Manuscript accepted on :30-03-2023
Published online on: 15-06-2023
Plagiarism Check: Yes
Reviewed by: Dr. Pratibha Kakadia
Second Review by: Dr. Dhruv Desai
Final Approval by: Dr. Patorn Promchai
Firew Elias1,3 , Sudhamani Muddada1
, Sudhamani Muddada1 , Diriba Muleta2*
, Diriba Muleta2* and Belachew Tefera3
and Belachew Tefera3
1Koneru Lakshmaiah Education Foundation, Department of Biotechnology, Guntur, India
2Environmental Biotechnology Unit, institute of Biotechnology, Addis Ababa University, Ethiopia.
3Animal Products Veterinary Drugs and Feed Quality Control Centre of Ethiopia
Corresponding Author E-mail: firew2010@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/2679
Abstract
Many bioactive secondary metabolites with intriguing antibacterial, antiviral, and anticancer properties have been produced by Streptomyces species. The objective of this work is to use conventional and statistical techniques to improve the antibiotic production medium of Streptomyces monomycini RVE129, which was isolated from rhizospheric soil in Hawassa, Ethiopia. The main media components were chosen using the one factor at a time method and the Plackett-Burman design, which was then, further, optimized using the Box-Behnken Design for increased antibiotic production. On ISP4 medium (10 g/L starch, 1 g/L NaCl, 1 g/L MgSO4.7H2O, 2 g/L (NH4) 2SO4, 2 g/L CaCO3and 1 g/L K2HPO4, 0.1 g/L FeSO4·7H2O, 0.1 g/L MnCl2·4H2O, 0.1 g/L ZnSO4·7H2O), S. monomycini RVE129 produced the greatest amount of antibiotics. Starch and soybean meal were found to be the best sources of carbon and nitrogen for the strainRVE129. During the eighth day of incubation under shaking conditions, the best conditions for antibiotic synthesis were determined at a temperature of 30°C and a pH of 7.5. Plackett-Burman design identified K2HPO4, starch, and soybean meal as having the highest influence on antibiotic synthesis with a confidence level above 95%. The yield of producing antibiotics increased by 24.30% when the concentration of critical variables was further improved by using the Box-Behnken Design of the Response Surface approach. The optimum concentration was 20 g/L starch, 7.5 g/L s oybean meal, and 1.25 g/L K2HPO4. To the best of our knowledge, this is the first investigation into medium optimization for the production of the antibiotic from S. monomycini RVE129.
Keywords
Antimicrobial metabolites; Biomass; Optimization; Streptomyces monomycini
Download this article as:| Copy the following to cite this article: Elias F, Muddada S, Muleta D, Tefera B. Improved Antibiotic Activity from Streptomyces monomycini strain RVE129 Using Classical and Statistical Design of Experiments. Biomed Pharmacol J 2023;16(2). |
| Copy the following to cite this URL: Elias F, Muddada S, Muleta D, Tefera B. Improved Antibiotic Activity from Streptomyces monomycini strain RVE129 Using Classical and Statistical Design of Experiments. Biomed Pharmacol J 2023;16(2). Available from: https://bit.ly/45ZUsMl |
Introduction
Among the actinomycetes, the genus Streptomyces contains largest number species and is the source of the most well-known secondary metabolites. They have also produced novel bioactive natural compounds, the majority of which are highly useful as antimicrobial, antiviral, anticancer, antiparasitic, and antihelminthic agents1, 2. In search of novel antibiotics with distinct modes of action to treat a variety of drug-resistant diseases and other disorders, research into new and novel bioactive metabolites produced by Streptomyces is still ongoing and has gained greater attention in recent years3. Hence, it is considered that Streptomyces sp. is a potential source for new antimicrobial substances with applications in biotechnology, medicine, and other fields4, 5.
Streptomyces species capacity to produce secondary metabolites is a dynamic phenomenon. The nutritional and cultural conditions of the cultivation medium can have a significant impact on the production of secondary metabolites due to their high complexity5. Minor change in the fermentation medium composition can have a significant impact on the yield and metabolic profile of microorganisms. Optimizing the nutritional and environmental aspects of the fermentation conditions is crucial to increase biomass and the production of antibiotics by the potent Streptomyces sp6. Successive experiments are required to provide the optimum condition for growing producer strains, including selecting the most suitable media, adjusting the level of carbon, nitrogen, and trace elements in the medium, and optimizing temperature, incubation time, growth pH, and other physical culture parameters7. The techniques used for the medium optimization of various fermentation parameters can be carried out by using a conventional approach, the one-variable-at-a-time approach method8, 9. After that, it is possible to identify important medium components for their relevant levels using Design-of-Experiments (DOE) techniques such the Plackett Burman Design (PBD) and Response Surface Methodology (RSM). For the purpose of screening the components of the production medium in shake flask fermentation, the PBD statistical technique is well-known and often used9, 10. RSM can then be used to optimize key fermentation media parameters and their levels using mathematical and statistical tools11, 12.
S. monomycini RVE129, a promising strain with a wide range of antibiotic action, was previously isolated from the central Rift Valley regions of Hawassa, Ethiopia as part of our search for novel antibiotic metabolites13. Through medium and growth condition optimization using one-variable-at-a-time and statistical tools that significantly improved antibiotic production of strain RVE129, this research aimed to determine the best growth conditions for enhancing the production of antibiotics by S. monomycini strain RVE129.
Materials and Methods
Test strains and their maintenance
An antibiotic-producing strain previously isolated from the rhizosphere soil collected from the Rift Valley region of Hawassa, Ethiopia, was used in this investigation13. The strain was identified and designated as Streptomyces monomycini RVE129. The strain was stored at 4°C as a slant culture using a tryptic soy agar medium and at -70 to -80°C in the tryptic soy broth (TSB) containing (15%, v/v) glycerol in the deep freezer14. The test microorganism, S. aureus ATCC-259233, was supplied by the Ethiopian Health and Nutrition Research Institute (EHNRI) and maintained under refrigerated conditions.
Inoculum preparation
To prepare a spore suspension of the fermentation inoculum of the strain, a colony was transferred from a seven-day-old culture grown on tryptic soya medium plates by suspending with 10 mL of sterile normal saline, and then the suspension of the strain was used as a seed culture14, 15. Conical flasks with a volume of 250 mL containing 100 mL of TSB broth medium were used for the experiments. The medium was inoculated with 5.0 mL of spore suspension at a density of 1×108spores/mL, at 150 rpm for 3 days at 30°C and then used as fermentation seed stock15, 16.
