Manuscript accepted on :09-09-2022
Published online on: 19-09-2022
Plagiarism Check: Yes
Reviewed by: Dr. B. Kirthika
Second Review by: Dr. Sohayla Mohamed Elsherbini Attalla
Final Approval by: Dr. Jihan Seid Hussein
Sachin Chaudhary1* , Abdel-Nasser El-Shorbagi1
, Abdel-Nasser El-Shorbagi1 , Anurag Chaudhary2
, Anurag Chaudhary2 , Garima Agarwal2
, Garima Agarwal2 , Prabhash Nath Tripathi2
, Prabhash Nath Tripathi2 , Shweta Dumoga2
, Shweta Dumoga2
1Department of Medicinal Chemistry, College of Pharmacy, University of Sharjah, Sharjah, United Arab Emirates.
2Department of Pharmaceutical Technology, Meerut Institute of Engineering and Technology, NH-58, Baghpat Road Crossing, Bypass Road, Meerut, India.
Corresponding Author E-mail: schaudhary@sharjah.ac.ae
DOI : https://dx.doi.org/10.13005/bpj/2456
Abstract
The coronavirus 2 illness (Covid-19) global pandemic has resulted in severe infection causing fever, cough, shortness of breath, pneumonia and even death. WHO is monitoring intimately in coordination with scientific experts and government agencies, the transmission rate of this virus and its neoteric variants identified worldwide. Since the start of year 2020, all the health authorities of each country are working in collaboration to explore the scientific information on coronavirus and providing essential guidelines to save humans. Undoubtedly, this pandemic impacted many lives regardless of all preventive steps followed to minimize its transmission. Currently, multiple anti Covid-19 vaccines are available everywhere and government authorities are monitoring and providing the guidelines for taking booster dose of vaccine to minimize and control the transmission of different variants of this virus. It is rational to state that some of the available anti Covid-19 vaccines may not be highly efficient against new emerging variants so further investigation and research are the need of hour. Therefore, the present review portrayed the features of the novel variants and mutations of coronavirus 2 (Covid-19) and therapeutic updates associated with the effectiveness of different vaccines against new strains reported worldwide.
Keywords
Coronavirus 2; Health; Mutation; Pandemic; Vaccine
Download this article as:| Copy the following to cite this article: Chaudhary S, El-Shorbagi A. N, Chaudhary A, Agarwal G, Tripathi P. N, Dumoga S. The Recent Updates on Neoteric Variants of Covid-19 Virus and Therapeutic Effectiveness of Vaccines against the Variants. Biomed Pharmacol J 2022;15(3). |
| Copy the following to cite this URL: Chaudhary S, El-Shorbagi A. N, Chaudhary A, Agarwal G, Tripathi P. N, Dumoga S. The Recent Updates on Neoteric Variants of Covid-19 Virus and Therapeutic Effectiveness of Vaccines against the Variants. Biomed Pharmacol J 2022;15(3). Available from: https://bit.ly/3DzzJD2 |
Introduction
The neoteric variant of Covid-19 virus (coronavirus 2), the contributing representative of Covid-19, that flare-up in year 2019 and was declared as global pandemic in March 2020. As of April 2022, there have been beyond 0.5 billion cases universally with exclusive of 6.19 million loss of lives.1,2 This Covid-19 pandemic has outcome to social, cognitive, and economical collapse. Meanwhile, numerous Covid-19 vaccines have been developed in a very short period by late 2020 for emergency use and are competent towards distinctive variants to diminish this pandemic.3,4 The approval for emergency use of different vaccines by the World Health Organization in less than a year is remarkable and a matter of pride in medical research. In addition to the fact that mortality and morbidity rate of this pandemic is very high resulting to further epidemic outbursts. Hence, development of further advanced therapeutic and preventive approaches is very crucial. Undoubtedly, primary detection in early phase and proper treatment will minimize the outbreak of Covid-19.5.6
However, by the end of 2020, the new variants of Covid-19 have been perceived and recognized in numerous regions. The mutation of genetic material of viruses is a common process, and recently around 4000 transmutations in the spike amino acid chain of coronavirus 2 have been reported that prompted new travel restrictions, lockdowns and border closings and ultimately massive mortality rate. Furthermore, a viral mutant can abscond the human defense mechanism and proliferate or duplicate more competently within the human body.7-10
Covid-19 Virus Variants and Mutations
