene p53 in Oral Squamous Cell Carcinoma and its Implication in Oral Carcinogenesis. Biomed Pharmacol J 2023;16(4). Available from: https://bit.ly/3uHunU4 |
Introduction
Oral squamous cell carcinoma is the highly prevalent malignancy of the oral mucosa accounting for about 90% of all head and neck cancers1. High mortality and morbidity rates associated with this disease is attributed to late diagnosis and intervention 2. Oral carcinogenesis is a multistep process that disrupts various Oncogenes and tumor suppressor genes3. Understanding the epi genetic insights of oral cancer would result in the discovery of a powerful diagnostic and prognostic tool. Results of the research studies conducted during the past several years with similar intentions are yet to be clinically translated4.
MicroRNAs are endogenous RNAs that possess enormous gene modulatory potential. Based on their sequence and complementarity, microRNAs combine with specific mRNAs and regulate protein synthesis either by target mRNA degradation or translation repression5, 6. In this manner, the human microRNAs can control about 60% of the human genome. Every cellular process such as cell differentiation, proliferation, mobility, and apoptosis are subject to microRNA- dependent regulation7. Expression of microRNAs is tightly controlled by various transcription factors. MicroRNA expression is heavily dysregulated in human cancers and compelling evidence has shown that they affect all the hallmarks of cancer8. The variations in the expression of many microRNAs have assisted as diagnostic and prognostic predictors in specific tumors and might become potential and promising therapeutic targets in cancer treatment in the future9.
MicroRNA 375 is a tumor-suppressive microRNA located in 2q3510. It is an islet-specific miRNA that regulates insulin secretion and glucose homeostasis. Anti-tumor effects of miR-375 are potentiated by modulating its oncogenic targets JAK2, IGF1R, AEG-1 and YAP1 genes that moderate many processes like apoptosis, invasion, and metastasis. Many studies reveal that miR-375 is widely present in other human tissues and is significantly downregulated in many malignancies such as melanoma, glioma, Gastric, Laryngeal, Esophageal, and hepatocellular carcinoma11,12.
The p53 is a tumor suppressor gene with pro-apoptotic function13. The p53 transcription factor is considered the “guardian of the genome” as it senses the DNA damage and mediates cell cycle arrest or promotes apoptosis14. Mutations in p53 are associated with genomic instability and increased susceptibility to cancer15. In normal cells, the half-life of p53 protein is very short lasting up to 5-20 minutes making it difficult to be detected in normal tissues16. However, a mutated p53 protein is not easily digested and has a prolonged half-life. Therefore, it accumulates inside the cancer cells and can be immunohistochemically detected in premalignant and malignant tissues17, 18. p53 gene regulates, and in turn, is also regulated by several microRNAs19. p53 has been reported as a gene target for miR-37520,21. There are no previous studies in the literature that studied the association between miR375 and p53 expression in oral cancer. This study analyzed the expression of miR-375 and its gene target p53 in OSCC tissues and correlated their expression to the clinical and pathological parameters of the study samples to understand their role in oral carcinogenesis.
Materials and Methods
The study was conducted in the Oral Pathology department of the University Dental Hospital. The Institutional Review Board approved the study((SRIHER)- IEC-N1/12/MAR/27) and all the patients signed informed consent before sample collection. The study population comprised histopathologically diagnosed cases of squamous cell carcinoma using incisional biopsy samples. Only the patients with primary tumors were included in the study. Recurrent tumors and cases with previous treatments such as radiotherapy and chemotherapy were excluded from the study. After exclusions about 22 cases were included in the study. Resected tumors and the corresponding adjacent normal mucosa tissues were obtained from OSCC patients. Normal mucosal tissue was obtained from the contralateral side of the lesion proper. Fresh tissue samples were preserved in RNAlater (sigma Aldrich) and stored at -40oC until PCR analysis. Tissue for the histopathological study was stored in formalin at room temperature and routinely processed.
