Manuscript accepted on :18-11-2022
Published online on: 02-12-2022
Plagiarism Check: Yes
Reviewed by: Dr. Hanadi Jasim
Second Review by: Dr Mario Vincenzo Russo
Final Approval by: Dr. H Fai Poon
Enas R. Abdelhamid1 , Amira S. El Refay1*
, Amira S. El Refay1* , Alshaimaa A. ElKhatib1
, Alshaimaa A. ElKhatib1 , Ayman F. Armaneous1
, Ayman F. Armaneous1 , Lobna S. Sherif 1
, Lobna S. Sherif 1 , Shahinaz M. Hussien2, Adel Hashish4
, Shahinaz M. Hussien2, Adel Hashish4 , Nayra Mehanna 3
, Nayra Mehanna 3
1Child Health, National Research Centre, El-Bohouth Street, Dokki, Cairo, Egypt.
2Pediatric department, El Azhar university, Cairo, Egypt.
3Food and Dairy Microbiology, Central Laboratory Network, National Research Centre (NRC), Bohouth street, Dokki, Giza, Egypt.
4Children with special needs, National Research Centre, Bohouth street, Dokki, Giza, Egypt
Corresponding Author E-mail: amirasayed.ak@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/2560
Abstract
Background: It is well known that allergy development is linked to alteration in microbiome and cytokines levels. colonization of children gut by wide array of microbes and bacteria is associated with mucosal and systemic immune responses as well as allergy development. Investigating the role of gut microbiota and serum cytokines clarifies the pathophysiology of the disease and enhance development of management plan. This study aimed to assess fecal microbiota in asthmatic children and correlate it with serum CXCL8 and p38 MAPK as potential asthma severity markers. Results: This case control study enrolled 56 asthmatic children aged 2-8 years, and 20 non asthmatic children of matched sex and age group as a control. Fresh stool samples were obtained from enrolled children for analysis of gut microbiota through DNA extraction and Real time PCR, using species-specific primers, serum CXCL8 and P38MAPK levels were estimated by ELISA. Higher level of Bifidobacterium and lower level of Lactobacillus was reported in asthmatic compared to non-asthmatic children, current results showed significant difference between asthmatic and non-asthmatic subgroups regarding CXCL 8 serum level. Study reported significant negative correlation between presence of asthma and serum markers CXCL8 and p38MAPK while significant positive correlation between presence of asthma and Bifidobacterium Log. Conclusions: the association between CXCL8 level, p38 MAPK and microbiome suggesting a link between gut bacteria and inflammatory status. Moreover, elevated CXCL8 , and p38 MAPK level increase symptoms severity. The alteration of microbiome level associated with elevated markers level suggesting the protective role of gut microbiome in asthma control.
Keywords
Asthma; Bifidobacterium; Children; Cytokines; CXCL8; Lactobacillus; Microbiota; p38 MAPK
Download this article as:| Copy the following to cite this article: Abdelhamid E. R, El-Refay S. A, ElKhatib A. A, Armaneous A. F, Sheriff L. S, Hussien S. M, Hashish A, Mehanna N. Microbiota and Potential Asthma Markers: A Case Control Study in a Sample of Egyptian Children. Biomed Pharmacol J 2022;15(4). |
| Copy the following to cite this URL: Abdelhamid E. R, El-Refay S. A, ElKhatib A. A, Armaneous A. F, Sheriff L. S, Hussien S. M, Hashish A, Mehanna N. Microbiota and Potential Asthma Markers: A Case Control Study in a Sample of Egyptian Children. Biomed Pharmacol J 2022;15(4). Available from: https://bit.ly/3h0feqd |
Introduction
Pediatric asthma is a common childhood chronic disease 1. It is a multifaceted illness that affects 14% of the world’s children 2. Approximately one-half of asthmatic children become symptomatic before the age of 3 years with repeated episodes of wheezing with or without cough activated by a viral upper respiratory infection (URI), physical activity, or climate changes 3.
The severity of asthma may be influenced by the underlying disease activity, and it is well known that immunological disfunction has a significant role in the asthma progress 4-5.