It is intrinsic capability of viruses to undergo mutation process continuously and resulted in variants. Covid-19 virus (β-coronavirus) consists of single-stranded positive ribonucleic acid (RNA) viruses (Figure 1). Therefore, likely to all RNA viruses, it is more susceptible to mutation by alternating its genetic code after replication in human body. This virus has integral structural RNA repair system, and hence its mutation process is relatively slow-going than almost further RNA viruses. The genome of Covid-19 virus investigated in month of October 2020 has nearly 20 mutations in relation to the first strain gnomically sequenced in January 2020. Subsequently, numerous mutations of Covid-19 virus have been detected around us and most of the genetic alterations in Covid-19 virus promoted extensive transmissibility and turned down susceptibility to natural immune system.11-15
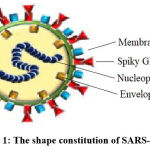 |
Figure 1: The shape constitution of SARS-CoV-2. |
Between the year 2020-2021, Covid-19 virus variants were recognized at a high rate that were more transmissible than currently reported strains and feasibly less permitted to neutralization by host antibodies. Additionally, it is a fact that further novel ‘variants of concern’ will be identified and reported in future. Hence it is highly recommended to develop a research agenda confirming rapid detection and characterization of variants of concern, to underpin swift and effective global human public health responses.16-18
COVID-19 Virus Variant Definition and Categorization
A variant is a viral genome containing intrinsic mutations. Subsequently, the group of variants with equivalent phenotypic alterations, such as a linkage or species of linkages, may be termed as VOC or VOI by the public health organizations around the world. Hence, a Covid-19 variant may have multiple mutations that distinguish it from other variants. Therefore, scientists of various countries are comparing the genetic differences to identify the variants and their relations.19-22
In a document released by World Health Organization on 25 February 2021, it is stated that, VOI is an isolate of Covid-19 virus that has genotypic and/or phenotypic modifications comparable to the standard reference genome and a VOC is a VOI that has remarkably more transmission rate, increase in virulence and/or ineffectively controlled by public health measures. The possible attributes of VOI are specific genetic markers affecting transmission rate, diagnosis, therapeutics, or immune escape. It requires sophisticated public health actions like increased surveillance, laboratory characterization, assessment of viral transmission rate and its severity and determination of efficacy of vaccines.23-25
A VOI (as interpreted above) is a VOC if, through a correlative analysis, it has been exhibited to be integrated with an escalation in transmission or destructive alterations in Covid-19 pandemic pattern26-30. The different variants of Covid-19 have been enlisted along with some effective approved vaccines in table 1.
Table 1: The summary of Coronavirus 2 (Covid-19) Variants and effectiveness of some approved vaccines.
| WHO Label | Linkage and additional mutation | First detected country | Spike mutations | Year of detection | Approved effective vaccines |
| Alpha | B.1.1.7 | United Kingdom | N501Y, D614G, P681H | September 2020 | BioNTech-Pfizer (BNT162b2) |
| Moderna (mRNA- 1273) |
|||||
| AstraZeneca (ChAdOx1-S/nCoV-19) | |||||
| Beta | B.1.351 | South Africa | K417N, E484K, N501Y, D614G, A701V | September 2020 | BioNTech-Pfizer (BNT162b2) |
| Moderna (mRNA- 1273) |
|||||
| Johnson & Johnson (Ad26.COV2.S) | |||||
| Gamma | P.1 | Brazil | K417T, E484K, N501Y, D614G, H655Y | December 2020 | Moderna (mRNA- 1273) |
| Johnson & Johnson (Ad26.COV2.S) |
|||||
| Delta | B.1.617.2 | India | L452R, T478K, D614G, P681R | December 2020 | Moderna (mRNA- 1273) |
| Omicron | B.1.1.529 | Multiple countries | N211del/L212I, Y145del, Y144del, Y143del, G142D, T95I, V70del, H69del, A67V | November 2021 | BioNTech-Pfizer (BNT162b2), |
| Moderna (mRNA-1273) |
COVID-19 VIRUS Discovered Variants
Alpha (B.1.1.7)
The Alpha variant (B.1.1.7) was the first identified variant, its trans ability and transmission rate was much higher than Covid-19 virus first strain. First time, it was detected in UK during Nov 2020. The rapid trans ability of this variant forced to think on the mutation in spiky glycoprotein of wild type Covid-19 virus and its different from the usual mutation has been observed, and it’s becoming in the category of VOC31. It is reported that a limited number of B.1.1.7 with E484K variations have been detected. Finally, on 31st May 2021, WHO declared that this VOC would be named as ‘Alpha’ for the public forum.32,33