Culture media selection for antibiotic activity and growth
The prepared inoculum of Streptomyces sp. was transferred separately into each of the eight different microbial growth media. The eight types of media used were: tryptone yeast extract broth (TYE), yeast extract-malt extract-dextrose (YMD), oatmeal broth (OM), starch inorganic salts broth (SIS), glycerol-asparagine broth (GA), starch casein broth (SC), tyrosine broth (TB), and glucose soybean meal broth (GSB), (with pH=7±0.2) for the selection of a suitable basal medium for the production of antibiotics and growth of mycelia biomass, and the optimum medium was screened for further production of antimicrobial compounds from RVE129. In 250 mL Erlenmeyer flasks, five mL (10%, v/v) of 1×108 spores/mL density were inoculated into 100 mL of various sterile media and cultured for 8 days at 30°C in a shaker at 150 rpm16.
Determination of antibiotic activity
The fermentation broth was taken aseptically and each culture broth of S. monomycini RVE129 was centrifuged at 10,000 rpm for 10 min, followed by filtration to separate the cell-free supernatant and the mycelia biomass to determine antibiotic activity and growth17. To obtain a crude extract, the culture broth supernatant was combined 1:1 with ethyl acetate, agitated for one hour, and then evaporated using a rotary vacuum evaporator. The extract was then bio-tested using the conventional disc diffusion method against Staphylococcus aureus ATCC 2592317, 18. On Mueller Hinton agar (MHA) plates, 0.2 ml of (0.5 McFarland) an overnight culture suspension of S. aureus ATCC 25923 having 1.5 108 CFU/mL was evenly and aseptically spread. A 100 L amount of antibiotic extract was poured on sterile discs with an agar plate diameter of 6 mm. As a negative control, sterile discs (6.0 mm in diameter) filled with ethyl acetate were used and incubated for 24 hours at 37°C. The diameter of the inhibition zone (ZI) was measured and recorded following incubation.
Determination of growth
Mycelium collected from the previous experiment was used to evaluate the strain’s growth. The mycelia were dried in a 70°C oven overnight, and the strain growth was calculated as the dry cell weight in g/L of culture medium19.
Optimization of nutritional conditions
The suitability of various sources of carbon and nitrogen supplemented to the basal medium for growth and antibiotic production by S. monomycini RVE129 was evaluated following standard procedures. Carbon sources such as glucose, fructose, galactose, lactose, sucrose, cellobiose, mannose, mannitol, and glycerol were added separately into the medium at a rate of 1% (w/v) while other parameters remained constant19, 20. Similarly, nitrogen sources like ammonium sulfate, ammonium chloride, malt extract, soya bean meal, peptone, yeast extract, and casein were individually supplemented in the production media at a 0.3% (w/v) level while other constituents remained constant21.
Optimization of growth conditions
To select the best incubation period for the growth and antibiotic production, the S. monomycini RVE129 strain was inoculated into 100 ml of the basal medium into each 250 mL conical flask and incubated for 1–14 days in a shaker at 150 rpm at 30°C using modified starch inorganic salts broth (SIS) medium at pH 7.5 with some modification22. During fermentation, 5 mL culture samples were taken aseptically at 24 h intervals, and the pellets were collected from the broth culture by centrifugation.
Similarly, the optimum pH for maximum antibiotic and biomass production was examined by changing the pH (4–11) of the basal medium. A 100 mL medium contained in a 250 mL Erlenmeyer flask was seeded with five mL of the spore suspension and incubated on a rotary shaker at 150 rpm at 30°C for eight days to determine antibiotic activity and growth. The S. monomycini RVE129 strain was inoculated into a modified SIS broth production medium and incubated for 8 days at various temperatures (20, 25, 30, 35, 40, and 45°C) in a shaking incubator (150 rpm) at pH 7.5. The antibiotic activity of crude extracts from mycelia-free culture filtrate was concentrated in a vacuum evaporator, and 50 μL was assayed to find the best incubation period, pH, and temperature for maximum antibiotic activity23. The dry cell weight of collected cells was reported as g/L of culture media and was used to determine growth.
Statistical optimization medium components
Plackett-Burman Design (PBD)
The most important goal of screening is to identify the major effects of significant nutritional factors. Based on the findings of OFT, soybean meal and starch were found to be excellent N and C sources for triggering the highest antibiotic activity used for screening purposes. Therefore, the already screened nitrogen and carbon sources were used along with other constituents for media optimization experiments. The PBD screening aids in identifying the most crucial media components which have a significant impact on the production of bioactive metabolites from a vast pool of available candidates24-26. The most crucial medium components for S. monomycini RVE129 to produce antibiotics were determined using a PBD. Minitab 18.0 (Minitab Inc., PA, USA) software, which was also utilized to analyze the experimental data, was used to design the trials. In the experimental design, a total of eight medium components (independent factors) were studied by showing them at two levels, low (-1) and high (1), with 12 trials. The studies were conducted in triplicate, and the result was recorded as the average antibiotic activity against S. aureus. The details of the PBD are shown in Table 1. The factors that had confidence levels above 95% were expected to have a significant effect on the production of the antibiotic and were selected for further optimization.
Table 1: Plackett Burman Design determining high and low levels of each variables.
|
Variables |
Media components (g/L) |
levels |
|
|
-1 |
1 |
||
|
A |
Starch |
5 |
20 |
|
B |
Soybean meal |
2.5 |
7.5 |
|
C |
Yeast extract |
2 |
8 |
|
D |
CaCO3 |
1 |
3 |
|
E |
K2HPO4 |
1 |
3 |
|
F |
NaCl |
1 |
3 |
|
G |
FeSO4.7H2O |
0.0001 |
0.003 |
|
H |
ZnSO4.7H2O |
0.0001 |
0.003 |
Response Surface Methodology (RSM)
The PBD experiments were used to determine the essential media ingredients that positively influence antibiotic synthesis, including starch, soybean meal, and K2HPO4. The optimum concentration of these elements was then determined in order to increase the production of antibiotics by RSM in S. monomycini RVE129.RSM optimization not only enables quick screening of a vast experimental domain but also considers the significance of each component25-26. The Box-Behnken Design (BBD) design matrix was used to optimize the concentration of the selected components. The remaining media components’ levels were set to 0.The observed values were the average of three replications. Each response was used to fit a distinct second-order polynomial model after being measured for each trial. Following the use of BBD, the regression shown in Eq. 1 below demonstrates an experimental association between the logarithmic values of antibiotic activity and the study factors.
