Manuscript accepted on :06-02-2023
Published online on: 18-09-2023
Plagiarism Check: Yes
Reviewed by: Dr. Cherry Bansal
Second Review by: Dr. Dito Anurogo
Final Approval by: Dr. Anton R Kiselev
Arnica F Lal1* , Pushpraj S Gupta1
, Pushpraj S Gupta1 and Pramod Kumar Yadav2
and Pramod Kumar Yadav2
1Department of Pharmaceutical Science, SHALOM Institute of Health and Allied Sciences, Sam Higginbottom University of Agriculture, Technology and Sciences, Naini, Prayagraj, India.
2Department of Computational Biology and Bioinformatics, JIBB Sam Higginbottom University of Agriculture, Technology and Sciences, Naini, Prayagraj, India.
Corresponding Author E-mail: arnica0594@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/2736
Abstract
Immunocompromised patients get Pseudomonas aeruginosa infections. P. aeruginosa's harmful effect is linked to quorum sensing (QS), which causes bacterial biofilm. Targeting QS is a promising novel method to treat P. aeruginosa infections, which are antibiotic-resistant. The Las system has garnered great interest due to LasR, the expedited gene during QS that regulates other virulence-associated genes. We used high-throughput virtual screening (VS) of Phenol Explorer to uncover a new category of LasR inhibitors that might be used as antagonists. Molecular docking-based VS against LasR (PDB: 2UV0) resulted in six best-scored compounds: Chrysin, Galangin, Coumestrol, 3',4',7- Trihydroxyisoflavanone, Dihydrodaidzein, Dihydroformononetin with docking score of -11.0 kcal/mol and a suitable ADMET profile. Six compounds were chosen for their lack of carcinogenicity in mice and rats, low molecular weight of 270 Da, and moderate to total solubility. Our current work shows that these six chemicals could block P. aeruginosa quorum sensing. Molecular dynamics investigations of a prospective therapeutic candidate (Chrysin) using Gromacs version 2022.2 demonstrated system stability; nonetheless, the antibiofilm assay showed a positive reaction against our in silico finding, suggesting a disturbance in quorum sensing regulating pathogenicity, i.e., biofilm formation. This study is the first to describe chrysin as a disruptor of quorum sensing signaling by inhibiting biofilm formation.
Keywords
Antimicrobial resistance; Antibiofilm; Dynamics; Network Pharmacology; Molecular docking; Quorum sensing; Pseudomonas aeruginosa
Download this article as:| Copy the following to cite this article: Lal A. F, Gupta P. S, Yadav P. K. In silico Study of Phenol Explorer Database as Potential Inhibitors of Quorum-Sensing Regulated Pathogenicity in Pseudomonas aeruginosa. Biomed Pharmacol J 2023;16(3). |
| Copy the following to cite this URL: Lal A. F, Gupta P. S, Yadav P. K. In silico Study of Phenol Explorer Database as Potential Inhibitors of Quorum-Sensing Regulated Pathogenicity in Pseudomonas aeruginosa. Biomed Pharmacol J 2023;16(3). Available from: https://bit.ly/46jXGtn |
Introduction
Healthcare-associated infections caused by P. aeruginosa (Pseudomonas aeruginosa) are still prevalent despite various preventive measures taken in hospitals that lead to increased mortality, and hospital stays resulting in increased treatment costs 1–3. Increasing multi-drug resistance among opportunistic pathogens is alarming and requires prompt management before further delay. One such pathogen is P. aeruginosa, which can be found in the environment and mainly affects people with compromised immune systems and ICU patients. They pose serious threats to human lives when contracted either through healthcare mishandling or after surgery, especially in the case of burns and wounds. They can cause infections in the body’s organs and fluids after surgery.
Several cases show that P. aeruginosa possesses a higher mortality rate than other bacterial infections and is susceptible to a limited number of antimicrobial agents. As per recent data, most antibiotics have become resistant to P. aeruginosa 4,5.
P. aeruginosa develops resistance, leading to altered expression and functional mechanisms of the bacteria 6. Quorum sensing (QS) in P. aeruginosa is a vital phenomenon for its pathogenicity and is well-studied to determine the target compounds/ligands that can curb infections and biofilm formation caused by the bacteria 7. As a result, QS inhibition is a new option for finding antivirulence and antibiofilm compounds to combat multidrug resistance by suppressing the genes responsible for these processes. P. aeruginosa has three QS systems: PQS, LasI–LasR, and RhlI–RhlR 8, which collectively contribute to biofilm production 9.
The polyphenol compounds are reasonably present in the plants as secondary metabolites, produced in response to defence mechanisms. These polyphenols are routinely consumed in the diet and have several health-related functions. Dietary polyphenols are reported to have various therapeutic effects. These days, the daily use of dietary polyphenols in terms of nutraceuticals is also catching the consumer’s interest. Several studies have found that natural products or polyphenols can be used as an adjuvant with antibiotics to produce a synergistic effect that can 10 reduce the burden on the antibiotic in terms of dose, dosage regimen, and overexploitation11,12.
The use of in silico-based approaches offers various tools for the identification of potential drug candidates against specific targets by studying the chemical and biological details of drug-macromolecular complexes. The molecular docking-based VS technique is quite popular for assessing ligands for the target protein to fit in the ligand binding domain. Structure-based virtual screening (SBVS) can be applied to find the top lead compounds that can act as QS inhibitors on the LasR receptor.