The microbiome considered as the largest immune system, making a very complex micro-ecological system that affects nutritional state, metabolic status, and immunological status6-8. Recently, Microbiome has linked to asthma pathophysiology. However, the relationship between the microbiome and loss of asthma control is still obscure 9.
Asthma symptoms are indorsed by Eosinophilic infiltration which is linked to cytokines secretions and immunological dysfunction10.
Interleukins (ILs) have an essential role in asthma; particularly CXCL-8; which is encoding protein for interleukin 8 and is highly linked with asthma severity 11. It is a main cytokine for enrolment of neutrophils to the inflammation area, “Cytokine-mediated interactions among the inflammatory cells may play a role in the pathogenesis of bronchial asthma”. It is a valuable indicator of disease activity and treatment effectiveness in bronchial asthma.12. p38MAPK is another potential marker which was linked recently with asthma severity and the inflammatory process13.
Despite the new tools available such as clinical approach, predicting tests, and a trial of bronchodilators or gluco-corticosteroids, diagnosing asthma in children remains a challenge, It is mainly symptom based 14 Though all advantages of serum biomarkers, it is well known that peripheral blood investigations often do not imitate airway biology, and therefore peripheral blood biomarkers might not signify physiologic mechanisms in the airways 15.Diagnosis of asthma by a appropriate biomarker is important for the early detection, which helps in affording a suitable mangment for the optimum treatment 16.
This study was designed as a case control study to assess Gut microbiota in asthmatic and non-asthmatic children and correlate it with CXCL8 as potential asthma marker.
Methods
A case control study was conducted on 56 asthmatic children aged 2-8 years from those diagnosed as bronchial asthma patients and enrolled to follow up in asthma clinic in El Hussein Hospital during the period from September 2019 to March 2020. Another 20 non asthmatic children of matched sex and age group were enrolled in the study as a control.
The present study got the approval of the National Research Centre ethics committee. Informed consent was collected and signed from legal guardian of the children enrolled in the study prior to participation. A designed questionnaire was verbally processed to legal guardian containing demographic and clinical data.
All the enrolled children were subjected to through detailed history and clinical examination to assess the severity of asthma, Bronchial asthma was diagnosed according to GINA guidelines 17.
Any child had diarrhea or intestinal disease within one month of stool sample collection was excluded from the study.
Categorizing Asthma Severity
Bronchial asthma was diagnosed by GINA guidelines 2019 and asthma severity scoring was calculated according to the Severity of Asthma score (SOA) [18] which include asthma symptoms degree, using of asthma medication as corticosteroids and or Inhaled b -agonists and if the child was hospitalized for asthma or not. The study was first designed to be continued for a whole year and to use Expert Panel II recommendations 19 for asthma symptoms and severity but due to the COVID circumstances and the suspension of most research activity during lockdown and the inability to perform lung function test to the participant children as all the pulmonary function clinics were closed, we adapted the SOA instead to assess the severity of asthma.
As our design was to enroll outpatient clinic visitors. We enrolled the studied cases from the outpatient clinic, so all the case were fall into mild and moderate severity.
Severity of Asthma score is based on a validated disease particular questionnaire that focus on asthma symptoms frequency, corticosteroids using, use of medication other than corticosteroids and hospitalization history and or tube insertion for asthma.
|
Item |
Description |
Score |
| Symptoms during the past 2 weeks (chose one only)
|
0-4
|
|
| .