𝑌=𝛽0 + Σ𝛽𝑖𝑋𝑖 + Σ𝛽𝑖𝑗𝑋𝑖𝑋𝑗 + Σ𝛽𝑖𝑖𝑋2𝑖 (1)
Y is the predicted response (antibiotic activity), β0 is the constant term coefficient, 𝛽𝑖 is the linear coefficient, 𝛽𝑖𝑗 is the quadratic coefficient, 𝛽𝑖𝑖 is the interaction coefficient, and XiXj represents the independent variables. Using Minitab 18.0 software a regression analysis of the collected data was carried out. An analysis of variance was used to determine the model’s statistical significance (ANOVA). Model values, Fisher’s F-test, and significance probability P (𝐹) were the essential calculations to determine the overall model significance. Regression models have a high degree of reliability when their F-and P-values are large. The coefficient of determination (R2) and adjusted R2 were used to statistically confirm the accuracy of the polynomial model equation27.The relationship between the responses and the experimental values of each independent variable was then depicted using three-dimensional response surface plots to illustrate the fitted polynomial equation.
Experimental validation
By cultivating S. monomycini strain RVE129 in both unoptimized and optimized production media in shaking flasks, the combination of various optimized factors that produced the maximum response was experimentally validated. The upper organic layer of the fermented broths was dried for further examination after the cell-free supernatant was collected and extracted with an equal volume of ethyl acetate18. The antibacterial activity was examined using an extracted antibiotic.
Results
Optimal nutrient medium selection
Different medium compositions, nutrition, and growing conditions all have an impact on a microorganism’s potential to produce antimicrobial metabolites. In this work, the medium utilized for cultivation of the S. monomycini RVE129 strain, nutrient sources (carbon, nitrogen, and minerals), and culture conditions (incubation temperature, pH of production medium, and duration of fermentation) were studied for increased antibiotic production. Antibiotic activity of the strain was examined by growing it in various production media (Fig. 1). Among eight different liquid media tested, SIS broth medium showed maximum antibiotic production with the highest inhibition zone diameter (27.04±0.26 mm), followed by YMD broth medium (25.3±0.54) and GSB broth medium (23±0 mm) against S. aureus by the S. monomycini RVE129 strain (Figure 1). Regarding cell growth, the highest biomass (3.8±0.23 mg/ml) was obtained with modified SIS, followed by the culture filtrate grown on SC (3.6±0.12 mg/mL). Other broth media investigated were TYE, OM, GA, and TB, which were found to have lower growth as well as the synthesis of the antibiotic compound. SIS medium containing 10 g/L starch, 1 g/L NaCl, 1 g/L MgSO4.7H2O, 2 g/L (NH4) 2SO4, 2 g/L CaCO3and 1 g/L K2HPO4, 0.1 g/L FeSO4·7H2O, 0.1 g/L MnCl2·4H2O, 0.1 g/L ZnSO4·7H2O, showed the highest antibiotic productivity. Therefore, it was selected for further experiments as the best basal medium for antibiotic production as well as the growth of S. monomycini RVE129 in batch fermentation.
 |
Figure 1: Influence of various culture media on antibiotic production by |
To enhance antimicrobial activity, attempts were made under optimized nutritional conditions to culture media by growing Streptomyces sp. As presented in Figure 2, the effects of different carbon sources on antibiotic production by S. monomycini RVE129 were investigated with modified SIS broth selected as the production medium. The strain was capable of producing antibiotics in all of the carbon sources tested, although the maximum biomass (3.766 mg/mL) was recorded in a medium supplemented with starch (Fig 2). Similarly, the medium treated with soluble starch as a carbon source produced the maximum antibiotic activity (a zone of inhibition of 27.56 mm). The carbon sources like glucose, lactose, glycerol, mannose, and galactose were recorded as comparatively remarkable antibiotic activity inhibition zones, ranging from 16.95 to 25.3 mm (Fig. 2). However, fructose and manitol supplemented with the production medium also favoured the growth of the strain RVE129. The antibiotic activity recorded was lower, ranging from 6.7 to 9.8 mm when compared with starch (Fig. 2).
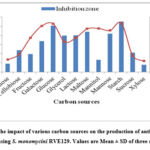 |
Figure 2: The impact of various carbon sources on the production of antibiotics and growth using S. monomycini RVE129. Values are Mean ± SD of three replicates. |
Similarly, different nitrogen sources supplemented in the media tested with S. monomycini RVE129 supported various levels of effects on antibiotic biosynthesis as well as biomass yields. As shown in Fig 3, of all the examined nitrogen sources, higher antibiotic activity and good growth were recorded with tryptone (inhibition zone 24±06 mm) followed by peptone (inhibition zone 21.18±1.2 mm). However, the highest antibiotic activity was found to be with soybean meal, which was shown as the suitable nitrogen source for maximum growth (3.65 mg/mL) as well as antibiotic production (inhibition zone 27.1±0.21) (Fig. 3). No antibiotic activity was recorded with urea.
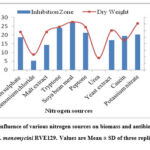 |
Figure 3: Influence of various nitrogen sources on biomass and antibiotic activity of S. monomycini RVE129. Values are Mean ± SD of three replicates. |
Antibiotic production and growth by many species of the genus Streptomyces are greatly influenced by optimal nutritional and cultural parameters. Suitability of incubation time for antibiotic production as well as growth was performed at a fixed range of time (1–14) days by cultivating S. monomycini RVE129 in production medium in shake flask condition (Fig. 4A). It is clear that the growth of the strain was detected only after two days of incubation, whereas little-noticed antibiotic production began on the third day and then increased until it reached a maximum 27.64 mm zone of inhibition on the eighth day of incubation. The antibiotic activity remained more or less stable until the 10th day, while the mycelia biomass and antibiotic activity started to decrease gradually (Fig. 4A). The temperature of the incubation has an effect on biomass and antibiotic activity. The results of various incubation temperatures on the biomass and antibiotic biosynthesis by the S. monomycini RVE129 strain are indicated in Figure 6. In this study, antibiotic activity was recorded at temperatures between 20 and 45 °C. The optimum growth (3.76 mg/mL), as well as antibiotic production (a zone of inhibition of 27.3 mm), was recorded at 30 °C (Fig. 4B).
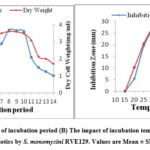 |
Figure 4(A): The impact of incubation period (B) The impact of incubation temperature on biomass and production of antibiotics by S. monomycini RVE129. Values are Mean ± SD of three replicates. |
An experiment was carried out to find out the influence of pH on growth and antibiotic production by the strain RVE129. The findings observed in Fig. 5 revealed that antibiotic production increased with increasing pH from 4.0 to 7.5, but maximum growth (3.78 mg/mL) and the highest antibiotic activity (27.66 mm zone of inhibition) were recorded at pH 7.5. Both lower and higher pH values were unfavorable and caused a decline in both growth and the level of antibiotic activity. The findings clearly state that there is almost a positive correlation between pH and antibiotic production by S. monomycini RVE129.