There are few reports where the use of a rational drug design approach to identify inhibitors of LasR has been attempted13. However, screening of the polyphenol database (Phenol-Explorer) against LasR has not yet been studied. In our study, a few new antagonists of LasR were identified by performing VS on Phenol Explorer, and, ADMET studies were performed to screen for the potency of compounds. The compounds with docking scores > -11.0 kcal/mol were selected for pharmacokinetic and toxicity profiling. Later, the molecular dynamics study was carried out using Gromacs version 2022.2 to study the stability of the complex. The finding was supported by the antibiofilm assay, which showed a positive response against P. aeruginosa. This research is the first, to our best knowledge, to report the pure chrysin contribution, selected from the Phenol Explorer database, as a quorum sensing inhibitor, unlike the research done by Singh et al., which showed the collective contribution of honey polyphenol components and not just a specific component against quorum sensing pathogenicity 14. Also, the research done by De et al. does not account for VS in the Phenol Explorer database 15. This research is the first to explore the potential of the Phenol Explorer16-18 database against quorum-sensing-related pathogenicity.
Materials and Methods
Culture conditions, bacterial strains, and chemicals used
P. aeruginosa PAO1 was used as a bacterial strain. Assay plates for antimicrobial activity, MIC, and antibiofilm activity included Muller Hilton Agar (MHA), Luria Bertani (LB) media, and Nutrient Broth media, which were purchased from Hi Media Laboratories Pvt. Ltd. Methanol was used as a negative control, purchased from Rankem Ltd. Chrysin and ciprofloxacin were purchased from Sigma-Aldrich.
Network pharmacology and target validation
Network pharmacology derived from the STRING database (Fig. 1) shows the genes and receptors responsible for the pathogenic phenotype response of P. aeruginosa. From all the nodes shown, the node LasR with STRING ID 287.DR97_592 functions as a LasB transcriptional activator that binds to the PAI autoinducer and has a good score of 0.999, while the node LasI with STRING ID 287.DR97_591 is required for the synthesis of PAI and has an autoinducer called C12-homoserine lactone that binds to the LasR receptor. Thus, considering the LasR and LasI relationships, further study of the nosocomial P. aeruginosa quorum sensing system is warranted. This prompted us to look for a suitable drug candidate for the 2UV0 protein, which has a similar binding affinity and is homologous to N-(3-oxododecanoyl)homoserine lactone (OHN) 19.
 |
Figure 1: Network depicting P. aeruginosa genes and protein. LasR is responsible for functions as transcriptional activation 19. |
Protein Structure
The domain of ligand binding in LasR, which binds to the autoinducer 3-oxo-C(12)-acyl-homoserine lactone, has been demonstrated in (Fig. 2) from pymol20. The structure seems to show a symmetrical dimer, with the cocrystallized agonist molecule OHN (N-(3-oxododecanoyl)homoserine lactone) at the ligand binding site, which forms four intermolecular hydrogen bonding interactions with residues ASP73, SER129, TYR56, and TRP60. The residues Trp60, Tyr56, Ser129, and Asp73 are the main protein-ligand interactions among LuxR homologs 21. OHN is a bacterial quorum-sensing signal molecule that is responsible for various pathogenic gene expressions in P. aeruginosa. Hydrogen bonds are the main participants in predicting the stability and precise binding of ligands to the active site of the receptor protein 22-24.
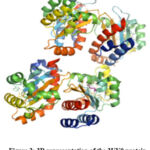 |
Figure 2: 3D representation of the 2UV0 protein |
The structure has a 175 amino acid chain with a theoretical weight of 19.69 KDa 21.
Selection of Drug Candidate from Docking Results
To identify the compounds showing interactions with LasR, VS was carried out for 752 polyphenols from the Phenol Explorer database. 35 compounds were selected among them and ranked according to their docking score Table 1. Finally, the docking analysis showed good scores for the compounds Chrysin, Galangin, Coumestrol, 3′, 4′, 7-Trihydroxyisoflavanone, Dihydrodaidzein, and Dihydroformononetin (Fig. 3), having docking lesser than -8.7 kcal/mol along with a good ADME and toxicity profile.In present circumstances, the technologies are advanced, and computer-aided drug design supports a variety of studies in discovering new molecules by using structure-based drug design techniques. Peculiarly, molecular docking studies help understand the conformation and main interactions of molecules/ligands with their receptors. The small molecules/ligands give information regarding binding to the residues of proteins at the atomic level, which disseminates target protein behavior. The likeliness of the drug, pharmacokinetic parameters, and ADMET data of 35 compounds were also listed 25 in supplementary Tables S1, S2, and S3.
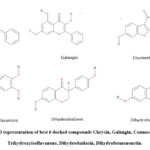 |
Figure 3: 2D representation of best 6 docked compounds Chrysin, Galangin, Coumestrol, 3′,4′,7-Trihydroxyisoflavanone, Dihydrodaidzein, Dihydroformononetin. |
Docking Studies
Molecular docking is an in silico technique to predict the best docking score of the ligand(s) with the macromolecule. This method repurposes an existing compound database to quickly identify the critical interactive amino acids that stabilise protein-ligand interactions. For docking analysis, preparation of proteins, generation of the appropriate grid, and docking of ligands to the selected grid were carried out. This method has been used in our previous studies26,27.