Quantitative Real-time (qRT-PCR) assay
The tumor and normal tissues were homogenized using mortar and pestle and extraction of total DNA was performed by the trizol method (Invitrogen, USA). Stem loop primers (table 1) and primescript RT reagent kit ((Takara Bio Inc) were used for synthesizing the Complementary DNA from the extracted RNA. Reverse transcriptase reactions included 7μl of RNA and 3 μl of master mix containing RT buffer, stem loop primers and RT enzyme. 10 μL aliquots were prepared and incubated for 15 min at 37°C, and 15 seconds at 85°C, followed by maintenance at 4°C. The primeScript SYBR Green PCR kit (Qiagen NV) was used to perform the qPCR analysis. One microliter of cDNA was used as a template in 10μL reactions. The master mix was prepared separately for the microRNAs and the control gene with the forward and reverse primers (Table 1), SYBR green dye, ROX dye and water. Gene expression was quantified using triplicates in the 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). The 10 μL aliquots PCR plates were denatured for 95℃ for 30 seconds, annealed at 95℃ for 30 seconds, 60℃ for 35 seconds (40 cycles), 95℃ for 15 seconds and extended for one minute at 60℃.
Table 1: Primer sequence for RT- PCR
|
GENE |
PRIMER SEQUENCE |
|
miR-375 stem loop RT primer |
5’-GTCGTATCCAGTGGAGGGTCCGAGGTATTCGCACTGG ATACGACTCACGCG-3’ |
|
RNU6 stem loop RT primer |
5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGG ATACGACTATGGAAC-3’ |
|
miR-375 Forward Primer |
5’-GCCCGCTTTGTTCGTTCGGCT-3’ |
|
RNU6 Forward Primer |
5’-GTGCTCGCTTCGGCAGCAC-3’ |
|
Universal Reverse Primer |
5’-GTGCAGGGTCCGAGGT-3’ |
Analysis of relative gene expression
The relative gene expression in the tumor and normal tissues was calculated using the 2-ΔΔCq method (Livak method) by normalizing it to the U6 housekeeping gene22. The first step in the analysis involved the determination of the threshold cycle (Ct) value of individual samples. Using the threshold cycle value of each microRNA and control gene the ΔCt value was calculated using the formula ΔCt= Ct miR375 – Ct U6. The ΔΔCt value was calculated from the from the ΔCt values using the formula (ΔΔCt= ΔCt Cancer – ΔCt Normal). Finally, the fold value (2-ΔΔCt value) was calculated. A Fold value more than 1, is considered as an upregulation of the gene and a fold value of less than 1 signifies a downregulation of the gene.
Immunohistochemistry (IHC)
The tumor and control tissues were routinely processed, and 4-um paraffin sections were taken on charged slides. The sections were dewaxed using xylene, dehydrated in graded alcohols, and subsequently rinsed thoroughly using distilled water. Endogenous peroxidase activity was blocked by applying peroxide block to the tissue sections for 5 minutes at 37℃. The pressure cooker method was used for antigen retrieval. Further p53 primary antibody (Mouse monoclonal, clone DO7, class-IgG1 -BioGenex Life sciences Pvt Ltd) was applied to the tissue sections for 1 hour. Further, the sections were thoroughly rinsed with citrate buffer and incubated with universal secondary antibody for 30 minutes (Super SensitiveTM Polymer HRP Kit Manual- BioGenex life sciences Pvt Ltd). Finally, the sections were thoroughly rinsed with distilled water and stained with the chromogen diaminobenzidine (DAB), counterstained with Harris hematoxylin, air dried, cleared and mounted with dibutyl phthalate xylene (DPX). Breast cancer tissues were used as a positive control for mutant p53. Internal negative controls were used to validate the staining.
All the slides were scored by two certified pathologists. Mutant p53 positivity was confirmed by the presence of brown staining of the nucleus of the cells. The sections were considered negative when <5% of the cells were stained for p53. Further positive sections were graded as mild positive (5-25%), Moderate positive (25-50%) and intense positive (> 50%) based on the number of cells that were immune-positive for mutant p53.
Data collection/statistical analysis
The clinical parameters of the study samples were obtained from the medical records session. The study sample comprised 16 men and 6 women. The mean age of the patients was 54.5. Eleven cases were excised from buccal mucosa, 5 from the tongue, 3 from alveolar mucosa, 2 from the lip, and one from the floor of the mouth. The revised International TMN Staging System was used to assess the tumor stage. 15/22 cases had nodal metastasis of the disease. However, none of the patients had distant organ metastasis confirmed by a PET scan. The date of the surgery was considered the beginning of the disease. Follow-up data was obtained for 5 years. Recurrence was noted in 2 of the 22 cases. The statistical analysis of the study results was performed using version 18 of SPSS software. Descriptive statistics were presented as numbers and percentages. Mean and SD were calculated for all the data. Statistical differences between clinicopathological parameters and the miRNA375 levels were evaluated using non-parametric tests. The difference in microRNA expression between the lesion and control tissues was calculated student T-test. A chi-square test was used for comparison between two attributes. Inter-observer reliability was calculated using Cohens Kappa. A p-value less than 0.05 was considered statistically significant.
