Manuscript accepted on :18-11-2022
Published online on: 30-11-2022
Plagiarism Check: Yes
Reviewed by: Dr. Rajat Goyal, Dr. Rajesh L. Dumpala
Second Review by: Dr. Jagdish Joshi
Final Approval by: Dr. Patorn Promchai
Imadeldin Elfaki1* , Rashid Mir2
, Rashid Mir2 , Faris J Tayeb2
, Faris J Tayeb2 , Jameel Barnawi2
, Jameel Barnawi2 , Adel Ibrahim Alalawy1
, Adel Ibrahim Alalawy1 , Hyder Mirghani3
, Hyder Mirghani3 , Naseh Algehainy2
, Naseh Algehainy2 , Sanad E Alshammari4
, Sanad E Alshammari4 and Pradeep Kumar Dabla5
and Pradeep Kumar Dabla5
1Department of Biochemistry, Faculty of Science, University of Tabuk, Tabuk 71491, Kingdom of Saudi Arabia.
2Prince and Fahd Bin Sultan Research Chair, Department of Medical Lab Technology, Faculty of Applied Medical Sciences, University of Tabuk, Tabuk 71491, Kingdom of Saudi Arabia.
3Internal Medicine and Endocrine, Medical Department, Faculty of Medicine, University of Tabuk, Tabuk 71491, Kingdom of Saudi Arabia.
4Department of Pharmacology and Toxicology, College of Pharmacy, Hail University, Hail, Saudi Arabia.
5 Department of Biochemistry, G.B.Pant Institute of Postgraduate Medical Education and Research (GIPMER),Associated Maulana Azad Medical College, Delhi, India.
Corresponding Author E-mail: elfakiimadeldin@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/2528
Abstract
The prevalence of diabetes mellitus (DM) is increasing worldwide including Saudi Arabia. DM increases mortality rate, morbidity and vascular complications, accompanied by poor general health status and low quality of life. CYP2C19*17 polymorphism in CYP2C19 gene is associated with the clinical outcome of drugs that are substrates of CYP2C19. CYP2C19*17 confers reduced susceptibility to certain illnesses. This research was conducted to develop a robust method to genotype the rs12248560 single nucleotide variation (SNV). We enrolled 206 subjects: 100 subjects were clinically confirmed cases of type 2 diabetes (T2D), and 106 subjects were healthy controls in this study. Samples from all subjects were screened for the CYP2C19 rs12248560 (c.-806C>T) by the amplification-refractory mutation system PCR (ARMS-PCR). The frequencies of CYP2C19*17 TT, CT, CC genotypes in T2D cases were 12%, 21%, and 67%, respectively whereas those in healthy controls were 70.75%, 26.41%, and 2.83%, respectively. The difference was significant (p < 0.035). T allele (fT) prevalence was found to be substantially greater in T2D cases compared to healthy controls (0.22 vs. 0.16). Results indicated that the CYP2C19*17 - TT genotype is associated with increased susceptibility to T2D with OR = 4.47, RR = 2.64, (p < 0.024). Moreover, the ARMS-based assay proved to be an easy method for the determination of CYP2C19*17 genotypes with reduced cost and good accuracy. In addition, this result helps in the detection and stratification of the individuals who are at risk for the development of T2D. Nevertheless, this finding needs to be validated in molecular genetic studies with increased specimen size and in different ethnicities.