Preparation of Protein
The LasR receptors (PDB code: 2UV0) [28] co-crystallized with C12-homoserine lactone (C16H27NO4), downloaded from the RCSB website. The file was finally converted to pdbqt format after the missing atoms were repaired and charges were added.
Ligand Preparation
ChemBioDraw Ultra 12.0 software was chosen for ligand minimization. AutoDock Vina 1.1.2 (open-source software) [29, 30] was employed for molecular docking and also for ligand and grid preparation. The roots were detected for setting torsions, and then aromaticity criteria were set at 7.5.
Ligand docking
The bound ligand was extracted for the docking protocol validation and again used for docking to generate the same docking pose as downloaded from the RCSB website in its co-crystallized form. After optimization, the ligands were docked on the LasR receptor, and docking scores and intermolecular interactions were taken as the basis for categorising the ligands.
Visualization
The interaction of bonds between ligands and protein residues was visualised using Pymol software, in an academic version20.
Molecular Dynamics Simulation of LasR and Chrysin (possible drug candidate)
The molecular dynamics simulation of LasR receptors (PDB ID code: 2UV0) was performed using Gromacs version 2022.2 31. The energy minimization step was employed to optimise the structure of the protein (LasR) and ligand (Chrysin). Before starting with the simulation, the Gromacs environment was set up, and topology files of the protein and selected ligand were created using the “pdb2gmx” command. The protein-ligand complex was solvated in a defined box through the “genbox” command at a distance of 1.0 nm from either edge of the box. Ions were added, and minimization was carried out using the steepest algorithm in 50000 steps. The influential force field of CHARMM36 was employed in the system, and sodiumions were added to stabilise it against folding and disorganization. An equilibrium run of water around the protein was performed, followed by a molecular dynamics simulation using a 100000 ps time scale and 50000000 steps at a temperature of 300 °K and a pressure of 1 atm. The Grace programme was employed to analyse the molecular dynamics simulation results.
Antimicrobial assay and MIC (minimum inhibitory concentration)
The antimicrobial activity was done against the P. aeruginosa PAO1 strain using the disc diffusion technique. The bacterial strains were grown on MHA media, and each 2 mm circular disc was loaded with drug (chrysin) concentrations of 0.25, 0.5, and 1 mg/ml and placed on the bacterially inoculated media. Methanol (a negative control) 32 and ciprofloxacin (a positive control) 33 were used. The plate was incubated for 24 hours at 37 °C.
MIC was carried out using a broth dilution technique with concentrations ranging between 0.5 to 1.5 mg/mL. The Muller Hinton nutrient broth was used in 8 test tubes consisting of six chrysin drug concentration ranges (0.5, 0.7, 0.9, 1.1, 1.3, and 1.5 mg/ml), growth control, and media control, respectively. The test tube was incubated at 37 °C for 48 hrs.
Anti-biofilm assay using crystal violet staining
The biofilm is a cascade of extracellular polymeric substances (EPS), in which bacteria aggregate and form a matrix 34. This biofilm produced results from the complex regulatory mechanisms of the Las, Rhl, and PQS genes of P. aeruginosa 35, contributing to quorum-sensing regulated pathogenicity. In our study, we have focused on the drug chrysin resulting from our in silico finding, as it is a cheap and easily accessible drug. The planktonic bacteria were grown on a microtiter plate in Luria Bertani media, to which the drug concentrations (0.7, 0.9, 1.1, 1.3, and 1.5 mg/ml) and control (100 µl of bacterial inoculums) were added. This microtiter plate was incubated for 48 hours at 37 °C; the media was then discarded. Crystal violet dye was added to see the biofilm formation, which was mostly formed on the walls of the plates. After a few seconds, the dye was discarded and washed. The process was repeated one more time to ensure adequate staining. Then the excess dye was washed with acetic acid, and an OD at 600 nm was taken. The readings were taken in triplicate to measure the standard deviation.
Statistical and graphical analysis
Investigations are carried out over three measurements, with the results expressed as a standard deviation and mean value. The variability between the test sample and the control was analysed using one-way ANOVA. Results having a P value of < 0.05 were considered significant.
Results
Docking studies, drug likeliness, and the ADMET result
A molecular docking study was performed on 752 compounds extracted from the Phenol Explorer database, with only 35 showing satisfactory docking scores of around -11.0 kcal/mol, which is lower than OHN (N-(3-oxododecanoyl)homoserine lactone; LasR indigenous autoinducer) (-8.7 kcal/mol) Table 1. Only six drug candidates, Chysin, Galangin, Coumestrol, 3′, 4′, 7-Trihydroxyisoflavanone, Dihydrodaidzein, and Dihydroformononetin—showed negative carcinogenic results on both mouse and rat. Tables 2a, 2b, and 2c, in conjunction with a good drug likeliness and ADMET profile carried out through open-source software SwissADME 25, indicate that out of 35 compounds (Supplemental Tables S1, S2, and S3), six compounds were selected. The amino acid residues and hydrogen bonds that are involved in the interaction of Chrysin, Galangin, Coumestrol, 3′, 4′, 7-Trihydroxyisoflavanone, Dihydrodaidzein, and Dihydroformononetin (Figure 4a-g) with the 2UV0 protein of P. aeruginosa are listed in Table 3 along with LasR autoinducer (OHN).