Results
The study analyzed the MicroRNA375 expression in OSCC and its paired normal tissues by using qRT-PCR. The tissues were also analyzed for the expression of p53 using immunohistochemistry. The microRNA and p53 expression were also compared to the clinical and pathological attributes of the tumor.
Expression of miR-375 in oral cancer and normal tissues
A significant difference was noted in the expression level of miR-375 between the tumor and adjacent normal tissues. Among 22 OSCC cases, 15(68.1%) tumor samples demonstrated downregulation of miR-375(p<0.05) (Figure1). A mean fold change of 83.96 was noted in the tumor tissues compared to the controls.
 |
Figure 1: Downregulation of miR-375 in OSCC. |
MicroRNA-375 expression correlation to clinico-pathological parameters
Table 2 displays miR-375 expression based on the clinicopathological parameters. All the cases below 40 years, 50% of cases between 40-60 years and 77.8% of cases above 60 years of age demonstrated miR-375 downregulation. Men showed significant downregulation of miR-375 compared to women (p <0.05). Cases with pan/gutkha chewing habits (77.8%) showed significant (p <0.05) miR-375 downregulation compared to cases without habits. Smokers and non-smokers did not exhibit any significant difference in miR-375 expression(P>0.05). 80% of the cases with N2 nodal involvement exhibited downregulation of miR-375. However, this finding was not statistically significant. The miR-375 expression levels did not reveal any statistically significant relationship with patient age, tumor size, location, nodal metastasis, histopathological grade, and survival of the patient. The Kaplan –Meier analysis could not yield a meaningful interpretation.
p53 Expression in OSCC
The positive control breast cancer tissue stained intensely with p53 antibody (Figure 2), and the internal negative controls did not exhibit any p53 positivity. The interobserver variation or the measurement of agreement between the observes was statistically analyzed and was highly significant (p <0.00) The kappa score was calculated to analyze the overall significance of p53 expression between the normal and OSCC sections and was statistically significant (p <0.00)
In our study mutant p53 expression was observed in the nuclei of the dysplastic epithelial cells of OSCC. Staining ranged from focal positivity in the tumor islands to intense positivity in sheets of dysplastic squamous cells in the underlying connective tissue (Figure 3-5). Among 22 tumor cases, 14 tissues (63.6%) exhibited immunopositivity for mutant p53 (4 – mild positive; 5-Moderate positive & 5- intense positive).
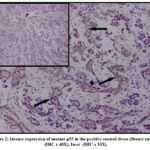 |
Figure 2: Intense expression of mutant p53 in the positive control tissue (Breast cancer) (IHC x 40X), Inset -(IHC x 10X). |
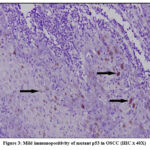 |
Figure 3: Mild immunopositivity of mutant p53 in OSCC (IHC X 40X). |
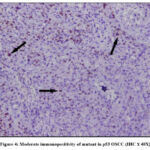 |
Figure 4: Moderate immunopositivity of mutant in p53 OSCC (IHC X 40X). |
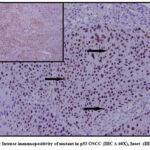 |
Figure 5: Intense immunopositivity of mutant in p53 OSCC (IHC X 40X), Inset -(IHC x 10X). |
Mutant p53 expression correlation to clinico‑ pathological parameters of OSCC
The expression of mutated p53 in the study samples is presented in Table 2. All the smokers exhibited mutant p53 expression compared to the nonsmokers (p < 0.05). Regarding the tumor size, the T1 tumors exhibited mild to moderate staining compared to the intense immunopositivity noted among T2 and T4 tumors (p <0.05). Intense mutant p53 staining was observed in moderately differentiated and poorly differentiated tumors compared to the well-differentiated OSCC tissues (p <0.05). Tumor cases with N2 nodal metastasis showed intense mutant p53 staining followed by the N1 cases. Whereas patients without nodal metastasis showed weak to moderate immunostaining (p <0.05). There was no significant association between the mutant p53 expression with age, gender, or the clinical location of the tumor (P>0.05).