Keywords
ARMS-PCR; CYP2C19*17 variation; Cytochrome P450; Diabetes
Download this article as:| Copy the following to cite this article: Elfaki I, Mir R, Tayeb F. M, Barnawi J, Alalawy A. I, Mirghani H, Algehainy N, Alshammari S. E, Dabla P. K. "Pharmacogenetics of CYP2C19*17: Functional and Clinical Implications of CYP2C19*17 - rs12248560 (c.-806C>T) in the Development of Type 2 Diabetes". Biomed Pharmacol J 2022;15(4). |
| Copy the following to cite this URL: Elfaki I, Mir R, Tayeb F. M, Barnawi J, Alalawy A. I, Mirghani H, Algehainy N, Alshammari S. E, Dabla P. K. "Pharmacogenetics of CYP2C19*17: Functional and Clinical Implications of CYP2C19*17 - rs12248560 (c.-806C>T) in the Development of Type 2 Diabetes". Biomed Pharmacol J 2022;15(4). Available from: https://bit.ly/3VVcCZR |
Introduction
In terms of the incidence rate of diabetes mellitus, the WHO reported that kingdom of Saudi Arabia (KSA) is number two in middle east and number seventh worldwide [1, 2]. It was reported that about seven million are diabetic and around three million individuals are in the pre-diabetic stage in KSA [2]. Diabetes mellitus is a metabolic disorder characterized by severe hyperglycemia in the patients. The cytochrome p450s (Cyp450s) are a member of a superfamily of enzymes that metabolizes endogenous substrates and catalyzes the biotransformation of xenobiotics, such as carcinogens and drugs [3]. The genes and pseudogenes of cytochrome P450 (CYP) are classified into 18 families and 44 subfamilies based on the similarity of their DNA sequence [3, 4]. Cyp450s members metabolize up to 80% of the drugs in clinical use [3]. Moreover, Cyp450s are important in the metabolism of endogenous substrates [3]. For instance, the fatty acid (e.g.arachidonic acid), steroid hormones (e.g. sex hormones), eicosanoids, prostaglandins, cholic and chenodeoxycholic acid are metabolized by Cyp450 family members [3, 5]. Cytochrome p450s gene is mainly expressed in hepatocytes and in intestinal cells, but also found in nervous system, kidney, placenta, lung, adrenal gland, pancreas, mammary gland and genitalia [3, 5]. CYP 2C19 enzyme is expressed in liver cells, and it is a protein with a molecular weight of 55.93 kDa and 490 amino acids, and catalyzes the metabolism of about ten percent of prescribed medicines or drugs such as citalopram, omeprazole and clopidogrel [6, 7]. There are interpersonal variations in response (e.g. toxicity or lack of response) to the drugs that are CYP 2C19 substrates [8]. These interpersonal variations are due to the highly polymorphic nature of CYP2C19 gene[8]. In terms of the response to the CYP 2C19 substrates, subjects can be classified into different phenotypes: extensive metabolizers or ultra rapid metabolizers, intermediate metabolizers and poor metabolizers[8]. CYP2C19*17 SNV increases enzyme catalytic activity and the expression of the CYP2C19 enzyme [4, 8].
T2D is a metabolic condition or disorder indicated by raised blood sugar due to the peripheral impaired insulin action (hepatocytes, fat tissues and muscles) and the pancreatic beta cell dysfunction [9]. The risk factors of T2D are mixtures of genetic and metabolic and environmental factors [9]. The metabolic risk factors include high-caloric diet containing large amounts of fats and carbohydrates, and obesity, while the environmental risk factors include decreased physical activity and smoking [9, 10]. Genome-wide association (GWA) studies have revealed the link of gene polymorphisms (SNPs) and mutations with the cause or development of diseases including cancers, Type 2 diabetes (T2D) and cardiovascular diseases [11]. Moreover, certain isoforms of Cyp450 SNPs have been shown to be associated with the development of diseases such as T2D [12], coronary artery disease (CAD) [13], and cancers[14]. Therefore the aim of our research study is to determine the association of CYP2C19*17 C>T with T2D development in Saudi population.
Materials, Methods and Subjects
Study subjects and criteria of inclusion and exclusion
This project received approval from the Ethics Committee of university of Tabuk and Ethics Committee of university of Taif (Code 229). The study population comprised of type 2 diabetes (T2D) patients visiting the hospitals routinely for checkup. T2D diagnosis was made based on the WHO criteria. The cases were citizens of KSA. All patients signed the patient information sheet as well as written consent form prior to the inclusion in this project. We excluded cases with type 1 diabetes or any other chronic disease. The healthy subjects were selected from the local population of Tabuk region. A standard physical examination was conducted (blood biochemistry tests and hematology). The subjects that were apparently healthy without significant illness were selected as healthy controls (HCs).
Sample collection and purification of the genomic DNA
About 3 ml blood specimen was collected by venipuncture in EDTA tube from each subject. The genomic DNA purification was performed by using DNA isolation from Qiagen, (DNeasy Blood Kit-Germany) according to the provided protocol. The purified DNA was evaluated by agarose gel electrophoresis and by using the NanoDrop Spectrophotometer and then placed at -20 °C until genotyping.