Table 1: Docking scores of the best 35 compounds.
|
S.No |
Polyphenol name |
Docking result on LasR in Kcal/mol |
|
1. |
Eriodictyol |
-12.1 |
|
2. |
Chrysin |
-11.1 |
|
3. |
Apigenin 7-O-glucoside |
-11.5 |
|
4. |
Apigenin 7-O-glucuronide |
-11.5 |
|
5. |
Geraldone |
-11.5 |
|
6. |
Hispidulin |
-11.4 |
|
7. |
Cirsimaritin |
-11.1 |
|
8. |
Chrysoeriol 7-O-glucoside |
-11.2 |
|
9. |
Galangin |
-11.1 |
|
10. |
Methylgalangin |
-11.5 |
|
11. |
Genistein |
-11.2 |
|
12. |
Coumestrol |
-11.2 |
|
13. |
6”-O-Acetyldaidzin |
-11.8 |
|
14. |
Daidzin |
-11.1 |
|
15. |
p-Coumaroyl tyrosine |
-11.7 |
|
16. |
Episesaminol |
-12.7 |
|
17. |
2′-Hydroxyformononetin |
-11.6 |
|
18. |
3′,4′,7-Trihydroxyisoflavanone |
-11.3 |
|
19. |
4′,6,7-Trihydroxyisoflavanone |
-11.1 |
|
20. |
4′,7-Dihydroxy-3′-methoxyisoflavan |
-11.1 |
|
21. |
5,6,7,4′-Tetrahydroxyisoflavone |
-11.3 |
|
22. |
Daidzein 4′-O-glucuronide |
-11.2 |
|
23. |
Dihydrobiochanin A |
-11.1 |
|
24. |
Dihydrodaidzein |
-11.3 |
|
25. |
Dihydroformononetin |
-11.3 |
|
26. |
Dihydrogenistein |
-11.6 |
|
27. |
Genistein 7-O-glucuronide |
-11.5 |
|
28. |
5,7-Dihydroxy-8,4′-dimethoxyisoflavone |
-11.1 |
|
29. |
3′,4′,5,7-Tetrahydroxyisoflavanone |
-11.7 |
|
30. |
Enterolactone |
-11.2 |
|
31. |
2′-Hydroxyenterolactone |
-11.2 |
|
32. |
6′-Hydroxyenterolactone |
-11.1 |
|
33. |
5-Hydroxyenterolactone |
-11.1 |
|
34. |
Urolithin B 3-O-glucuronide |
-11.8 |
|
35. |
3-O-Methylrosmarinic acid |
-11.2 |
|
36. |
OHN (native/indigenous Ligand) |
-8.7 |
Table 2a: Six best compounds with good Drug likeliness
|
LIPINSKI PARAMETER |
|||||||||||
|
S. No |
Polyphenol |
MWa |
n-rotbb |
n-ONc |
nOH, |
Mre |
TPSAf |
iL |
Lipinski |
Lead |
Synthetic |
|
1. |
Chrysin |
254.24 |
1 |
4 |
2 |
71.97 |
70.67 |
2.27 |
0 |
1 |
2.93 |
|
2. |
Galangin |
270.24 |
1 |
5 |
3 |
73.99 |
90.9 |
2.08 |
0 |
0 |
3.12 |
|
3. |
Coumestrol |
268.22 |
0 |
5 |
2 |
73.81 |
83.81 |
1.8 |
0 |
0 |
3.16 |
|
4. |
3′,4′,7-Trihydroxyiso |
258.27 |
1 |
4 |
3 |
71.15 |
69.92 |
1.87 |
0 |
0 |
3.01 |
|
5. |
Dihydro |
256.25 |
1 |
4 |
2 |
69.55 |
66.76 |
1.52 |
0 |
0 |
3.03 |
|
6. |
Dihydro |
270.28 |
2 |
4 |
1 |
74.02 |
55.76 |
2.2 |
0 |
0 |
3.13 |
Table 2b: Six best compounds with good ADME profile.
|
PHARMACOKINETIC PARAMETER |
||||||||
|
S.No. |
Polyphenol |
Ali |
Silicos-IT class |
Absorption by GIT |
Permeation through the blood-brain barrier |
Pgp substrate |
CYP1A2 inhibitor |
CYP2C19 inhibitor |
|
1. |
Chrysin |
Moderately Soluble |
Moderately Soluble |
↑ |
Y |
N |
Y |
N |
|
2. |
Galangin |
Soluble |
Moderately Soluble |
↑ |
N |
N |
Y |
N |
|
3. |
Coumestrol |
Moderately Soluble |
Moderately Soluble |
↑ |
N |
N |
Y |
N |
|
4. |
3′,4′,7- |
Soluble |
Soluble |
↑ |
N |
Y |
N |
N |
|
5. |
Dihydro |
Soluble |
Soluble |
↑ |
Y |
Y |
Y |
N |
|
6. |
Dihydro |
Soluble |
Moderately Soluble |
↑ |
Y |
N |
Y |
Y |
‘Y’ = Yes; ‘N’ = No; ‘ ↑ ’ = High; ‘ ↓ ’ = Low.