1 |
None
Minimal Occasional Most days or nights Everyday of night |
0
1 2 3 4 |
| Systemic corticosteroids use
Choose one or more |
||
| 2 | Ever used | 0 or 2 |
| 3 | Used in the past year | 0 or 2 |
| 4 | Used > 3 months within past 2 years | 0 or 3 |
| 5 | Inhaled B agonist SABA/LABA
None <2 > 2 puffs |
0 or 1 or 2 |
| 6 | ICS
None < 20 > 20 puffs |
0 or 1 or 2 |
| 7 | CroolynL/nedoeromil | 0 or 1 |
| 8 | Anti-cholinergic therapy | 0 or 1 |
| 9 | Theophylline, oral B-agonist, or leukotriene modifier | 0 or 1 |
| 10 | Antihistaminic, decongestant, or intra-nasal spray | 0 or 1 |
| 11 | use of home nebulizer | 0 or 1 |
| 12 | Hospital admission because of asthma | 0 or 1 |
| 13 | Ever intubated for asthma | 0 or 1 |
| Total score | 0-25 |
Severity of asthma score predicts clinical outcome
Laboratory investigations
Sample was obtained and centrifuged at 700-1,000 x g for ten minutes at 4°C. Serum was separated and frozen then kept at -80°C. CXCL8[20] and P38MAPK level were estimated by ELISA 21.
For analysis of gut microbiota, fresh stool samples were collected from enrolled children in sterile clean cups then stored at -80°C. Detection of Bifidobacterium and Lactobacillus species were based on 16S rRNA gene sequences identification through Target organism Primer Set Sequence. In brief extraction of DNA was done by lysis of the bacterial cells within the fecal material purification of the DNA and stored at −20 °C. DNA extract amplification followed by testing the different strains by real-time PCR bacterial quantification 22.
Statistical analysis
Statistical analysis was presented using the SPSS statistical package software for windows version 21 (SPSS Inc, Pennsylvania, USA). Quantitative data were explained as mean± standard deviation (SD). Qualitative data were explained as frequency and percentage. Non-parametric data was described by median and range. Data were analyzed to test statistically significant difference between groups. Differences between parametric variables were assessed using 2-tailed unpaired t-test. Pearson’s correlation coefficients were utilized to calculate correlations between the data displaying parametric allocation. P value < 0.05 was considered significant difference and p < 0.005 was measured highly significant discrepancy, at a confidence interval of 95%.
Results
The descriptive data of the studied group are described in table 1. The mean age for the asthmatic and control groups were (4.33 ± 2.041) and (4.289 ± 1.53) respectively. As regards the severity of asthma, all the children were from mild to moderate asthma.
Table 1: Descriptive data of the asthmatic and non-asthmatic children.
| Groups | N (%) | ||
|
Gender |
Asthmatics children | Male | 25 (44.6) |
| Female | 31 (55.4) | ||
| Non asthmatic | Male | 10 (47.6) | |
| Female | 11 (52.4) | ||
| Mean ± SD | Range | ||
| Age
|
Asthmatics children | 4.33 ± 2.041 | 1-9 |
| Non asthmatic | 4.289 ± 1.53 | 2-8 | |
| Severity of asthma score (SOA) | Asthmatics children | 16.36 ± 2.652 | 5-21 |
| Non asthmatic | NA | NA |
NA: non-applicable
Laboratory finding were illustrated in table 2. A significant difference between CXCL8levels between asthmatic and non-asthmatic group was reported P value < 0.001. As regard Microbiome log, a significant difference between fecal lactobacillus log was documented between the studied groups P <.001 while no significance difference was reported between the studied groups in Fecal Bifidobacterium Log (Table 3) .
Table 2: Lab findings and microbiota study in the studied groups
| Asthmatic group
Mean ± SD |
Non asthmatic
Mean ± SD |
t | P value | |
| CXCL8 (Pg/ml) | 293.52±34.3 | 223.24±8.9 | 6.700 | <.001 |
| P38MAPK (Pg/ml) | 186.13±7.13 | 161.73±5.76 | 10.601 | <.001 |
| Fecal Lactobacillus Log | 6.01±0.624 | 6.97± 0.805 | -3.675 | <.001 |
| Fecal Bifidobacterium Log | 5.76±0.658 | 5.860±0.785 | -0.347
|
0.731 |
Table 3: Correlations between lab markers and microbiota
| Pearson Correlation | Sig. (2-tailed) | |
| CXCL8 (Pg/ml) & Lactobacillus Log | -0.293 | 0.070 |
| CXCL8 (Pg/ml) & Bifidobacterium Log | -0.161 | 0.326 |
| p38MAPK (Pg/ml) & Lactobacillus Log | -0.343* | 0.033 |
| p38MAPK (Pg/ml) Bifidobacterium Log | -0.188 | 0.252 |
The quantities of Bifidobacterial and Lactobacillus in the fecal samples were measured (Fig. 1). The quantities of the two bacteria verified that the amount of Bifidobacterial in asthmatic children was higher than that in the non-asthmatic group whereas the amount of Lactobacillus in non-asthmatic was higher than asthmatic children.