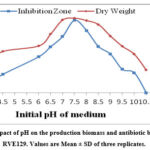 |
Figure 5: The impact of pH on the production biomass and antibiotic by S. monomycini RVE129. Values are Mean ± SD of three replicates. |
A statistical approach for optimization of the fermentation medium
Screening of significant medium ingredients by PBD
Based on the findings of the antibiotic assay, Table 2 shows the levels of selection, an evaluation component, and antibacterial activity for each experiment. A trial is represented by each column representing a single variable (medium components), while each row represents one trial, either high (1) or low (-1), within each experimental trial. The antibacterial activity (mm) for each experimental design was considered the response value.
Table 2: PB design matrix and its experimental results obtained for S. monomycini RVE129
|
Run Order |
Variables |
Antibiotic Activity (mm) |
|||||||
|
A |
B |
C |
D |
E |
F |
G |
H |
||
|
1 |
1 |
1 |
1 |
-1 |
1 |
1 |
-1 |
1 |
27.00± 1.54 |
|
2 |
-1 |
1 |
-1 |
-1 |
-1 |
1 |
1 |
1 |
25.00± 1.63 |
|
3 |
-1 |
-1 |
-1 |
-1 |
-1 |
-1 |
-1 |
-1 |
9.86±2.11 |
|
4 |
1 |
1 |
-1 |
1 |
-1 |
-1 |
-1 |
1 |
33.82± 0.77 |
|
5 |
1 |
-1 |
1 |
-1 |
-1 |
-1 |
1 |
1 |
16.80± 0.00 |
|
6 |
-1 |
-1 |
-1 |
1 |
1 |
1 |
-1 |
1 |
6.83±1.55 |
|
7 |
1 |
1 |
-1 |
1 |
1 |
-1 |
1 |
-1 |
29.30±1.54 |
|
8 |
-1 |
1 |
1 |
-1 |
1 |
-1 |
-1 |
-1 |
17.36±0.61 |
|
9 |
1 |
-1 |
1 |
1 |
-1 |
1 |
-1 |
-1 |
27.61±0.00 |
|
10 |
1 |
-1 |
-1 |
-1 |
1 |
1 |
1 |
-1 |
14.63±0.00 |
|
11 |
-1 |
1 |
1 |
1 |
-1 |
1 |
1 |
-1 |
26.30±0.63 |
|
12 |
-1 |
-1 |
1 |
1 |
1 |
-1 |
1 |
1 |
6.87± 2.77 |
The effect, standard error, +value, -value, and confidence level of each component are shown in Table 3. The three medium components, starch (A), soybean meal (B), and K2HPO4 (E), were found to be the most significant in the production of antibiotics based on the low P-values (p<0.001), which was evident from their confidence levels above 99% (Table 3). It was further supported by the Pareto chart of the standardized effects (Figure 7), which shows that the highest effects are shown for the upper fields while the minimal impacts are shown for the lower fields, with close to zero in the upper portion. The relationship between the t-value (effect) and ranks was shown by using a horizontal reference line with the statistical significance (t=3.18) (Figure 7). For S. aureus, any effect that exceeds this reference line is regarded as significant.
Table 3: Statistical analysis of the data generated by the PBD
|
Coded variable |
Medium component |
Effect |
SE |
t-Value |
P-value |
Confidence level (%) |
|
A |
Starch |
12.217 |
0.357 |
17.09 |
0.000 |
* |
|
B |
Soybean meal |
9.323 |
0.357 |
13.04 |
0.001 |
* |
|
C |
Yeast extract |
1.127 |
0.357 |
1.58 |
0.213 |
NS |
|
D |
CaCO3 |
0.853 |
0.357 |
1.19 |
0.318 |
NS |
|
E |
K2HPO4 |
-8.797 |
0.357 |
-12.31 |
0.001 |
* |
|
F |
NaCl |
1.163 |
0.357 |
1.63 |
0.202 |
NS |
|
G |
FeSO4.7H2O |
1.877 |
0.357 |
2.63 |
0.079 |
NS |
|
H |
ZnSO4.7H2O |
2.207 |
0.357 |
3.09 |
0.054 |
NS |
Note: *; significant at the 0.001 level, NS; not significant at the 0.001 level
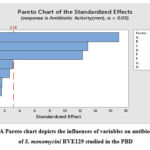 |
Figure 6: A Pareto chart depicts the influences of variables on antibiotic activity of S. monomycini RVE129 studied in the PBD |
Optimization of selected medium components by BBD
The most significantly positively affecting independent variables obtained for antibiotic production in the PBD studies were starch, soybean meal, and K2HPO4. To find out the optimal concentrations of selected media components, such as starch (A), soybean meal (B), and K2HPO4 (C), for improving the antibiotic activity of S. monomycini RVE129, RSM was used with BBD. The selected medium components (independent variables) were examined at three distinct concentrations, (−), (0), and (+) for less, moderate, and high, respectively (Table 4).
Table 4: Coded values (low, moderate, and high) levels of the BBD experimental variables for RSM
|
Variables |
Code |
Level (g/L) |
||
|
Starch |
A |
+ |
0 |
− |
|
Soybeanmeal |
B |
+ |
0 |
− |
|
K2HPO4 |
C |
+ |
0 |
− |
The optimal concentrations and interaction effects of selected media components were found in a quadratic model with 17 experimental trials and five replicas of the center point used as controls to estimate experimental error (Table 5). Following a batch of experiments using the BBD, the experimental design and the observed responses of the variables to the antibiotic activity are summarized along with the predicted value (Table 5).
The experimental BBD data (Table 5) and regression analysis (Table 6) were used to develop the quadratic polynomial equation (Eq. 2) that evaluates the relationship between the response and three variables.
𝑌 (Antibiotic activity) =27.11+0.497A+0.61B-13.08 C- 0.0394A2– 0.029 B2 +3.15 C2 + 0.2318 A*B+0.050 A*C-0.408 B*C…………………………………………………. (2)
Antibiotic activity is represented by the letter Y, whereas the codes for starch, soybean meal, and K2HPO4 are A, B, and C, respectively.