Table 2c: Six best compounds with good toxicity profiles.
|
TOXICITY |
|||||||||
|
S. No |
Polyphenol Name |
TA100 10RLI |
TA100 NA |
TA1535 10RLI |
TA1535 NA |
Ames test |
Carcino Mouse |
Carcino Rat |
hERG Inhibition |
|
1. |
Chrysin |
+ |
+ |
– |
– |
Mutagen |
– |
– |
Medium Risk |
|
2. |
Galangin |
+ |
+ |
– |
– |
Mutagen |
– |
– |
Medium Risk |
|
3. |
Coumestrol |
+ |
+ |
– |
– |
Mutagen |
– |
– |
Medium Risk |
|
4. |
3′,4′,7-Trihydroxyisoflavanone |
+ |
– |
– |
– |
Mutagen |
– |
– |
Medium Risk |
|
5. |
Dihydrodaidzein |
+ |
– |
– |
– |
Mutagen |
– |
– |
Medium Risk |
|
6. |
Dihydroformononetin |
+ |
– |
– |
– |
Mutagen |
– |
– |
Medium Risk |
‘+’ = Positive; ‘-’ = Negative
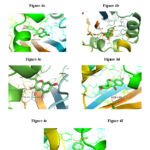 |
Figure 4: 2UV0 receptor’s amino acids interaction with the ligands (OHN (a), Chrysin (b), 3′,4′,7-Trihydroxyisoflavanone (c), Coumestrol (d),Galangin |
Table 3: Drug-Protein Interaction data of the best 6 compounds along with the LasR autoinducer.
|
Compounds |
Docking Score |
Hydrogen Bonds |
Hydrogen Bond interaction with LasR residues 2UV0 |
|
OHN (N-(3- oxododecanoyl)homoserine lactone) |
-8.7 |
4 |
ASP73, SER129, TYR56, TRP60 |
|
Chrysin |
-11.1 |
3 |
SER129, THR75, ARG61 |
|
3′,4′,7-Trihydroxyisoflavanone |
-11.3 |
1 |
SER129 |
|
Coumestrol |
-11.2 |
5 |
SER129, THR115, THR75, TYR93, ARG61 |
|
Galangin |
-11.1 |
4 |
SER129, THR115, THR75, ARG61 |
|
Dihydrodaidzein |
-11.3 |
1 |
SER129 |
|
Dihydroformononetin |
-11.3 |
1 |
SER129 |
From the visualization, it is revealed that OHN, being the indigenous ligand, shows interactions with TRP60, SER129, ASP73, and TYR56 residues of protein 2UV0; also, the selected six drug candidates showed interactions with some of these residues (Fig. 4a–g). From the above data, it is inferred that the most polar contacts with protein 2UV0 were formed by Chrysin, Galangin, and Coumestrol with hydrogen numbers 3, 4, and 5, respectively. Chrysin forms polar contacts with 2UV0 protein residues SER129, THR75, and ARG61, while Galangin forms contacts with SER129, THR115, THR75, and ARG61, and Coumestrol forms contacts with SER129, THR115, THR75, TYR93, and ARG61. The polar contacts shown in Table 3 were also formed by the rest of the compounds.
Result of MDS using Gromacs
The 3D structure of the LasR protein (PDBID: 2UV0) was studied using Gromacs version 2022.2. Before performing MDS, the input files were created, and then the solvation was done in a dodecahedron water box. After that, equilibration at a defined temperature and pressure of 300 K and 1 atm, respectively, was carried out. The minimization of the structure was carried out using the steepest descent minimization method in 50000 steps at 100000 ps to get a stable conformation. The average potential energy of the system was calculated using the CHARMM36 force field and was found to be -1.91084e + 06 KJ/mol. At first, the potential energy of the system was -1.90808e+06 KJ/mol with little convergence (Fig. 5a), showing system stability. Eight sodium ions were added to neutralise the net charge. Upon completion of two equilibration steps, i.e., temperature (NVT) and pressure (NPT), the system becomes well equilibrated at the desired temperature and pressure and is now ready for releasing the position restraint, i.e., solvent moving. The MDS was carried out at a scale of 100000 ps scale and 50000000 steps (iterations) at a temperature of 300 °K and a pressure of 1 atm. The obtained trajectories from the simulation were analysed using the ‘Grace’ program. The average radius of gyration (Rg) was found to be 1.62 nm, showing the compactness of the structure (Fig. 5b). Root mean square deviations (RMSD) increased rapidly at the beginning of the simulation but stabilised around 0.5 nm (Fig. 5c). Root mean square fluctuation (RMSF) was found between 0.14 nm to 0.4 nm for most of the amino acid residues, indicating structure stability (Fig. 5d).
 |
Figure 5a: Potential Energy |
 |
Figure 5b: Radius of gyration |
 |
Figure 5c: RMSD |
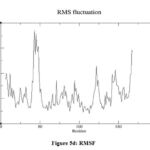 |
Figure 5d: RMSF |
Antimicrobial assay and MIC
The zone of inhibition was found to have a 9 mm diameter at 1 mg/mL of chrysin drug concentration, while the other concentrations (0.25 and 0.5 mg/mL) did not show any zone of inhibition, while the positive control (ciprofloxacin) had an 11 mm diameter of the inhibition zone. Ciprofloxacin, considered a standard in antimicrobial susceptibility analysis against P. aeruginosa 33. This indicates that there might be a possibility of exploring the bacterial activity of chrysin, particularly for quorum-sensing-related pathogenicity at a concentration range of around 1 mg/ml (Fig. 6). The minimum inhibitory concentration (MIC) of the drug Chrysin was calculated and found to be 0.9 mg/mL.