Non-significant negative correlation was found between CXCL8 and both Lactobacillus Log and Bifidobacterium Log
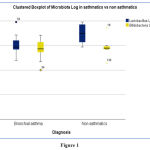 |
Figure 1 |
Discussion
Asthma in childhood is a heterogeneous disease with variable clinical manifestations, which depends on age, gender, genetic and environmental factors 23. Early childhood considered a critical phase for vulnerability to environmental exposures that introduced to the growing immune system through complex host environment interactions 24-25.
Immune microbe interactions in early life affect the risk of allergy, bronchial asthma, and other inflammatory diseases 26. Science birth, humans are exposed to various exposures continuously that effect microbiome ecosystem. Microbes colonize the gut directly after birth 27 In Childhood, children are colonized by extensive collection of microbes and bacteria in their gut, gut being the most heavily and extremely colonized organ 28 these microbes are associated with systemic and mucosal immune responses and allergy development late 29-30. It is well known that the microbial flora changes in its structure because of different lifestyles and diet in modern cultures which in turn, plays a role in the higher prevalence of allergy 31 .
Many previous epidemiologic studies suggested that early events in gut microbial exposure and colonization may have important roles in development of disease at sites remotely from the gastrointestinal tract.32.
The current study added to the previous research about the difference in the structure of the intestinal flora between allergic and non-allergic children. As it showed higher level of Bifidobacterium and lower level of Lactobacillus in asthmatic compared to non-asthmatic children
This agreed with Simonyté Sjödin et al., 2019 33 study which reported that Lactobacillus was underrepresented in allergic compared with nonallergic children; while, Bifidobacterium, was overrepresented in allergic children.
Similarly, studies of Björkander et al., 2020 34 and Johansson et al., 2011 reported that early Lactobacilli colonization seems to decrease the risk for allergy, the latter explained this protective role of Lactobacilli by its impact as probiotic supplementation early in life which reduces eczema and/or IgE sensitization 35
In contrast, Studies of Liwen et al., 2018, Fujimura et al., 2016, Kalliomäki et al., 2007, and Björkstén et al., 2001, all came with different results, as they reported lower colonization and lower relative abundance of bifidobacterial in stool samples of allergic and atopic infants and children. They suggested that colonization with bifidobacterial appear to be associated with protection against allergy, as these microorganisms are common to initiate a TH1-type immune response. Study of Stokholm et al., 2018 came with same results, as it suggested that the development of constant asthma in childhood may be mediated through a skewed maturation of the gut microbiome in the first year of life, predominantly in children born to asthmatic mothers36.
Although, our quantities results of Bifidobacterium were higher in asthmatic children than non-asthmatic, it agreed with other studies have linked the high concentration of Bifidobacterium in acute exacerbation of wheezy child only 37.Other found an associations between microbiota and immunologic features in subjects mild asthma 38 As our cases were fall in the mild or moderate asthma during the enrollment which explains the low level of Bifidobacterium. However, other studies didn’t detect significant changes in the prevalence of Bifidobacterium classes between fecal samples from healthy and allergic children 39. Hence, the differences in the amounts of fecal Bifidobacterium in children with wheezing diseases during the acute aggravation phase is still not clear.
Whereas a study of Arrieta et al., 2015 reported that gut community composition did not differ substantially among allergic and non-allergic children, as shown in 3-month and 1-year stool samples. Moreover, the study did not disclose any significant differences in variety between different allergic phenotypes 40.