Table 5: Box-Behnken experimental design, experimental response, and predicted response (antibiotic activity).
|
Run Order |
Factors |
Antibiotic Activity(mm) ± SEM |
|||
|
Starch |
soybean meal |
K2HPO4 |
Observed |
Predicted |
|
|
1 |
0 |
0 |
0 |
25.27±0.94 |
24.67 |
|
2 |
− |
0 |
− |
24.67±1.54 |
25.16 |
|
3 |
0 |
+ |
− |
35.33±0.27 |
36.27 |
|
4 |
0 |
− |
− |
20.21±00 |
20.11 |
|
5 |
0 |
0 |
0 |
24.56±1.54 |
24.67 |
|
6 |
− |
0 |
+ |
19.65±0.81 |
19.22 |
|
7 |
0 |
0 |
0 |
23.82±2.6 |
24.67 |
|
8 |
0 |
− |
+ |
20±2.6 |
20.12 |
|
9 |
0 |
0 |
0 |
23.82±0.94 |
24.67 |
|
10 |
+ |
0 |
− |
33.28±0.81 |
33.41 |
|
11 |
− |
− |
0 |
19±0.27 |
19.13 |
|
12 |
+ |
0 |
+ |
29.27±1.54 |
29.3 |
|
13 |
0 |
0 |
0 |
25.27±0.81 |
24.67 |
|
14 |
+ |
− |
0 |
20±0.32 |
20.22 |
|
15 |
− |
+ |
0 |
19.65±0.00 |
19.31 |
|
16 |
+ |
+ |
0 |
34.56±0.32 |
35.05 |
|
17 |
0 |
+ |
+ |
32.23±0.00 |
32.11 |
Mean±SD where n=3
The response surface quadratic regression model was also statistically examined using an analysis of variance (ANOVA), and the findings are shown in Table 6. The model F value of 12.10 suggests that the proposed model is significant. The model variables with the codes A, B, C, AB, BC, and C2 are significant when the value of “prob F” is less than 0.05 (Table 6). The lower calculated F-value of 0.516, which shows that the lack-of-fit is insignificant in comparison to the pure error, shows that the statistical insignificance of the lack-of-fit value also supported the model equation and was sufficient to determine the antibiotic activity (Table 6).
Table 6: Results of ANOVA for quadratic polynomial model and regression equation.
|
Source |
DF |
SS |
MS |
𝐹 value |
𝑃 value Probability> F |
|
Model |
9 |
459.36 |
51.04 |
12.10 |
0.002 |
|
A |
1 |
145.01 |
145.01 |
34.39 |
0.001 |
|
B |
1 |
202.00 |
202.00 |
47.90 |
0.000 |
|
C |
1 |
20.23 |
20.23 |
4.80 |
0.045 |
|
A*B |
1 |
59.68 |
59.68 |
14.15 |
0.007 |
|
A*C |
1 |
0.59 |
0.59 |
0.14 |
0.021 |
|
B*C |
1 |
2.91 |
2.91 |
0.69 |
0.432 |
|
A2 |
1 |
1.28 |
1.28 |
0.30 |
0.599 |
|
B2 |
1 |
0.49 |
0.49 |
0.12 |
0.743 |
|
C2 |
1 |
28.00 |
28.00 |
6.64 |
0.037 |
|
Residual |
7 |
2.62 |
0.73 |
|
|
|
Lack-of-Fit |
3 |
27.395 |
9.132 |
17.37 |
0.516 |
|
Pure Error |
4 |
2.103 |
0.526 |
|
|
|
Total |
16 |
548.520 |
|
|
|
The coefficient of variation result (CV% = 0.75) provided additional evidence of the model’s accuracy and reliability. The second-order polynomial model Eq. 2 might indicate 99.53% variation in the response, as shown by the determination coefficient of R2 (0.9953) and adjusted coefficient of determination (0.9823), which can further demonstrate accuracy and reliability.
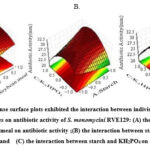 |
Figure 7: 3D response surface plots exhibited the interaction between individual and combined influences of variables on antibiotic activity of S. monomycini RVE129: |
Fig. 7 shows the interaction among the components. Response surface 3D plots showed the combined pair-wise amounts of the three factors: starch, soybean meal, and K2HPO4, while the remaining components were maintained at the middle level. The plots clearly show that higher starch and soybean meal concentrations and lower K2HPO4 concentrations favor higher antibacterial activity (Fig. 7A). With the increase in starch concentration from 15 to 20 g/L (coded values, -1 to +1), the antibacterial activity gradually increased to a maximum at a low concentration of KH2PO4 (coded values, +1 to 0.0) (Fig. 7B). The same trend was observed for the increasing concentration of soybean meal when its concentration increased from 2.5 to 10 g/L (coded values, –1 to +1) (Fig. 7C). However, as the KH2PO4 level in the fermentation medium increased, the antibacterial activity significantly decreased. Consequently, it is clear that the fermentation process was significantly impacted by the medium composition.
Experimental validation
The model and regression equation performed validation testing of the statistical results in triplicate using the optimum medium for shake flask fermentation. The maximum antibiotic activity was experimentally obtained, as predicted by a numerical optimization method, at 35.33 mm, when the optimum values of independent components in the coded units were starch (20 g/L), soybean meal (7.5 g/L), and K2HPO4 (1 g/L), respectively. The maximal antibacterial activity against S. aureus was observed to be 31.49, indicating that the experimental and predicted values were in reasonable agreement. The antibacterial activity was increased from 27.0 to 35.33 mm (S. aureus) by optimizing the medium components. This result revealed the suitability of the model for predicting the antibiotic production by S. monomycini strain RVE129.
Discussion
Many bacteria have been examined for their capacity to produce antimicrobial metabolites at their optimum. The optimal accessibility of primary metabolites as precursors determines the efficiency of antibiotic production, which in turn directs the expression of antibiotic-producing genes to activate the required metabolic pathways6. Hence, improving culture conditions is a fundamental necessity for increasing the output of secondary metabolites. Many studies have revealed that the optimum physical and nutritional characteristics of the growing conditions are necessary for both the growth of Streptomyces species and the improvement of antibiotic production 8–12.
In light of the aforementioned information, the current study was conducted to examine the effects of different culture mediums and their components (carbon, nitrogen, and mineral sources) on the efficient production of antibiotics by Streptomyces RVE129.Incubation time, temperature, and pH of the culturing conditions were examined for their effects on biomass production and antibiotic activity.The effects of incubation time, temperature, and pH on antibiotic activity and biomass production were investigated. The selection of basal medium is a crucial step in improving medium formulation and medium component optimization to increase antibiotic synthesis by Streptomyces sp. Results showed that among the culture media tested, the modified SIS broth was the optimal culture medium for improved antibiotic synthesis and growth. It was utilized as the basal media to select appropriate carbon and nitrogen sources for batch fermentation of S. monomycini RVE129. The strain that produced the maximum growth and antibiotics when grown in modified SIS media could be attributable to the fact that this composite medium contained all essential nutrients and growth factors for cellular proliferation and antibiotic production. The production of metabolites is significantly influenced by the composition of the growth medium15. Although there is no universal medium that works for all microorganisms, actinomycetes of Nonomuraea sp. JAJ18 found starch inorganic salts (ISP4) broth fermentation medium to be the best antimicrobial metabolite production and growth medium16. Moreover, noted an increase in antibiotic activity in the medium used to produce starch inorganic salts (ISP4) from Streptomyces sp. AS1120. This conclusion was found to be in line with our findings.