CYP2C19 rs12248560 (c.-806C>T) genotyping
We used the amplification-refractory mutation system-PCR primers that were previously used for CYP2C19*17 C>T (rs12248560) genotyping [15]. Optimization of an amplification-refractory mutation system PCR was performed by gradient PCR by using arms tetra-primers specific for CYP2C19*17 C>T (rs12248560) genotyping. In order to bring the temperature near to that of its counter primer, the inner primers were altered. The Tm difference between primer combinations needed to be less than or equal to 2 °C in order for the PCR reaction to produce effective amplicons. i.e., Fo, Ro, FI, RI (Table 1, Figure 1). The PCR reactions were carried out in a reaction volume of 25 µL containing four ARMS primers, Fo -0. 25 µL, Ro -0. 25 µL, FI-0. 25 µL, and RI -0. 25 µL (25 pmol of each primer) and 12 µL of Green PCR Master Mix (2X) (GoTaq® Green Master Mix (Cat No M7122) (Promega, Madison, Wisconsin, USA) . The final volume of 23 µL was adjusted by adding nuclease-free ddH2O. Finally 2 ul (50 ng) of DNA was added from each subject.
Table 1: Primers used for genotyping the CYP2C19*17 – rs12248560 SNP.
| Gene direction | Allele | Sequence | PCR product size | Tmo C |
| CYP2C19*17 FO | 5′-GAGATCAGCTCTTCCTTCAGTTACAC-3′ | 462 bp | 56 | |
| CYP2C19*17 RO | 5′-CACCTTTACCATTTAACCCCCTAAAAA-3′ | |||
| CYP2C19*17 FI | T allele | 5′-TTTTTCAAATTTGTGTCTTCTGTTCTCAAATT-3′ | 227 bp | 56 |
| CYP2C19*17 RI | C allele | 5′-GCGCATTATCTCTTACATCAGAGCTG-3′ | 292 bp | 56 |
| Abbreviations: FI, forward primer; FO, forward outer primer; RI, reverse inner primer; RO, reverse outer primer. | ||||
 |
Figure 1: The annealing sites of the primers used for genotyping are shown in the DNA sequence of the CYP2C19 gene. The annealing sites and the SNV site (T) are highlighted in gray and bold font. |
PCR conditions
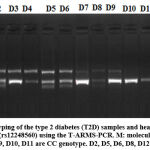
The PCR settings were 95 °C for 6 min of initial denaturation, followed by 30 cycles of 95 °C for 30 secs, 56 °C for 35 secs and 72 °C for 40 secs and with the final extension lasting 5 min at 72 °C. After that, the amplification products were processed at 120 volts for roughly 30 minutes on a 2 percent agarose gel (Figure 2).
Statistical analyses
With the aid of the SPSS 16.0 program, a statistical analysis of the genotyping of the CYP2C19*17 C>T (rs12248560) gene in T2D cases and healthy controls was conducted. To compare the single nucleotide variants of CYP2C19*17 C>T (rs12248560) with various clinic-pathological characteristics, the Chi-square test and Fisher exact analysis test were used. The Chi-square and Fisher exact test were performed to compare the CYP2C19*17 C>T gene polymorphism with the clinic-pathological aspects of the T2D patients. We looked for any deviations from Hardy-Weinberg equilibrium in the genotyping distribution of the healthy controls. By comparing risk ratios (RRs), and odds ratios (ORs) with 95% confidence intervals, multivariate analyses were utilized to investigate the connection between SNP and T2D susceptibility (CIs).
Results
Hardy-Weinberg equilibrium (HWE)
The distributions of the CYP2C19*17 -rs12248560 C>T genotype and allele frequencies showed no deviation from HWE (all P values > 0.05) in the healthy controls.
The CYP2C19*17 -rs12248560 C>T was significantly different between healthy controls and T2D cases
In T2D cases, the CC, CT and TT genotype frequencies were 67%, 21% and 12%, respectively, whereas in healthy controls CC, CT and TT genotype frequencies were 70.75%, 26.41%, and 2.83%, respectively (Table 2). The distribution of CYP2C19*17 -rs12248560 C→T genotypes observed between T2D cases and healthy controls was significant (P < 0.035). Moreover, the frequency of T allele (fT) was found to be significantly higher among T2D cases than in HCs (0.22 vs. 0.16) (Table 2).