The relative expression of mutant p53 was compared with miR-375 expression in OSCC cases and about 73.3% (11/15) of miR-375 down regulated cases showed mutant p53 expression in OSCC demonstrating a very significant association within the genes. (p < 0.05).
Table 2: Clinco-pathological Correlation of OSCC patients to miR-375 and p53 expression.
|
|
|
miR375 |
|
p53 expression |
|
||||
|
Characteristics |
n |
Downregulation |
p-value |
Weak |
Moderate |
Intense |
Negative |
p-value |
|
|
Age in years |
|||||||||
|
< = 40 |
3 |
3 |
0.191 |
1 |
2 |
0 |
0 |
0.411 |
|
|
41 – 60 |
10 |
5 |
2 |
1 |
3 |
4 |
|||
|
> 60 |
9 |
7 |
1 |
2 |
2 |
4 |
|||
|
Gender |
|||||||||
|
Male |
16 |
13 |
*0.032 |
3 |
4 |
5 |
4 |
0.158 |
|
|
Female |
6 |
2 |
1 |
1 |
0 |
4 |
|||
|
Location |
|||||||||
|
Buccal mucosa |
11 |
9 |
0.471 |
0 |
4 |
3 |
4 |
0.27 |
|
|
Tongue |
5 |
3 |
2 |
0 |
2 |
1 |
|||
|
Lip |
2 |
1 |
1 |
0 |
0 |
1 |
|||
|
Alveolus |
3 |
1 |
1 |
0 |
0 |
2 |
|||
|
Floor of mouth |
1 |
1 |
0 |
1 |
0 |
0 |
|||
|
Habits- Pan chewing |
|||||||||
|
Present |
18 |
14 |
*0.040 |
4 |
4 |
4 |
6 |
0.762 |
|
|
Absent |
4 |
1 |
0 |
1 |
1 |
2 |
|||
|
Smoking |
|||||||||
|
Smokers |
9 |
8 |
0.083 |
1 |
3 |
5 |
0 |
0.003* |
|
|
Non-Smokers |
13 |
7 |
3 |
2 |
0 |
8 |
|||
|
Tumor size |
|||||||||
|
T1 |
11 |
6 |
0.323 |
3 |
4 |
0 |
4 |
0.05* |
|
|
T2 |
9 |
7 |
1 |
1 |
3 |
4 |
|||
|
T4 |
2 |
2 |
0 |
0 |
2 |
0 |
|||
|
Nodal status |
|||||||||
|
N0 |
7 |
5 |
0.717 |
2 |
2 |
0 |
3 |
0.037* |
|
|
N1 |
10 |
6 |
2 |
2 |
1 |
5 |
|||
|
N2 |
5 |
4 |
0 |
1 |
4 |
0 |
|||
|
Tumor grade |
|||||||||
|
WDOSCC |
12 |
7 |
0.33 |
4 |
4 |
0 |
4 |
0.035* |
|
|
MDOSCC |
8 |
7 |
0 |
0 |
4 |
4 |
|||
|
PDOSCC |
2 |
1 |
0 |
1 |
1 |
0 |
|||
*p < 0.05 Statistically Significant, p > 0.05 Non Significant
Discussion
MicroRNAs are molecular regulators of physiological cellular processes. MicroRNA dysregulation has been noted in various diseases including cancer8. In current study analyzed miR-375 expression in 22 OSCC tissues and the corresponding normal tissues. The results reveal that MicroRNA-375 expression is considerably downregulated in OSCC compared to non-tumor tissue which is supported by the previous studies23-29. In the current study miR-375 was 83.9 %-fold downregulated compared to the adjacent paired normal tissues. MicroRNA studies have shown a 10 to 22-fold downregulation of miR-375 in OSCC and a 32-fold downregulation in HNSCCs24,26,87,30. This very high level of downregulation of miR-375 in our study is attributed to a single case of poorly differentiated squamous cell carcinoma which showed a very high Ct value in the PCR analysis.