Carbon and nitrogen supply are critical aspects of culture growth media for increasing Actinomycetes bioactive metabolite production22. Our findings confirmed the effect of different carbon and nitrogen sources on the yield of antibiotics produced by S. monomycini RVE129.In this study, starch supported the test strain’s biomass growth and antibiotic synthesis the most, while other carbon sources only moderately and weakly supported biomass growth and antimicrobial activity. Similarly, several studies21, 22 found starch to be an efficient carbon source for increased antibiotic production. This result was consistent with that of Streptomyces rimosus NRRL 2455, which used starch as an effective source of carbon to produce the antibiotic paromomycin23. Other carbon sources provided moderate-to-low antimicrobial activity. Producing antimicrobial metabolites from a carbon source that has been completely consumed during growth would be difficult. A carbon source that is only partially utilized during biomass growth, on the other hand, may be better suited for subsequent antibiotic production 28. However, each Sterptomyces sp. has different needs for carbon sources.
In addition to carbon, the regulation of antibiotic synthesis in microbes via complicated mechanisms of glutamate synthetases depends on the assimilation of nitrogen sources29. The requirements of nitrogen sources vary depending on the type of microorganism24-28. The present study revealed that S. monomycini RVE129 grown in a medium containing soybean meal as a nitrogen source produced maximum cell growth and enhanced antibiotic production as compared to other organic and inorganic nitrogen sources. Similar results were found for soybean meal as the optimal source to produce the improved antibiotics by S. sannanensis strain SU1183, S. Albidoflavus12, S. tanashiensis A2D 21, S. violates 22, and S. rimosus NRRL 245523. Soy flour and soybean meal as protein-rich raw materials suitable for antibiotic fermentations as reported by29. Hence, soybean meal was selected as the best nitrogen source in the basal medium.
The production of bioactive metabolites by Streptomyces spp. is influenced by culture conditions such as incubation temperature, time, and pH18. After three days of growth, the antibiotic activity of the strain S. monomycini RVE129 in a modified SIS broth medium was observed. It reached its maximum on the eighth day of incubation and then remained stable for three days under optimal conditions. On the eleventh day, both the biomass and antibiotic activity began to slightly decline. As a result, the time course of 8 days was preferred as the optimum incubation period for the production of antibiotics by S. monomycini RVE129. Similar results were also obtained when they optimized various growth conditions for Streptomyces sp. production of antibiotics20. According to their findings, antibiotic production began on day three of incubation, while the highest antibiotic production was obtained on the eighth day by isolates R3 and Y8 of the genus Streptomyces.
After 8 days of incubation, antibiotic synthesis may have decreased due to the decrease in nutrients available to the microorganism or the buildup of toxic byproducts and metabolites. Therefore, although more secondary metabolites may be produced as time passes, this does not necessarily suggest that more antibiotics are being synthesized. It could produce additional toxins that hinder the synthesis of antimicrobial molecules30.
Temperature also has an influence on biomass and antibiotic production. S. monomycini RVE129 was grown and showed antimicrobial activity at temperatures ranging between 20–40 °C, and the production of antibiotics and cell biomass was reported to be maximal at a growth temperature of 30 °C. Temperatures below 20 °C or above 40 °C had a negative impact on the isolate’s growth and the amount of the antibiotic compound it could produce. Higher or lower temperatures inhibit the metabolic processes of the microbe by denaturing enzymes, transport proteins, and other proteins, which results in minimum biomass and secondary metabolite production. Similar findings were previously reported by several researchers20–22, 30.
S. monomycini RVE129 displayed its best growth and antibiotic activity at pH 7.5, which then steadily declined as the pH moved either toward an acidic or basic range. Previous research showed that Streptomyces sp. had its maximum cell growth and antibiotic activity at an initial pH of 7.521–24. This strain may therefore be classified as neutrophilic and was found to be capable of producing antibiotics at neutral pH levels between 7.0 and 7.5. Our results are in line with those of 25, who found that the optimal pH needed for Streptomyces aureusBG03 to produce biomass and antibiotics was 7.5, and a value of this factor greater than or less than 8 was unfavorable for antibiotic production. So, it was concluded that the amount and frequency of antibiotic synthesis are both impacted by pH value changes28.
Secondary metabolite biosynthesis by Streptomyces sp. can be significantly enhanced with minor variations in the fermentation medium composition27-30. In order to increase the synthesis of antibiotic metabolites, it is essential to determine the constituents of the medium employed in the fermentation process. In order to detect, alter, and optimize crucial medium components, several researchers working on antibiotic discovery combined PBD and RSM design as mathematical and statistical methodologies. Streptomyces nogalater (NIIST A30) was able to produce antibacterial metabolites in the best possible conditions when used statistical techniques like PBD and RSM to optimize the medium31. They found that antibiotic activity increased by 86.66% as a result. Streptomyces sp. AS432 produced 10 times as much antibiotics using the PBD and RSM procedures, as demonstrated by32. In this work, the statistically significant media components were optimized by PBD and RSM to identify the important factors and determine the best concentration and levels of those factors in the culture medium for increased antibiotic production by Streptomyces sp. against S. aureus. Our results exhibited that the compelling media components that facilitated maximum antibiotic production by S. monomycini RVE129 were starch, soybean meal, and K2HPO4. Similar findings were reported by31, 32, who identified starch as a crucial medium component for antibiotic synthesis. Streptomyces viridochromogenes uses soybean meal in the manufacture of antibiotics, according33. According to a recent study by34, soluble starch, soybean cake powder, and K2HPO4 significantly increased the synthesis of antibiotics in Streptomyces alfalfae XN-04 culture medium.The experiment’s findings were validated in order to assess the models’ accuracy and reliability in predicting the optimum responses. The most widely used statistical tool for examining the significance of a model is an analysis of variance (ANOVA), which offers a better understanding of the causes of variation32-34. The significance of the corresponding factor increases as the P-value decreases and the sum of squares increases34. Results of the p-value analysis (P <0.05) revealed that the interaction of starch, soybean meal, and K2HPO4 indicated that the model is significant. The fact that “Prob>F” is less than 0.05 indicates that the model terms developed in this investigation were significant and may be used to explain antibiotic synthesis by fermentation33. Alternately, for the model to work well with the experimental design, the lack of fit should be non-significant. According to our findings, the model is trustworthy for the current study because the lack of fit implied by the p-value of 0.0869 implies a non-significant lack of fit. The value coefficient variation (CV) indicates the level of accuracy used to compare the treatments, and as the CV rises, model dependability often declines22-25.Our low CV value suggests that the experimental data are sufficiently reliable and accurate23. Moreover, the model’s validity can be assessed using the R2 coefficient determination.The better the model predicts the response, the closer the R2 value is to one26–28. The obtained model R2 of 0.9953 demonstrates a close agreement between the experimental and predicted values. The model R2 of 0.9953 indicated that the model equation could represent a 99.53% variation in the response. In our study, the predicted R2 and adjusted R2 in the regression model had a high correlation and were in fair agreement with each other, and the measured R2 value is comparable with the earlier reports27-29. As a result, the validation experiments showed that the predicted and observed experimental findings were in agreement, and they were also determined to be trustworthy for optimizing the important media components.