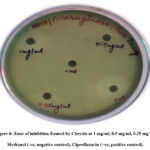 |
Figure 6: Zone of inhibition formed by Chrysin at 1 mg/ml, 0.5 mg/ml, 0.25 mg/ml, Methanol (-ve, negative control), Ciprofloxacin (+ve, positive control). |
Antibiofilm assay
Biofilm formation is an indication of the virulence trait of P. aeruginosa, which is formed by complex regulation of genes 35. In our study, chrysin showed a positive response in biofilm inhibition, with an increase in the concentration range from 0.7 to 1.3 mg/ml along with the control depicted in Fig. 7a-c. It depicted that until 0.9 mg/ml of chrysin concentration, the biofilm could be seen on the wall of a microtiter plate stained with crystal violet more clearly, but as we increased the chrysin concentration, the biofilm formation diminished. This study’s findings suggest a strong link between biofilm inhibition and the inhibition of quorum-sensing-related pathogenicity. The data from the study done on the antibiofilm assay are shown in graph form in Graph 1.
 |
Figure 7: (a-c) Rows of microtiter plates stained with crystal violet demonstrating biofilm adherence on the wall at concentrations ranging from 0.7 to 1.5 mg/ml of pure drug Chrysin and control. |
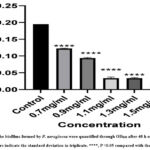 |
Graph 1: The biofilms formed by P. aeruginosa were quantified through OD600 after 48 h of incubation. Error bars indicate the standard deviation in triplicate. ****, P <0.05 compared with the control. |
Discussion
QS inhibition, adopted by the majority of bacterial pathogens, is a novel approach for identifying the compounds that would prove to be promising therapeutics for fighting antibiotic resistance. Plant bioactives have shown positive responses to infections for decades, and phytochemicals such as garlic 36, curcuma longa 37, and caffeine38 show potent responses to QS inhibition. Our study aimed to probe the potential of polyphenols to inhibit QS-regulated pathogenicity (biofilm). From the molecular docking study, carried out by Autodock Vina 1.1.2 [29, 30], results revealed that Chrysin, Galangin, Coumestrol, 3′, 4′, 7-Trihydroxyisoflavanone, Dihydrodaidzein, and Dihydroformononetin showed good docking scores among 752 compounds, extracted from the Phenol Explorer database [16–18]. The docking scores according to the order of binding affinity to the LasR receptor were found to be Dihydrodaidzein = 3′, 4′, 7-Trihydroxyisoflavanone = Dihydroformononetin (-11.3 kcal/mol) > Coumestrol (-11.2 kcal/mol) > Chrysin = Galangin (-11.1 kcal/mol) Table 3, with a lesser docking score than the indigenous LasR ligand OHN (-8.7 kcal/mol). As a result, as the affinity for the receptor increases, so do the pharmacological responses. The drug likeliness and ADMET profile were checked through the online server SwissADME [25], and these six compounds (Chrysin, Galangin, Coumestrol, 3′, 4′, 7-Trihydroxyisoflavanone, Dihydrodaidzein, and Dihydroformononetin) showed lesser toxicity, good drug likeliness and pharmacokinetic profile among 35 compounds Table 2a, b, c. From these six compounds, chrysin was purchased due to its easy availability, to assess the in vitro study on P. aeruginosa PAO1 for QS-related pathogenicity (biofilm formation). Research carried out by Singh et al., showed the collective effect of honey polyphenol components that included apigenin, pinocembrin, chrysin, pinobanksin, quercetin, caffeic acid, and kaempferol14. Our study concentrated on a single effective component (chrysin) retrieved from the Phenol Explorer database after screening 752 compounds for QS-related pathogenicity. Also, the research done by De et al. does not account for VS in the Phenol Explorer database 15. Our study is the first to explore the potential of the Phenol Explorer16-17 database against QS-related pathogenicity (biofilm formation). The molecular dynamics study was also carried out using Gromacs version 2022.2 of the LasR receptor protein and chrysin (possible lead compound), the result showed a positive response in the stability of the system, particularly after 50 ns from the RMSD graph. Potential energy, the radius of gyration, and the RMSF graph also supported our finding. The antimicrobial assay (Fig. 6) of chrysin against P. aeruginosa PAO1 showed effective results and led to exploring the bacterial activity of the chrysin drug, particularly for QS-related pathogenicity at a concentration range of around 1 mg/ml. The MIC value of chrysin was found to be 0.9 mg/ml, and an antibiofilm assay (Fig. 7a-c) was performed starting from higher sub-lethal concentrations to increasing concentrations of chrysin (0.7, 0.9, 1.1, 1.3, and 1.5 mg/ml). As shown in Graph 1, increasing the concentration of the drug Chrysin inhibited biofilm formation, which is most likely due to QS inhibition and its effect on P. aeruginosa PAO1 growth. This study supports that chrysin could be used alone or in combination, as a possible lead compound for QS inhibition in P. aeruginosa PAO1.