Identified as a tumor suppressor gene, miR375 suppresses the malignant properties of cancer cells. miR-375 loss has been noted in various human malignancies including gastric, Pharyngeal, esophageal, hepatic and breast cancers 11. Lian et al demonstrated that miR-375 targets tyrosine kinase Janus kinase 2 (JAK2) and inhibits cell proliferation in gastric cancer31. Further miR375 could suppress rapid cancer cell growth by inhibiting aerobic glycolysis via the PI3K-Akt signaling pathway. On the other hand, miR-375 upregulation results in cell cycle arrest at the G0 / G1 phase in oral cancer cell lines thereby inhibiting tumor growth26. MicroRNA-375 downregulation is also a contributing factor for the progression of potentially malignant disorders to oral cancer32, 33. Our study results show that miR-375 downregulation in oral cancer showed a positive association with paan chewing habits. Our findings along with the literature support provide evidence that miR-375 loss in oral cancer could contribute to oral carcinogenesis.
Men demonstrated significant downregulation of miR-375 versus women in our study. This distribution could be attributed to a higher number of men involved in the study compared to the women. miR-375 expression did not correlate to the oral cancer disease progression in our study. By contrast, Siow MY et al revealed that miR‐375 downregulation is associated with tumor size and disease progression in oral cancer27. Zhang B et al reported reduction in miR-375 levels correlates with increased lymph node metastasis and reduced survival rate in OSCC patients26.
The p53 gene is one of the most frequently mutated genes in human cancers13. About 63.3% of the tumor tissues demonstrated mutant p53 immunopositivity in this study which is consistent with the results of Patil NN et al (61%) Dave KV et al (62%) and Ghanghoria S et al (63%) in OSCC cases34-36. Further previous immunohistochemical studies have documented significant variability in the expression of mutant p53 in oral cancer, spanning from 36% to 80% 37-39. Detection of p53 in oral cancer confirms the mutation of p53 in oral cancer tissues. However, the presence of p53 beyond the basal layer of epithelium denotes an early sign of initiation of oral carcinogenesis, thereby demonstrating the fact that the genomic mutations take place well in advance of the observable morphological alterations in the affected tissue40. Thus, the deactivation of the p53 protein or alteration in its coding gene might have a significant impact on the development of oral cancer.
Mutant P53 expression was noted in all the smokers involved in this study. Santos FD et al also recorded an association between p53 expression and smoking41. A wide spectrum of p53 mutations were diagnosed in cancers reported among both active and former smokers. Exposure to cigarette carcinogens causse a variety of gene mutations that are closely related to tumorigenesis42,43. Langdon JD’s findings indicate that p53 mutations were frequently detected in tumors from individuals who had a history of heavy smoking. This suggests that genetic damage to p53 could be a contributing factor in this patient group38. Among 9 smokers included in the study 8 cases (88.9%) showed miR-375 downregulation. However, this finding was not statistically significant. It can be hypothesized that nicotine associated carcinogens might downregulate miR-375. Further, miR-375 dysregulation might initiate carcinogenesis by inactivating the p53 pathway. Further molecular studies are required to substantiate the above statement.
The current study results also demonstrated a greater intensity of expression of mutant p53 with increasing tumor size, tumor grade and nodal metastasis. This finding indicates the role of p53 in tumor progression. Mutated p53 gene forfeits its ability to inhibit cancer and behaves like an oncogene and promotes tumor growth by stimulating cell division44. The findings of the present study show that expression of mutant p53 was noted in 66.7% of miR-375 down regulated cases of OSCC demonstrating a very significant association within the genes. Liu Y et al study results revealed that microRNA-375 directly binds to the 3′-UTR regions of p53 and mediates down-regulation of the gene. Studies have shown that miR-375 overexpression leads to inactivation of the p53 pathway and reduces the p53 protein levels in gastric cancer cells21. Further, it aids in the evasion of apoptosis after damage to the DNA20. Song L et al demonstrated that miR-375 regulates radio resistance of cervical cancer cells through the p53 pathway45. Based on our findings, and literature evidence miR-375 downregulation may induce mutation in the p53 gene which might further result in the initiation of oral carcinogenesis.
Conclusion
In summary, MicroRNA-375 is a tumor-suppressive microRNA and its significant down-regulation in oral cancer highlights its association with oral carcinogenesis. miR-375 favors oral carcinogenesis by targeting the p53 gene, which is the frequently mutated gene in oral cancer. Further studies are required to analyze the molecular pathways involved in the interaction between these genes. Loss of miR-375 in oral cancer holds diagnostic implications and should be evaluated further as early diagnostic markers for OSCC. Targeting miR-375 could provide a promising strategy for oral cancer intervention in the future.