This diversity in qualitative and quantitative pattern in microbiome log may be explained by the fact that microbial flora changes in its composition as a consequence of altered lifestyles different ecological factors and diet pattern 31
It is well known that allergy development is linked to immune dysregulation including alteration in microbiome and cytokines levels. As childhood allergy development is preceded by altered production of interleukins by stimulated peripheral blood mononuclear cells (PBMC) links with immunoglobulin (Ig)E‐sensitization and allergy 41 .
Immune responses are activated by chemokines and cytokines secreted by airway epithelial cells. Bronchial epithelial cells produce CXCL8in response to interleukin-4 (IL-4) and interleukin-13 presences, which are increased in asthmatics. IL8 is a potent chemotactic cytokine that trigger inflammatory cells by engaging mast cells, mononuclear phagocytes T lymphocytes, and neutrophils to the inflammation site 42.
The current results showed significant difference between asthmatic and non-asthmatic subgroups regarding CXCL8serum level. This could be explained by its vital role in the disease pathogenesis, as it is secreted by airway epithelium cells to help the host body fight infection and disease. In the same context, studies of Tang & Chen, 2000, and of Pukelsheim et al., 2010 reported that the mean serum CXCL8level was significantly higher in asthmatic children with acute aggravation or in controlled asthma compared to the control group 12-43.
Study of Wei et al., 2021 and Nakagome et al., 2012 assessed CXCL8level in sputum of asthmatic children and revealed increased level in sputum of children with exacerbated neutrophilic asthma. it was suggested that IL-8-stimulated neutrophils which could cause accumulation of eosinophils in the airways of asthmatic patients 44-45.
In contrast, Arzu et al., 2012 in his study on asthmatic adults, approved that the levels of CXCL8, did not differ between asthma patients whether newly diagnosed or severe persistent cases and control 46.
Recently, the relation between microbiome and level of cytokines and / or inflammatory markers have gained the attention but still unclear. As Many literatures have reported that Bifidobacterium were effective in inhibiting allergic airway response and suppressing autoimmune responses. These effects are related to the ability of Bifidobacterium to reduce the number of activated DCs and CD4+ T cells and the production cytokines. Moreover, Bifidobacterium longum dampen host pro-inflammatory responses.
P38MAPK is an inflammatory marker that induces the differentiation and activation of Th2 cells, which in turn promote the release of Th2 cytokines. Many studies assessed the cytokines level as CXCL8 in sputum in asthmatic children and found to be high in those children suggestion that high level of cytokines stimulate neutrophils which could lead to eosinophils accumulation in the airways of asthmatic patients. Other studies documented no difference between asthma patients whether newly diagnosed or severe persistent cases and control.
In our study we documented significant negative correlation between presence of asthma and serum markers CXCL8 and P38MAPK and a significant positive correlation between presence of asthma and Bifidobacterium Log.
Conclusions
We reported an association between CXCL8 level, P38MAPK and microbiome suggesting the link between the gut bacteria and inflammatory status. Moreover, the more the elevated CXCL8 level the more the P38 level which in turn will lead to more symptoms. The alteration of microbiome level associated with elevated markers level suggest the protective role of gut microbiome in asthma control.
Ethics approval and consent to participate
This study was approved by The Medical Research Ethics Committee of the National Research Centre. All the participated children were subjected to written informed consent from the legal guardians after explaining the aim and the procedures of the study.
the current study are available from the corresponding author on reasonable request.
Conflict of Interest
The authors declare that they have no competing interests
References
- Asher, I. and N. Pearce, Global burden of asthma among children. Int J Tuberc Lung Dis, 2014. 18(11): p. 1269-78.
- Azmeh, R., et al., Update in Pediatric Asthma: Selected Issues. Dis Mon, 2020. 66(4): p. 100886.
- Devonshire, A.L. and R. Kumar, Pediatric asthma: Principles and treatment. Allergy Asthma Proc, 2019. 40(6): p. 389-392.
- Taylor, D.R., et al., A new perspective on concepts of asthma severity and control. European Respiratory Journal, 2008. 32(3): p. 545.