Table 2: The CYP2C19*17 (rs12248560) C>T SNP in T2D cases and controls.
| Subjects | N= | CC | CT | TT | Df | X2 | C | T | P value |
| Cases | 100 | 67(67%) | 21(21%) | 12(12%) | 2 | 6.68 | 0.78 | 0.22 | 0.035 |
| Controls | 106 | 75(70.75%) | 28(26.41%) | 03(2.83%) | 0.84 | 0.16 |
The CYP2C19*17 -rs12248560 was associated with T2D susceptibility
To determine the relationship between CYP2C19*17 CT genotypes and risk of T2D, we performed a multivariate analysis based on logistic regression. Odds ratios (OD) and risk ratios (RR) with 95% confidence intervals (CI) were calculated for each group. It is reported that the CYP2C19*17 – TT genotype is linked with raised T2D susceptibility (odd ratio 4.47 and Risk ratio 2.64, p < 0.024) (Table 3). However the CYP2C19*17 – CT genotype (heterozygosity) was not associated with T2D susceptibility with OR = 0.83(95%), CI = (0.4362 to 1.6158), RR = 0.92(0.6930 to 1.2328), and P < 0.60 (Table 3). This study results indicated that in the recessive model, the CYP2C19*17 -TT vs (CC+CT) genotype was correlated with increased T2D susceptibility with OR = 4.68 (95%), CI = (1.2801 to 17.1236), RR = 1.09 (0.8104 to 1.4671) and P < 0.019 (Table 3).
Table 3: Association of CYP2C19*17 (rs12248560) C→T genotypes with T2D susceptibility.
| Genotypes | Healthy
controls |
CAD
cases |
OR (95% CI) | Risk Ratio(RR) | P-Val |
| (N = 106) | (N = 100) | ||||
| Co-dominant inheritance model | |||||
| CYP2C19*17-CC | 75 | 67 | 1(Ref.) | 1(Ref.) | |
| CYP2C19*17-CT | 28 | 21 | 0.83(0.4362 to 1.6158) | 0.92(0.6930 to 1.2328) | 0.60 |
| CYP2C19*17-TT | 03 | 12 | 4.47(1.2113 to 16.55) | 2.64(0.948 to 7.353) | 0.024 |
| Dominant inheritance model | |||||
| CYP2C19*17-CC | 75 | 67 | 1(ref.) | 1(ref.) | |
| CYP2C19*17 (CT+TT) | 31 | 33 | 1.19(0.6601 to 2.1511) | 1.09(0.8104 to 1.4671) | 0.65 |
| Recessive inheritance model | |||||
| CYP2C19*17-(CC+CT) | 103 | 88 | 1(ref.) | 1(ref.) | |
| CYP2C19*17-TT | 03 | 12 | 4.68(1.2801 to 17.1236) | 1.09(0.8104 to 1.4671) | 0.019 |
| Allele | |||||
| CYP2C19*17-C | 178 | 155 | 1(ref.) | 1(ref.) | |
| CYP2C19*17-T | 34 | 45 | 1.51(0.9268 to 2.4927) | 1.24(0.9455 to 1.6315) | 0.09 |
Association of CYP2C19*17 (rs12248560) C>T genotypes with lipid profile biomarkers
Association with HBA1c%
A significant correlation was reported between CYP2C19*17 (rs12248560) C>T genotypes and HBA1c in T2D cases (p < 0.042) (Table 4).
Correlation with gender
Results showed that the CYP2C19*17 (rs12248560) C>T genotypes were significantly correlated with gender in T2D cases (p < 0.013) (Table4).
Correlation with total cholesterol (mg/dL)
The statistical analysis showed that there was a significant association between CYP2C19*17 (rs12248560) C>T genotypes and blood cholesterol (mg/dL) levels in T2D patients (p < 0.0001) (Table 4).
Correlation with LDL-C (mg/dL)
Moreover, the result of the analyses showed that there is no statistically significant association between CYP2C19*17 (rs12248560) C>T genotypes and LDL-C (mg/dL) of T2D cases (p < 0.27) (Table 4).
Correlation with serum HDL-C (mg/dL)
A strong statistically significant association was established between CYP2C19*17 (rs12248560) C>T genotypes and HDL-C (mg/dL) of T2D patients (p < 0.029) (Table 4).
Association with serum triacylglycerol (TG) (mg/dL)
Our results indicated an association between CYP2C19*17 (rs12248560) C>T genotypes and TG. A significant difference was reported between cases and cases with hypertriglyceridemia (p < 0.0001) (Table 4).