Acknowledgments
We extend our sincere thanks and gratitude to Prof. Ganesh Venkatraman and Prof Rayala Suresh Kumar for providing laboratory access to conduct our PCR experiments.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest regarding the publication of this paper.
Funding Sources
No funding resources to be declared.
References
- Lauritano D, Lucchese A, Contaldo M, Serpico R, Lo Muzio L, Biolcati F, Carinci F. Oral squamous cell carcinoma: diagnostic markers and prognostic indicators. J Biol Regul Homeost Agents. 2016 Apr 1; 30 (2 Suppl 1):169-76.
- Cervino G, Fiorillo L, Herford AS, Romeo U, Bianchi A, Crimi S, D’Amico C, De Stefano R, Troiano G, Santoro R, Laino L. Molecular biomarkers related to oral carcinoma: clinical trial outcome evaluation in a literature review. Disease markers. 2019 Mar 25; 2019. doi: 10.1155/2019/804036.
CrossRef - Williams HK. Molecular pathogenesis of oral squamous carcinoma. Molecular Pathology. 2000 Aug; 53(4):165. doi: 10.1136/mp.53.4.165.
CrossRef - Warnakulasuriya S. Prognostic and predictive markers for oral squamous cell carcinoma: the importance of clinical, pathological and molecular markers. Saudi Journal of Medicine and Medical Sciences. 2014 Jan 1; 2(1):12.
CrossRef - Arif KM, Elliott EK, Haupt LM, Griffiths LR. Regulatory mechanisms of epigenetic miRNA relationships in human cancer and potential as therapeutic targets. Cancers. 2020 Oct; 12(10):2922. doi: 10.3390/cancers12102922.
CrossRef - Oliveto S, Mancino M, Manfrini N, Biffo S. Role of microRNAs in translation regulation and cancer. World J Biol Chem. 2017 Feb 26;8(1):45-56. doi: 10.4331/wjbc.v8.i1.45.
CrossRef - Menini M, De Giovanni E, Bagnasco F, Delucchi F, Pera F, Baldi D, Pesce P. Salivary micro-RNA and oral squamous cell carcinoma: A systematic review. Journal of Personalized Medicine. 2021 Feb 4;11(2):101.
CrossRef - Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal transduction and targeted therapy. 2016 Jan 28; 1(1):1-9. doi: 10.1038/sigtrans.2015.4.
CrossRef - Wang S, Claret FX, Wu W. MicroRNAs as therapeutic targets in nasopharyngeal carcinoma. Frontiers in oncology. 2019 Aug 13; 9:756. doi: 10.3389/fonc.2019.00756.
CrossRef - Guo Y, An R, Zhao R, Sun Y, Liu M, Tian L. miR-375 exhibits a more effective tumor-suppressor function in laryngeal squamous carcinoma cells by regulating KLF4 expression compared with simple co-transfection of miR-375 and miR-206. Oncology reports. 2016 Aug 1; 36(2):952-60. doi: 10.3892/or.2016.4852. Epub 2016 Jun 3.
CrossRef - Yan JW, Lin JS, He XX. The emerging role of miR‐375 in cancer. International journal of cancer. 2014 Sep 1; 135(5):1011-8. doi: 10.1002/ijc.28563.
CrossRef - Osan C, Chira S, Nutu AM, Braicu C, Baciut M, Korban SS, Berindan-Neagoe I. The Connection between MicroRNAs and Oral Cancer Pathogenesis: Emerging Biomarkers in Oral Cancer Management. Genes (Basel). 2021 Dec 15;12(12):1989. doi: 10.3390/genes12121989. PMID: 34946938; PMCID: PMC8700798
CrossRef - Solomon H, Madar S, Rotter V. Mutant p53 gain of function is interwoven into the hallmarks of cancer. The Journal of pathology. 2011 Dec; 225(4):475-8. doi: 10.1002/path.2988.
CrossRef - Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J Pathol. 2011 Jan;223(2):116-26. doi: 10.1002/path.2784. Epub 2010 Oct 25. PMID: 21125670.
CrossRef - Singh RD, Patel KR, Patel PS. p53 mutation spectrum and its role in prognosis of oral cancer patients: A study from Gujarat, West India. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2016 Jan 1; 783:15-26. doi: 10.1016/j.mrfmmm.2015.12.001.