Conclusions
From our study, it is inferred that chrysin can be used as a lead compound alone or in combination to inhibit QS-related pathogenicity. Chrysin showed an affinity for binding with the 2UV0 protein of P. aeruginosa with closeness to residue positions 20–147 on the E chain protein and a Vander Waals force of interactionthat will interfere with the LasR-mediated QS signalling pathway responsible for the bacterial pathogenicity and biofilm formation. From the antibiofilm assay of Graph 1 and Fig. 7a, b, c, it is inferred that chrysin can be employed in pre-clinical studies and clinical trials against assessing QS-mediated actions caused by P. aeruginosa, which is an alarming pathogen, especially in community-acquired infections that have become almost resistant to most of the antibiotics 39.
Acknowledgement
The author thanks Dr. Pramod Kumar Yadav from the Department of Computational Biology & Bioinformatics of Sam Higginbottom University of Agriculture, Technology and Sciences, Naini, Prayagraj for his help in lab for molecular dynamic simulation. Arnica F Lal has done all the computational and biological work that was checked by Dr. Pushpraj S Gupta.
Conflict of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Funding Sources
The authors alone are responsible for the content and writing of the paper
References
- Spagnolo A.M, Sartini M, Cristina M.L. Pseudomonas aeruginosa in the healthcare facility setting. Rev. Med. Microbiol., 2021;32(3):169-75.
CrossRef - Hernandez-Jimenez P, Lopez-Medrano F, Fernandez-Ruiz M, Silva J.T, Corbella L, San-Juan R, Lizasoain M, Díaz-Reganon J, Viedma E, Aguado J.M. Risk Factors and Outcomes for Multidrug Resistant Pseudomonas aeruginosa Infection in Immunocompromised Patients. Antibiotics, 2022; 11(11):1459.
CrossRef - Kothari A, Jain N, Kishor Kumar S, Kumar A, Kaushal K, Kaur S, Pandey A, Gaurav A, Omar BJ. Potential Synergistic Antibiotic Combinations against Fluoroquinolone-Resistant Pseudomonas aeruginosa. Pharmaceuticals, 2022;15(2):243.
CrossRef - Bassetti M, Vena A, Croxatto A, Righi E, Guery B. How to manage Pseudomonas aeruginosa infections. Drugs in context. 2018; 7.
CrossRef - Morales E, Cots F, Sala M, Comas M, Belvis F, Riu M, Salvado M, Grau S, Horcajada J.P, Montero M.M, Castells X. Hospital costs of nosocomial multi-drug resistant Pseudomonas aeruginosa acquisition. BMC Health. Serv. Res., 2012; 12(1): 1-8.
CrossRef - Langendonk R.F, Neill D.R, Fothergill J.L. The building blocks of antimicrobial resistance in Pseudomonas aeruginosa: implications for current resistance-breaking therapies. Front. Cell. Infect. Microbiol., 2021; 11:665759.
CrossRef - Lal A.F, Singh S, Franco F.C, Bhatia S. Potential of polyphenols in curbing quorum sensing and biofilm formation in Gram-negative pathogens. Asian Pac. J. Trop. Biomed., 2021; 11(6): 231.
CrossRef - Yan S, Wu G. Can biofilm be reversed through quorum sensing in Pseudomonas aeruginosa? Front. Microbiol., 2019; 10: 1582.
CrossRef - Mulcahy L.R, Isabella V.M, Lewis K. Pseudomonas aeruginosa biofilms in disease. Microb. Ecol., 2014; 68(1): 1-12.
CrossRef - Miklasinska-Majdanik M, Kepa M, Wojtyczka R.D, Idzik D, Wasik T.J. Phenolic compounds diminish antibiotic resistance of Staphylococcus aureus clinical strains. Int. J. Environ. Res. Public. Health., 2018; 15(10): 2321.
CrossRef - Coppo E, Marchese A. Antibacterial activity of polyphenols. Curr. Pharm. Biotechnol., 2014; 15(4): 380-390.
CrossRef - Tomas-Menor L, Barrajon-Catalan E, Segura‐Carretero A, Marti N, Saura D, Menendez J.A, Joven J, Micol V. The promiscuous and synergic molecular interaction of polyphenols in bactericidal activity: an opportunity to improve the performance of antibiotics?. Phytother. Res., 2015; 29(3): 466-473.
CrossRef - Ferreira L.G, Dos Santos R.N, Oliva G, Andricopulo A.D. Molecular docking and structure-based drug design strategies. Molecules., 2015; 20(7): 13384-421.
CrossRef - Singh B.R, Shoeb M, Sharma S, Naqvi A.H, Gupta V.K, Singh B.N. Scaffold of selenium nanovectors and honey phytochemicals for inhibition of Pseudomonas aeruginosa quorum sensing and biofilm formation. Front. Cell. Infect. Microbiol., 2017; 7: 93.
CrossRef - De Marco S, Piccioni M, Pagiotti R, Pietrella D. Antibiofilm and antioxidant activity of propolis and bud poplar resins versus Pseudomonas aeruginosa. Evid-based Complement. Altern. Med., 2017.
CrossRef - Neveu V, Perez-Jimenez J, Vos F, Crespy V, du Chaffaut L, Mennen L, Knox C, Eisner R, Cruz J, Wishart D, Scalbert A. Phenol-Explorer: an online comprehensive database on polyphenol contents in foods. Database., 2010. doi: 10.1093/database/bap024.