- Oriss, T.B., et al., IRF5 distinguishes severe asthma in humans and drives Th1 phenotype and airway hyperreactivity in mice. JCI Insight, 2017. 2(10).
- Lee, N. and W.-U. Kim, Microbiota in T-cell homeostasis and inflammatory diseases. Experimental & Molecular Medicine, 2017. 49(5): p. e340-e340.
- Hooper, L.V., D.R. Littman, and A.J. Macpherson, Interactions between the microbiota and the immune system. Science, 2012. 336(6086): p. 1268-73.
- Haarman, M. and J. Knol, Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl Environ Microbiol, 2006. 72(4): p. 2359-65.
- Zhou, Y., et al., The upper-airway microbiota and loss of asthma control among asthmatic children. Nature Communications, 2019. 10(1): p. 5714.
- Du, C.L., et al., [Cytokine profiles of CD4(+) T memory cells in asthma and their relationship with asthma severity]. Zhonghua Yi Xue Za Zhi, 2017. 97(30): p. 2333-2337.
- Majumdar, S., A. Ghosh, and S. Saha, Modulating Interleukins and their Receptors Interactions with Small Chemicals Using In Silico Approach for Asthma. Curr Top Med Chem, 2018. 18(13): p. 1123-1134.
- Tang, R.B. and S.J. Chen, Evaluation of serum interleukin-8 as a marker of disease activity in acute asthma in children. J Asthma, 2000. 37(5): p. 409-13.
- Lea, S., et al., P38 MAPK and glucocorticoid receptor crosstalk in bronchial epithelial cells. Journal of Molecular Medicine, 2020. 98(3): p. 361-374.
- Pedersen, S.E., et al., Global strategy for the diagnosis and management of asthma in children 5 years and younger. Pediatric pulmonology, 2011. 46(1): p. 1-17.
- Kunc, P., et al., Biomarkers of bronchial asthma. Physiological research, 2020. 69: p. S29-S34.
- Zhang, L., et al., Detection of interleukin‐8 on microgapped dual electrodes for measuring asthma complication. Biotechnology and Applied Biochemistry, 2020.
- Reddel, H.K., et al., GINA 2019: a fundamental change in asthma management. Treatment of asthma with short-acting bronchodilators alone is no longer recommended for adults and adolescents, 2019. 53(6): p. 1901046.
- Eisner, M.D., A. Yegin, and B. Trzaskoma, Severity of asthma score predicts clinical outcomes in patients with moderate to severe persistent asthma. Chest, 2012. 141(1): p. 58-65.
- Expert Panel Report 3 (EPR-3): Guidelines for the Diagnosis and Management of Asthma-Summary Report 2007. J Allergy Clin Immunol, 2007. 120(5 Suppl): p. S94-138.
- Tang, R.-B. and S.-J. Chen, Evaluation of Serum Interleukin-8 as a Marker of Disease Activity in Acute Asthma in Children. Journal of Asthma, 2000. 37(5): p. 409-413.
- Fernandez-Bustamante, A., et al., Brief Glutamine Pretreatment Increases Alveolar Macrophage CD163/Heme Oxygenase-1/p38-MAPK Dephosphorylation Pathway and Decreases Capillary Damage but Not Neutrophil Recruitment in IL-1/LPS-Insufflated Rats. PLoS One, 2015. 10(7): p. e0130764.
- Johansson, M.A., et al., Early colonization with a group of Lactobacilli decreases the risk for allergy at five years of age despite allergic heredity. PLoS One, 2011. 6(8): p. e23031.
- Dharmage, S.C., J.L. Perret, and A. Custovic, Epidemiology of Asthma in Children and Adults. Frontiers in Pediatrics, 2019. 7(246).
- Prescott, S.L., Early-life environmental determinants of allergic diseases and the wider pandemic of inflammatory noncommunicable diseases. Journal of Allergy and Clinical Immunology, 2013. 131(1): p. 23-30.
- Bäckhed, F., et al., Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe, 2015. 17(5): p. 690-703.