CrossRef - Midgley CA, Lane DP. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997 Sep; 15(10):1179-89. doi: 10.1038/sj.onc.1201459.
CrossRef - Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes & development. 2012 Jun 15;26(12):1268-86. doi: 10.1101/gad.190678.112.
CrossRef - Hashmi AA, Hussain ZF, Hashmi SK, Irfan M, Khan EY, Faridi N, Khan A, Edhi MM. Immunohistochemical over expression of p53 in head and neck Squamous cell carcinoma: clinical and prognostic significance. BMC research notes. 2018 Dec;11(1):1-5. doi: 10.1186/s13104-018-3547-7
CrossRef - Rokavec M, Li H, Jiang L, Hermeking H. The p53/miR-34 axis in development and disease. Journal of molecular cell biology. 2014 Jun 1;6(3):214-30. doi: 10.1093/jmcb/mju003.
CrossRef - Fan K, Spassova I, Gravemeyer J, Ritter C, Horny K, Lange A, Gambichler T, Ødum N, Schrama D, Schadendorf D, Ugurel S. Merkel cell carcinoma-derived exosome-shuttle miR-375 induces fibroblast polarization by inhibition of RBPJ and p53. Oncogene. 2021 Feb; 40(5):980-96. doi: 10.1038/s41388-020-01576-6.
CrossRef - Liu Y, Xing R, Zhang X, Dong W, Zhang J, Yan Z, Li W, Cui J, Lu Y. miR-375 targets the p53 gene to regulate cellular response to ionizing radiation and etoposide in gastric cancer cells. DNA repair. 2013 Sep 1; 12(9):741-50. doi: 10.1016/j.dnarep.2013.06.002.
CrossRef - Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2− ΔΔCT method. methods. 2001 Dec 1;25(4):402-8.. DOI: 10.1006/meth.2001.1262.
CrossRef - Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clinical Cancer Research. 2009 Apr 15;15(8):2850-5. doi: 10.1158/1078-0432.CCR-08-3131
CrossRef - Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O’Sullivan B, Waldron J, Gullane P. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clinical cancer research. 2010 Feb 15;16(4):1129-39. doi: 10.1158/1078-0432.CCR-09-2166.
CrossRef - Nohata N, Hanazawa T, Kikkawa N, Mutallip M, Sakurai D, Fujimura L, Kawakami K, Chiyomaru T, Yoshino H, Enokida H, Nakagawa M. Tumor suppressive microRNA-375 regulates oncogene AEG-1/MTDH in head and neck squamous cell carcinoma (HNSCC). Journal of human genetics. 2011 Aug;56(8):595. doi: 10.1038/jhg.2011.66.
CrossRef - Zhang B, Li Y , Hou D, Shi Q, Yang S, Li Q. MicroRNA-375 inhibits growth and enhances radiosensitivity in oral squamous cell carcinoma by targeting insulin like growth factor 1 receptor. Cellular Physiology and Biochemistry. 2017;42(5):2105-17. doi: 10.1159/000479913.
CrossRef - Siow MY, Karen Ng LP, Vincent Chong VK, Jamaludin M, Abraham MT, Abdul Rahman ZA, Kallarakkal TG, Yang YH, Cheong SC, Zain RB. Dysregulation of mi R‐31 and mi R‐375 expression is associated with clinical outcomes in oral carcinoma. Oral diseases. 2014 May;20(4):345-51. doi: 10.1111/odi.12118.
CrossRef - Lajer CB, Nielsen FC, Friis-Hansen L, Norrild B, Borup R, Garnaes E, Rossing M, Specht L, Therkildsen MH, Nauntofte B, Dabelsteen S. Different miRNA signatures of oral and pharyngeal squamous cell carcinomas: a prospective translational study. British journal of cancer. 2011 Mar;104(5):830. doi: 10.1038/bjc.2011.29
CrossRef - Wiklund ED, Gao S, Hulf T, Sibbritt T, Nair S, Costea DE, Villadsen SB, Bakholdt V, Bramsen JB, Sørensen JA, Krogdahl A. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS One. 2011 Nov 22;6(11): e27840. doi: 10.1371/journal.pone.0027840.