CrossRef - Rothwell J.A, Urpi-Sarda M, Boto-Ordoñez M, Knox C, Llorach R, Eisner R, Cruz J, Neveu V, Wishart D, Manach C, Andres-Lacueva C, Scalbert A. Phenol-Explorer 2.0: a major update of the Phenol-Explorer database integrating data on polyphenol metabolism and pharmacokinetics in humans and experimental animals. Database., 2012. doi: 10.1093/database/bas031.
CrossRef - Rothwell J.A, Pérez-Jiménez J, Neveu V, Medina-Ramon A, M’Hiri N, Garcia Lobato P, Manach C, Knox K, Eisner R, Wishart D, Scalbert A. Phenol-Explorer 3.0: a major update of the Phenol-Explorer database to incorporate data on the effects of food processing on polyphenol content. Database., 2013. doi: 10.1093/database/bat070.
CrossRef - Szklarczyk D, Gable A.L, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva N.T, Morris J.H, Bork P, Jensen L.J. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic. Acids. Res., 2019. 47(D1): D607-D613.
CrossRef - Schrodinger L and DeLano W. PyMOL, 2020. Available at: http://www.pymol.org/pymol. [Assessed July 2021].
- Chowdhury N and Bagchi A. Molecular insight into the activity of LasR protein from Pseudomonas aeruginosa in the regulation of virulence gene expression by this organism. Gene. 2016; 580(1):80-7.
CrossRef - Qiu S, Azofra L.M, MacFarlane D.R, Sun C. Hydrogen bonding effect between active site and protein environment on catalysis performance in H 2-producing [NiFe] hydrogenases. Phys. Chem. Chem. Phys., 2018; 20(9): 6735-6743.
CrossRef - Pace C.N, Fu H, Lee Fryar K, Landua J, Trevino SR, Schell D, Thurlkill R.L, Imura S, Scholtz J.M, Gajiwala K, Sevcik J. Contribution of hydrogen bonds to protein stability. Protein Sci., 2014; 23(5):652-61.
CrossRef - Hung C.L, Kuo Y.H, Lee S.W, Chiang Y.W. Protein Stability Depends Critically on the Surface Hydrogen-Bonding Network: A Case Study of Bid Protein. J. Phys. Chem. B. 2021;125(30):8373-82.
CrossRef - Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific reports., 2017; 7(1): 1-13.
CrossRef - Lal A.F, Giri S, Bhatia S, Singh S, Sonia M. Identification of ellagitannins as natural inhibitors of spike proteins of COVID19 virus: An in silico-based study for drug development. Afr J Health Sci., 2020; 33(5): 78-97.
- Bhatia S, Giri S, Lal A.F, Singh S. Identification of potential inhibitors of dietary polyphenols for SARS-CoV-2 M protease: An in silico study. One Health Bull., 2020; 1: 21-29.
- Kim H.S, Lee S.H, Byun Y, Park H.D. 6-Gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition. Scientific reports., 2015; 5(1): 1-11.
CrossRef - Forli S, Huey R, Pique M.E, Sanner M.F, Goodsell D.S, Olson A.J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc., 2016; 11(5):905-19.
CrossRef - Chatterjee A, Roy U.K, Haldar D. Case Study and Performance Analysis of AutoDock VS AutoDock Vina for Stable Drug Design, 2018; 7. doi = {10.24214/jecet.B.7.2.15779}.
CrossRef - Abraham M.J, Murtola T, Schulz R, Pall S, Smith J.C., Hess B, Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX., 2015;1:19-25.
CrossRef - Bansal A, Srivastava N, Nagpal K. Development and Validation of UV Spectrophotometric Method for Determination of Chrysin and Its Solubility Studies. J. Appl. Spectrosc., 2022; 89(1): 150-8.
CrossRef - Yasir M, Dutta D, Willcox M.D. Activity of antimicrobial peptides and ciprofloxacin against Pseudomonas aeruginosa biofilms. Molecules., 2020; 25(17):3843.
CrossRef - Kordmahaleh F.A, Shalke S.E. Bacterial biofilms: microbial life on surfaces. J. Biol., 2013; 2(5): 242-8.
- Elnegery A.A, Mowafy W.K, Zahra T.A, Abou El-Khier N.T. Study of quorum-sensing LasR and RhlR genes and their dependent virulence factors in Pseudomonas aeruginosa isolates from infected burn wounds. Access. Microbiol., 2021; 3(3).
CrossRef - Farrag H.A, Hosny A.E, Hawas A.M, Hagras S.A, Helmy O.M. Potential efficacy of garlic lock therapy in combating biofilm and catheter-associated infections; experimental studies on an animal model with focus on toxicological aspects. Saudi Pharmaceutical Journal. 2019; 27(6):830-40.
CrossRef - Packiavathy I.A, Priya S, Pandian S.K, Ravi A.V. Inhibition of biofilm development of uropathogens by curcumin–an anti-quorum sensing agent from Curcuma longa. Food chem. 2014; 148: 453-60.
CrossRef - Norizan S.N, Yin W.F, Chan K.G. Caffeine as a potential quorum sensing inhibitor. Sensors. 2013; 13(4): 5117-29.
CrossRef - Gellatly S.L and Hancock R.E. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis., 2013; 67(3):159-73.
CrossRef