The 3D plots can precisely identify the factors’ optimum levels by reflecting the impact of various levels of the factors on the response29-34. These plots and a numerical optimization function were used to determine the optimal conditions for maximum antibiotic production, which resulted in a maximum ZI diameter of 35.67±1.5 mm against S. aureus and was substantially identical to the value predicted by the model, demonstrating the model’s validity. The fact that the yield of antibiotic production in the current study was significantly higher on the optimized medium (35.67±1.5 mm) than on the unoptimized medium (27±1.5 mm) after optimization through RSM, with a 21.30% increase, strongly suggests that the quantity and quality of media components affect the production of antibacterial metabolites. This finding was found to be in perfect agreement with the preceding findings27-34. The current work serves as a great reminder that S. monomycini RVE129 can produce increased amounts of antibiotics by applying traditional and statistical methods of medium optimization and fermentation conditions.
Conclusion
The major goal of the study was to improve the production of antibiotics by S. monomycini strain RVE129 as a function of the different nitrogen and carbon sources and their concentrations in the basal medium. The results support the use of the traditional method for choosing appropriate nitrogen and carbon sources, as well as the fact that the PBD and RSM developed were highly efficient and reliable in identifying medium components for the production of antibiotics by S. monomycini RVE129. This is the first study to use the S. monomycini strain RVE129 to increase antibiotic activity through the use of traditional and statistical experimental methods for medium and fermentation parameters. The synthesis of antibiotics can therefore be increased by manipulating several nutritional aspects of the production medium using statistical and classical experimentation. This will be useful in developing a large-scale fermentation to increase antibiotic production from the S. monomycin strain RVE129.
Acknowledgement
The authors gratefully thank Koneru Lakshmaiah Education Foundation, Department of Biotechnology for supporting the laboratory facilities for this study.
Conflicts of Interest
The authors declare that they have no competing interests related to this work.
Funding Sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- Al-Dhabi NA, Esmail GA, Duraipandiyan V, Valan Arasu M, Salem-Bekhit MM. Isolation, identification and screening of antimicrobial thermophilic Streptomyces sp. Al-Dhabi-1 isolated from Tharban hot spring, Saudi Arabia. Extremophiles. 2016 Jan 20(1):79-90. DOI:10.1007/s00792-015-0799-1
CrossRef - Abd-Elnaby H, Abo-Elala G, Abdel-Raouf U, Abd-elwahab A, Hamed M. Antibacterial and anticancer activity of marine Streptomyces parvus: optimization and application. Biotechnology & Biotechnological Equipment. 2016 Jan 2;30(1):180-91.DOI:10.1080/1310 2818.2015.1086280.
CrossRef - Singh LS, Sharma H, Talukdar NC. Production of potent antimicrobial agent by actinomycete, Streptomyces sannanensis strain SU118 isolated from phoomdi in Loktak Lake of Manipur, India. BMC microbiology. 2014 Dec; 14(1):1-3.https://doi.org/10.11 86/s12 866-014-0278-3.
CrossRef - Munaganti RK, Muvva V, Konda S, Naragani K, Mangamuri UK, Dorigondla KR, Akkewar D. Antimicrobial profile of Arthrobacter kerguelensis VL-RK_09 isolated from Mango orchards. brazilian journal of microbiology. 2016 Oct;47:1030-8. DOI: 10.1016/j.bjm.2016.07.010.
CrossRef - Prashanthi R, GK S. Isolation, characterization, and molecular identification of soil bacteria showing antibacterial activity against human pathogenic bacteria. Journal of Genetic Engineering and Biotechnology. 2021 Dec;19(1):1-4.doi.org/10.1186/s43141-021-00219-x.
CrossRef - Nanjundan J., Ramasamy R. and Ponnusamy M. Optimization of culture conditions for antimicrobial metabolites production by Streptomyces sp. against bacterial leaf blight pathogen Xanthomonas oryzaepv. oryzae. International Journal of Chemical Studies; 2019; 7(3): 1187-1191.
- Oskay M. Effects of some Environmental Conditions on Biomass and Antimicrobial Metabolite Production by Streptomyces Sp., KGG32. International Journal of Agriculture & Biology. 2011 Jun 1;13(3).
- Al Farraj DA, Varghese R, Vágvölgyi C, Elshikh MS, Alokda AM, Mahmoud AH. Antibiotics production in optimized culture condition using low cost substrates from Streptomyces sp. AS4 isolated from mangrove soil sediment. Journal of King Saud University-Science. 2020 Mar 1;32(2):1528-35. https://doi.org /10.1016/j.jksus.2019.12.008
CrossRef - Wang L, Zhang M, Li Y, Cui Y, Zhang Y, Wang Z, Wang M, Huang Y. Application of response surface methodology to optimize the production of antimicrobial metabolites by Micromonospora Y15. Biotechnology & Biotechnological Equipment. 2017 Sep 3;31(5):1016-25. https://doi.org/10.1080/13102818. 2017.1356689
CrossRef - Augustine SK, Bhavsar SP, Kapadnis BP. Production of a growth dependent metabolite active against dermatophytes by Streptomyces rochei AK 39. Indian J Med Res. 2005 Mar 1;121(3):164-70.
- Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrika. 1946 Jun 1;33(4):305-25.
CrossRef - Narayana KJ, Vijayalakshmi M. Optimization of antimicrobial metabolites production by Streptomyces albidoflavus. Res J Pharmacol. 2008;2(1):4-7.
- Elias F, Muddada S, Muleta D, Tefera B. Antimicrobial potential of Streptomyces spp. isolated from the rift valley regions of Ethiopia. Advances in Pharmacological and Pharmaceutical Sciences. 2022 Jun 13;2022.DOI:10.1155/2022/1724906
CrossRef - Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical streptomyces genetics. Norwich: John Innes Foundation; 2000.