CrossRef - Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clinical Cancer Research. 2009 Apr 15;15(8):2850-5. doi: 10.1158/1078-0432.CCR-08-3131.
CrossRef - Ding L, Xu Y, Zhang W, et al: MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res 2010; 20: 784 – 793.
CrossRef - Shi W, Yang J, Li S, Shan X, Liu X, Hua H, Zhao C, Feng Z, Cai Z, Zhang L, Zhou D. Potential involvement of miR-375 in the premalignant progression of oral squamous cell carcinoma mediated via transcription factor KLF5. Oncotarget. 2015 Nov 24;6(37):40172. doi: 10.18632/oncotarget.5502.
CrossRef - Harrandah AM, Fitzpatrick SG, Smith MH, Wang D, Cohen DM, Chan EK. MicroRNA-375 as a biomarker for malignant transformation in oral lesions. Oral Surgery, Oral Medicine, Oral Pathology and Oral Radiology. 2016 Dec 1;122(6):743-52.
CrossRef - Patil NN, Wadhwan V, Chaudhary M, Nayyar AS. KAI-1 and p53 expression in oral squamous cell carcinomas: Markers of significance in future diagnostics and possibly therapeutics. Journal of oral and maxillofacial pathology: JOMFP. 2016 Sep;20(3):384. doi: 10.4103/0973-029X.190908.
CrossRef - Dave KV, Chalishazar M, Dave VR, Panja P, Singh M, Modi TG. Immunohistochemical expression of p53 and its clinicopathological correlation with modified Anneroth’s histological grading system. Journal of oral and maxillofacial pathology: JOMFP. 2016 Jan;20(1):29. doi: 10.4103/0973-029X.180922
CrossRef - Ghanghoria S, Ghanghoria A, Shukla A. p53 Expression in Oral cancer: A study of 50 cases. Journal of Pathology of Nepal. 2015 Mar 27;5(9):747-51.
CrossRef - Carlos de Vicente J, Junquera Gutierrez LM, Zapatero AH, Fresno Forcelledo MF, Hernández‐Vallejo G, Lopez Arranz JS. Prognostic significance of p53 expression in oral squamous cell carcinoma without neck node metastases. Head & Neck: Journal for the Sciences and Specialties of the Head and Neck. 2004 Jan;26(1):22-30. doi: 10.1002/hed.10339.
CrossRef - Langdon JD, Partridge M. Expression of the tumour suppressor gene p53 in oral cancer. British Journal of Oral and Maxillofacial Surgery. 1992 Aug 1;30(4):214-20. doi: 10.1016/0266-4356(92)90263-i.
CrossRef - Pillay M, Vasudevan DM, Rao CP, Vidya M. p53 expression in oral cancer: observations of a South Indian study. Journal of Experimental and Clinical Cancer Research. 2003 Sep 1;22(3):447-52.
- Cruz IB, Snijders PJ, Meijer CJ, Braakhuis BJ, Snow GB, Walboomers JM, van der Waal I. p53 expression above the basal cell layer in oral mucosa is an early event of malignant transformation and has predictive value for developing oral squamous cell carcinoma. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland. 1998 Apr;184(4):360-8. doi: 10.1002/(SICI)1096-9896(199804)184:4<360::AID-PATH1263>3.0.CO;2-H.
CrossRef - Toyooka S., Tsuda T., Gazdar A.F. The TP53 gene, tobacco exposure, and lung cancer. Hum. Mutat. 2003; 21:229–239. doi: 10.1002/humu.10177. doi: 10.1002/humu.10177.
CrossRef - Vahakangas K.H., Bennett W.P., Castren K., Welsh J.A., Khan M.A., Blomeke B., Alavanja M.C., Harris C.C. p53 and K-ras mutations in lung cancers from former and never-smoking women. Cancer Res. 2001;61:4350–4356.
- Li Y, Zhang J. Expression of mutant p53 in oral squamous cell carcinoma is correlated with the effectiveness of intra-arterial chemotherapy. Oncology letters. 2015 Nov 1;10(5):2883-7.
CrossRef - Song L, Liu S, Zeng S, Zhang L, Li X. miR-375 modulates radiosensitivity of HR-HPV-positive cervical cancer cells by targeting UBE3A through the p53 pathway. Medical science monitor: international medical journal of experimental and clinical research. 2015; 21:2210. doi: 10.12659/MSM.893859
CrossRef