- Henrick, B.M., et al., Bifidobacteria-mediated immune system imprinting early in life. Cell, 2021. 184(15): p. 3884-3898.e11.
- Milani, C., et al., The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev, 2017. 81(4).
- Chunxi, L., et al., The Gut Microbiota and Respiratory Diseases: New Evidence. J Immunol Res, 2020. 2020: p. 2340670.
- Zheng, D., T. Liwinski, and E. Elinav, Interaction between microbiota and immunity in health and disease. Cell Research, 2020. 30(6): p. 492-506.
- Collado, M.C., et al., Microbial ecology and host-microbiota interactions during early life stages. Gut microbes, 2012. 3(4): p. 352-365.
- Björkstén, B., et al., Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol, 2001. 108(4): p. 516-20.
- McLoughlin, R.M. and K.H. Mills, Influence of gastrointestinal commensal bacteria on the immune responses that mediate allergy and asthma. J Allergy Clin Immunol, 2011. 127(5): p. 1097-107; quiz 1108-9.
- Simonyté Sjödin, K., et al., Temporal and long-term gut microbiota variation in allergic disease: A prospective study from infancy to school age. Allergy, 2019. 74(1): p. 176-185.
- Björkander, S., et al., Childhood allergy is preceded by an absence of gut lactobacilli species and higher levels of atopy-related plasma chemokines. Clin Exp Immunol, 2020. 202(3): p. 288-299.
- Huurre, A., et al., Mode of delivery – effects on gut microbiota and humoral immunity. Neonatology, 2008. 93(4): p. 236-40.
- Stokholm, J., et al., Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun, 2018. 9(1): p. 141.
- Murray, C.S., et al., Fecal microbiota in sensitized wheezy and non-sensitized non-wheezy children: a nested case-control study. Clin Exp Allergy, 2005. 35(6): p. 741-5.
- Durack, J., et al., Distinct associations of sputum and oral microbiota with atopic, immunologic, and clinical features in mild asthma. J Allergy Clin Immunol, 2020. 146(5): p. 1016-1026.
- Akay, H.K., et al., The relationship between bifidobacteria and allergic asthma and/or allergic dermatitis: A prospective study of 0–3 years-old children in Turkey. Anaerobe, 2014. 28: p. 98-103.
- Arrieta, M.C., et al., Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med, 2015. 7(307): p. 307ra152.
- Björkander, S., et al., Childhood allergy is preceded by an absence of gut lactobacilli species and higher levels of atopy-related plasma chemokines. Clinical and experimental immunology, 2020. 202(3): p. 288-299.
- Mohamed, S.F., et al., Relation between Interleukin 8 and Bronchial Asthma in Children. The Egyptian Journal of Hospital Medicine, 2021. 85(2): p. 3621-3623.
- Pukelsheim, K., et al., Cytokine profiles in asthma families depend on age and phenotype. PLoS One, 2010. 5(12): p. e14299.
- Wei, Q., et al., Relationship between Th17-mediated immunity and airway inflammation in childhood neutrophilic asthma. Allergy Asthma Clin Immunol, 2021. 17(1): p. 4.
- Nakagome, K., S. Matsushita, and M. Nagata, Neutrophilic inflammation in severe asthma. Int Arch Allergy Immunol, 2012. 158 Suppl 1: p. 96-102.
- Yalcin, A.D., A. Bisgin, and R.M. Gorczynski, IL-8, IL-10, TGF-β, and GCSF levels were increased in severe persistent allergic asthma patients with the anti-IgE treatment. Mediators Inflamm, 2012. 2012: p. 720976.
List of abbreviations
CXCL8 : C-X-C Motif Chemokine Ligand 8
CD4 : cluster of differentiation 4
DNA : Deoxyribonucleic acid
ELISA: The enzyme-linked immunosorbent assay
PCR: polymerase chain reaction
URT : Upper respiratory tract
SPSS: statistical package for social science
Th2: T helper cell 2
p38MAPK : p38 mitogen-activated protein kinase
(Visited 210 times, 1 visits today)