Table 4: Association of CYP2C19*17 (rs12248560) C>T gene variation with clinical features of T2D cases.
| Clinical feature | N= | AA | AG | GG | X2 | DF | P-value | |
| Association with gender | 100 | 67 | 21 | 12 | ||||
| Male | 80 | 59 | 14 | 7 | 8.57 | 2 | 0.013 | |
| Female | 20 | 8 | 7 | 5 | ||||
| Association with age | ||||||||
| >40 | 78 | 55 | 15 | 8 | 2.08 | 2 | 0.35 | |
| <40 | 22 | 12 | 6 | 4 | ||||
| Association with fasting glucose mg/dl | ||||||||
| <100 | 28 | 15 | 8 | 5 | 3.2 | 2 | 0.199 | |
| >100 | 72 | 52 | 13 | 07 | ||||
| Association with HBA1c% | ||||||||
| >6 | 78 | 57 | 13 | 8 | 6.2 | 2 | 0.04 | |
| <6 | 22 | 10 | 8 | 4 | ||||
| Association with triglycerides mg/dl | ||||||||
| <200 | 26 | 10 | 13 | 3 | 18.14 | 2 | 0.0001 | |
| >200 | 74 | 57 | 8 | 9 | ||||
| Association with cholesterol mg/dl | ||||||||
| <200 | 65 | 56 | 6 | 3 | 30.47 | 2 | 0.0001 | |
| >200 | 35 | 11 | 15 | 9 | ||||
| Association with LDL-C mg/dl | ||||||||
| <100 | 40 | 25 | 8 | 7 | 2.61 | 2 | 0.27 | |
| >100 | 60 | 37 | 18 | 5 | ||||
| Association with HDL-L mg/dl | ||||||||
| <55 | 30 | 20 | 3 | 7 | 7.06 | 2 | 0.029 | |
| >55 | 70 | 47 | 18 | 5 | ||||
Discussion
Diabetes mellitus (DM) is a health concern all over the world, and KSA is not an exception. It has a major socioeconomic impact due to its serious complications on patients, such as blindness and limps amputation [16]. Furthermore, DM has serious impact on the budget that is spent on the health care and the treatment of diabetes and its complications[16]. The ARMS is a standard method for genotyping of single nucleotide variations [15]. Nevertheless, its optimization is laborious, tedious and takes time[15]. In our lab a few modifications were made in the reagent concentrations, which positively influence the ARMS PCR – particularly MgCl2. The most important step in ARMS PCR is balancing of the inner primers (FI/Ro). During optimization the inner primers’ band (FI/Ro) was faint; then after increasing the concentration of MgCl2, band strength was enhanced. Different annealing temperatures were used in gradient PCR, and fewer cycles were used (25 to 30 cycles). According to earlier studies, the optimization was accomplished by the gradient PCR in a set of PCR experiments over the course of a single run [17]. Through gradient PCR, the temperature at which the primers anneal was tuned to range from 56 °C to 62 °C.
At an annealing temperature of 56 °C, we achieved the best results. The utilization of Tetra ARMS-PCR meets the demands of cutting-edge genomic science and enables speedy, reliable, and straightforward examinations of the CYP2C19*17 (rs12248560) C>T SNP. T2D constitutes more than 90% of all diabetes cases[18]. The promoter polymorphism CYP2C19 rs12248560 (c.-806C>T) results in increased expression of the CYP2C19 [19]. Our findings showed that T2D was related with the TT genotype of the CYP2C19*17 (rs12248560) C>T gene (Table 2).