- Ju Y, Son KH, Jin C, Hwang BS, Park DJ, Kim CJ. Statistical optimization of culture medium for improved production of antimicrobial compound by Streptomyces rimosus AG-P1441. Food science and biotechnology. 2018 Apr;27(2):581-90..https://doi.org/10.1007/s10068-017-0257-1.
CrossRef - Arul Jose P, Jebakumar SR. Successive nonstatistical and statistical approaches for the improved antibiotic activity of rare actinomycete Nonomuraea sp. JAJ18. BioMed Research International. 2014 Sep 3;2014. DOI: 10.1155/2014/906097.
CrossRef - Jorgensen JH, Turnidge JD. Susceptibility test methods: dilution and disk diffusion methods. Manual of clinical microbiology. 2015 May 15:1253-73.
CrossRef - Elias F, Muddada S, Muleta D, Tefera B. Purification and Characterization of Bioactive Metabolite from Streptomyces monomycini RVE129 Derived from the Rift Valley Soil of Hawassa, Ethiopia. BioMed Research International. 2022 Dec 20;2022.https://doi.org/10.1155/2022/7141313.
CrossRef - Singh LS, Mazumder S, Bora TC. Optimisation of process parameters for growth and bioactive metabolite produced by a salt-tolerant and alkaliphilic actinomycete, Streptomyces tanashiensis strain A2D. Journal de mycologie médicale. 2009 Dec 1;19(4):225-33.https://doi.org/10.1016/j.mycmed.2009.07.006
CrossRef - Bundale S, Begde D, Nashikkar N, Kadam T, Upadhyay A. Optimization of culture conditions for production of bioactive metabolites by Streptomyces spp. isolated from soil. Advances in Microbiology. 2015;5(06):441.DOI: 10.4236/aim.2015.56045
CrossRef - Al-Ansari M, Kalaiyarasi M, Almalki MA, Vijayaraghavan P. Optimization of medium components for the production of antimicrobial and anticancer secondary metabolites from Streptomyces sp. AS11 isolated from the marine environment. Journal of King Saud University-Science. 2020 Apr 1;32(3):1993-8..https:// doi.org/10.1016/j.jksus.2020.02.005.
CrossRef - Singh C, Parmar RS, Jadon P, Kumar A. Optimization of cultural conditions for production of antifungal bioactive metabolites by Streptomyces spp. isolated from soil. International Journal of Current Microbiology and Applied Sciences. 2017;6(2):386-96.DOI:10.20546/ijcmas.2017.602.043
CrossRef - El-Naggar MY, Hassan MA, Said WY, Samy A EA. Effect of support materials on antibiotic MSW2000 production by immobilized Streptomyces violatus. The Journal of General and Applied Microbiology. 2003;49(4):235-43. DOI:10.2323/jgam.49.235
CrossRef - Ibrahim AA, El-Housseiny GS, Aboshanab KM, Yassien MA, Hassouna NA. Paromomycin production from Streptomyces rimosus NRRL 2455: statistical optimization and new synergistic antibiotic combinations against multidrug resistant pathogens. BMC microbiology. 2019 Dec;19(1):1-5.https://doi.org /10. 1186/s12866-019-1390-1.
CrossRef - Muthukumar R, Rajeswari E, Kalaiselvi T. Optimization of Cultural Conditions for the Antimetabolites Production by Streptomyces aureus strain BG03. Madras Agricultural Journal. 2019 Dec 20;106(1):1-3.DOI:10.29 321/MAJ.2019.000226
CrossRef - Osman ME, Khattab OH, Zaghlol GM, El-Hameed RM. Optimization of some physical and chemical factors for lovastatin productivity by local strain of Aspergillus terreus. Australian Journal of Basic and Applied Sciences. 2011;5(6):718-32..
- Wu JY, Huang JW, Shih HD, Lin WC, Liu YC. Optimization of cultivation conditions for fungichromin production from Streptomyces padanus PMS-702. Journal of the chinese Institute of Chemical Engineers. 2008 Jan 1;39(1):67-73..DOI:10.1016/j.jcice.2007.11.006
CrossRef - Mobeen SK, Sankar GG. Bioprocess development employing design of experiments for antibiotic production from Streptomyces parvulus strain sankarensis-A10. Indian Journal of Pharmaceutical Sciences. 2018 Oct 31;80(5):911-20. DOI:10.4172/pharmaceutical-sciences.1000438
CrossRef - Ju Y, Son KH, Jin C, Hwang BS, Park DJ, Kim CJ. Statistical optimization of culture medium for improved production of antimicrobial compound by Streptomyces rimosus AG-P1441. Food science and biotechnology. 2018 Apr;27:581-90.https://doi.org/10.1007/s10068-017-0257-1.
CrossRef - Sharma P, Ranghar S, Baunthiya M. Identification and Optimization of Fermentation Medium for Production of Antibacterial Compounds from Endophytic Streptomyces sp. GBTPR-167. International Journal of Current Microbiology and Applied Sciences. 2020 Jun;9(6):2594-608..DOI:10.20546/ijcmas.2020.906.316
CrossRef - Jacob J, Rajendran RU, Priya SH, Purushothaman J, Saraswathy Amma DK. Enhanced antibacterial metabolite production through the application of statistical methodologies by a Streptomyces nogalater NIIST A30 isolated from Western Ghats forest soil. PLoS One. 2017 Apr 24;12(4):e0175919. https://doi.org/10.1371/journal.pone.0175919.
CrossRef - Al Farraj DA, Varghese R, Vágvölgyi C, Elshikh MS, Alokda AM, Mahmoud AH. Antibiotics production in optimized culture condition using low cost substrates from Streptomyces sp. AS4 isolated from mangrove soil sediment. Journal of King Saud University-Science. 2020 Mar 1;32(2):1528-35.https://doi.org/10.1016/j.jksus.2019.12.008 1018-3647/2019.
CrossRef - Vu TH, Nguyen QH, Le TT, Chu-Ky S, Phi QT. Optimal fermentation conditions for antibiotic production by endophytic Streptomyces cavourensis YBQ59 isolated from Cinnamomum cassia Presl. Vietnam Journal of Science and Technology. 2019 Nov 12;57(3B):144-52.doi:10.15625/2525-2518/57/3B/14501.
CrossRef - Chen J, Lan X, Jia R, Hu L, Wang Y. Response Surface Methodology (RSM) Mediated Optimization of Medium Components for Mycelial Growth and Metabolites Production of Streptomyces alfalfae XN-04. Microorganisms. 2022 Sep 16;10(9):1854. https://doi.org/ 10.3390/microorganisms10091854.
CrossRef