The genotype distribution of rs12248560 in cases with normal HBA1c% levels and those with increased HBA1c% levels differed significantly, according to the findings (Table 4). These findings may be consistent with that of Hoyo-Vadillo et al., 2010, who indicated the association of the CYP2C19 genotype with T2D, in a Mexican population[20]. This outcome was also consistent with our previous study in which we have shown that the CYP2C19*3 (rs4986893) is probably associated with T2D in a Saudi population [21]. According to reports, CYP2C19 is controlled by the glucocorticoid receptor and the constitutive androstane receptor (CAR) [22]. The glucocorticoid receptor and CAR may be involved in energy metabolism, insulin resistance and T2D [20, 23, 24]. It has been reported that the glucocorticoid receptor increases the influence of glucocorticoids in metabolism and that the glucocorticoid receptor regulates genes involved in glucose metabolic pathways [24]. Additionally, the findings demonstrated a substantial difference between T2D individuals with normal lipid profiles and those with aberrant lipid profiles (Table 4). This outcome is in line with the most current research by Bai et al. [25], who discovered a link between Chinese population CYP2C19 gene polymorphisms and lipid metabolism [25]. Moreover, it has been reported that the single nucleotide variations in CYP2C19 gene such as CYP2C19 rs4244285 is associated with the risk of metabolic syndrome development in a population of South Portuguese[26]. Cardiovascular disease and type 2 diabetes are linked to the metabolic syndrome[26] . It has been reported that CYP2C19 is involved in the metabolism of many vital endogenous compounds and substrates[26]. The epoxyeicosatrienoic acids (EETs) compounds exhibit properties of vasodilation, anti-inflammation, anti-apoptosis, anti-thrombosis, fibrinolysis and cardiac protection [27]. The EETs influence the vascular tone and changes the blood pressure, and it has a role in enhancing the fibrinolysis[25].
Conclusion
It was concluded that CYP2C19 rs12248560 (c.-806C>T)-TT genotype was strongly linked with increased susceptibility to T2D (OR = 4.47 RR = 2.64, p < 0.024) in Saudi population. These findings assist in the detection and stratification of the individuals that are at risk for T2D development. The ARMS-PCR can be used robustly to detect the CYP2C19 rs12248560 (c.-806C>T) polymorphism, and it is an accurate, simple and inexpensive method. These results need to be verified in further population-based studies with more sample sizes and different ethnicities.
Acknowledgement
This study was approved by the Research and Studies Department, Directorate of Health Affairs, Taif, Approval No. 229, and by the Research Ethics Committee of the Armed Forces hospitals, Northwestern region, Tabuk, Approval No. R and REC2016-115.
Conflict of Interest
The authors declare that no conflicts of interest exist.
Funding Sources
This project is funded by the University of Tabuk, Deanship of Scientific Research (DSR), project No 0012-1441-S for IE and colleagues.
References
- Al Mansour MA: The Prevalence and Risk Factors of Type 2 Diabetes Mellitus (DMT2) in a Semi-Urban Saudi Population. Int J Environ Res Public Health 2019, 17(1).
CrossRef - Robert AA, Al Dawish MA, Braham R, Musallam MA, Al Hayek AA, Al Kahtany NH: Type 2 Diabetes Mellitus in Saudi Arabia: Major Challenges and Possible Solutions. Curr Diabetes Rev 2017, 13(1):59-64.
CrossRef - Zanger UM, Schwab M: Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 2013, 138(1):103-141.
CrossRef - Li-Wan-Po A, Girard T, Farndon P, Cooley C, Lithgow J: Pharmacogenetics of CYP2C19: functional and clinical implications of a new variant CYP2C19*17. Br J Clin Pharmacol 2010, 69(3):222-230.
CrossRef - Elfaki I, Mir R, Almutairi FM, Duhier FMA: Cytochrome P450: Polymorphisms and Roles in Cancer, Diabetes and Atherosclerosis. Asian Pac J Cancer Prev 2018, 19(8):2057-2070.
- Brown SA, Pereira N: Pharmacogenomic Impact of CYP2C19 Variation on Clopidogrel Therapy in Precision Cardiovascular Medicine. J Pers Med 2018, 8(1).
CrossRef - Sanford JC, Guo Y, Sadee W, Wang D: Regulatory polymorphisms in CYP2C19 affecting hepatic expression. Drug Metabol Drug Interact 2013, 28(1):23-30.
CrossRef - Lee SJ: Clinical Application of CYP2C19 Pharmacogenetics Toward More Personalized Medicine. Front Genet 2012, 3:318.
CrossRef - Galicia-Garcia U, Benito-Vicente A, Jebari S, Larrea-Sebal A, Siddiqi H, Uribe KB, Ostolaza H, Martin C: Pathophysiology of Type 2 Diabetes Mellitus. Int J Mol Sci 2020, 21(17).
CrossRef - Hameed I, Masoodi SR, Mir SA, Nabi M, Ghazanfar K, Ganai BA: Type 2 diabetes mellitus: From a metabolic disorder to an inflammatory condition. World J Diabetes 2015, 6(4):598-612.
CrossRef - McCarthy MI, Zeggini E: Genome-wide association studies in type 2 diabetes. Curr Diab Rep 2009, 9(2):164-171.
CrossRef - Zarish Noreen, Christopher A. Loffredo, Attya Bhatti, Jyothirmai J. Simhadri, Gail Nunlee-Bland, Thomas Nnanabu, Peter John, Jahangir S. Khan, Somiranjan Ghosh.Transcriptional Profiling and Biological Pathway(s) Analysis of Type 2 Diabetes Mellitusin a Pakistani Population.Int J Environ Res Public Health. 2020 Aug; 17(16): 5866
CrossRef - Sirotina S, Ponomarenko I, Kharchenko A, Bykanova M, Bocharova A, Vagaytseva K, Stepanov V, Churnosov M, Solodilova M, Polonikov A: A Novel Polymorphism in the Promoter of the CYP4A11 Gene Is Associated with Susceptibility to Coronary Artery Disease. Dis Markers 2018, 2018:5812802.
CrossRef - Li M, Li A, He R, Dang W, Liu X, Yang T, Shi P, Bu X, Gao D, Zhang N et al: Gene polymorphism of cytochrome P450 significantly affects lung cancer susceptibility. Cancer Med 2019, 8(10):4892-4905.
CrossRef - Ye S, Dhillon S, Ke X, Collins AR, Day IN: An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res 2001, 29(17):E88-88.
CrossRef - Hill J, Nielsen M, Fox MH: Understanding the social factors that contribute to diabetes: a means to informing health care and social policies for the chronically ill. Perm J 2013, 17(2):67-72.
CrossRef - Lucy Darakjian, Malavika Deodhar, Jacques Turgeon, Veronique Michaud, Chronic Inflammatory Status Observed in Patients with Type 2 Diabetes Induces Modulation of Cytochrome P450 Expression and Activity. Int J Mol Sci. 2021 May; 22(9): 4967
CrossRef - Deshpande AD, Harris-Hayes M, Schootman M: Epidemiology of diabetes and diabetes-related complications. Phys Ther 2008, 88(11):1254-1264.
CrossRef - Scott SA, Martis S, Peter I, Kasai Y, Kornreich R, Desnick RJ: Identification of CYP2C19*4B: pharmacogenetic implications for drug metabolism including clopidogrel responsiveness. Pharmacogenomics J 2012, 12(4):297-305.
CrossRef - Hoyo-Vadillo C, Garcia-Mena J, Valladares3 A, Venturelli CR, Wacher-Rodarte N, Kumate J, Cruz M: Association of CYP2C19 genotype with type 2 diabetes. 2010, 2(10):1184-1190.
CrossRef - Janice Forster, Jessica Duis, Merlin G. Butler.Pharmacogenetic Testing of Cytochrome P450 Drug Metabolizing Enzymes in a Case Series of Patients with Prader-Willi Syndrome.Genes (Basel) 2021 Feb; 12(2): 152
CrossRef - Chen Y, Ferguson SS, Negishi M, Goldstein JA: Identification of constitutive androstane receptor and glucocorticoid receptor binding sites in the CYP2C19 promoter. Mol Pharmacol 2003, 64(2):316-324.
CrossRef - Yan J, Chen B, Lu J, Xie W: Deciphering the roles of the constitutive androstane receptor in energy metabolism. Acta Pharmacol Sin 2015, 36(1):62-70.
CrossRef - Kokkinopoulou I, Diakoumi A, Moutsatsou P: Glucocorticoid Receptor Signaling in Diabetes. Int J Mol Sci 2021, 22(20).
CrossRef - Bai Y, Huang R, Wan L, Zhao R: Association between CYP2C19 gene polymorphisms and lipid metabolism in Chinese patients with ischemic stroke. J Int Med Res 2020, 48(7):300060520934657.
CrossRef - Gaio V, Nunes B, Fernandes A, Mendonca F, Horta Correia F, Beleza A, Gil AP, Bourbon M, Vicente A, Dias CM et al: Genetic variation at the CYP2C19 gene associated with metabolic syndrome susceptibility in a South Portuguese population: results from the pilot study of the European Health Examination Survey in Portugal. Diabetol Metab Syndr 2014, 6(1):23.
CrossRef - Oni-Orisan A, Alsaleh N, Lee CR, Seubert JM: Epoxyeicosatrienoic acids and cardioprotection: the road to translation. J Mol Cell Cardiol 2014, 74:199-208.
CrossRef