This variant is acknowledged by various names like UK variant, English variant, and British variant as it was first identified in the UK and for UK it was called Kent variant because first sample was identified in Kent34-35. For science community, first time it was referred in the category of (VUI-202012/01) in late 2020. This variant was also called variant 20I/501Y.V1 by Nextstrain. The mutated form of alpha variant with E484K was named as VOC-202102/02. According to COG-UK Consortium, 4000 mutations has been identified in the alpha variant’s spiky glycoprotein.36
As compared to existing Covid-19 virus variants, the transmission rate of B.1.1.7. variant was 43 to 90%. According to COG-UK report, the transmissibility rate of this variant is 50-100% higher and another report said that it was 75% more transmissible between Oct-Nov 2020 in UK. Thereafter, later studies revealed that previous estimation of transmissibility was overestimated.37-38 The Dutch Ministry of Health reported that transmissibility of this variant fluctuated between 28%-47% higher than wild type in first six weeks of 2021. Thereafter, many studies and reports revealed the same transmissibility rate of this variant in many countries.39-41
The most observable mutation in descendant B.1.1.7 is N501Y, which is the modification of asparagine to tyrosine at 501 orientations. This position is present specifically in the spiky glycoprotein’s RBM, which is segment RBD interacts with ACE242-43. The mutations in RBD is difficult to recognize by antibody and easily bind to ACE2 which lead to more virus infection.44
Beta (B.1.351/ 501Y.V2)
The Beta version, (B.1.351/501Y.V2), was first discovered in last month of the year 2020 in southern part of African region, sparking a second COVID-19 outbreak before being transmitted worldwide. It was observed that between October 2020 and January 2020 in South Africa, Covid-19 positive cases were increased from 2,000 to over 20,000 in a day45-46. Beta variant was characterized VOC by Centers for Disease Control (CDC), and as of June 2021, the transmission rate of Beta variant was estimated to be beyond 50% in numerous areas in African continent. The Beta variant has three spike protein variations namely (K417N, E484K, and N501Y) as well as five N-terminal domain (NTD) alterations, one of which is a loss within the NTD supersite at position 242-244. K417N, which helps the virus adhere to human cells more tightly, E484K, helps the virus in escaping some antibodies, N501Y helps the viral genome to attach firmly to human cellular component.47
The competency of vaccine BNT162b against the Beta strain was assessed at around 72% in previously mentioned Qatar cohort trial, which was 15% lower than the Alpha version. In March 2022, the number of cases with beta variant were still the highest in the African continent with South Africa leading with 7169 cases, Europe and France had 3426 cases, US had 3154 cases and Canada had 1484 cases, India had 312 cases and the United Arab Emirates had no cases of the beta variant. The Beta variation has been a major source of worry for the Covid-19 immunization effort in terms of vaccine-induced immunity escape. Several authorized vaccines, including mRNA-1273, BNT162b2, BBIBP-CorV, ChAdOx1nCoV-19, NVX-CoV2373, and mRNA-1273 have exhibited lower serum neutralization.48-50
Gamma (P.1)
The Gamma variant (P.1) of Covid-19 virus, was designated as VOC in January, 2021. The Brazilian variant or Brazil variant are some of the other names for this variant as Brazil was the first place where this variant was discovered. The variant has demonstrated a high level of transmissibility. Descendant P.1 is a Covid-19 virus variation with modifications in amino acid peptide chain, 10 of which are in the spiky glycoprotein, of which 3 namely N501Y, E484K, and K417T are of exceptional interest. The agency NIID located in Japan, first discovered this strain of Covid-19 virus on January 6, 2021. The Gamma variation is made up of two sub derived variants, 28-AM-1/2, each includes the K417T, E484K, and N501Y mutations. Gamma is distinct from the Zeta form (descendant P.2), which was also widely distributed in Brazil.51-53 Zeta, an instance, only possesses the E484K mutation and none of the other two dangerous variants, N501Y and K417T. According to initial reports, P.1 and P.2 were two distinct offspring of the Brazilian descendant B.1.1.248. B.1.1.248 was reclassified to B.1.1.28 after losing its identity as a separate descendant. P.1 has also been referred to as B.1.1.28.1, and P.2 has been referred to as B.1.1.28.2 or VUI-202101/01. Because the PANGO descendant method of naming allows only three sub-levels, B.1.1.28.1 is assigned to P.1 and B.1.1.28.2 is assigned to P.2., following its discovery GISAID received genomic data for four samples of the novel variation. 54-56
Covid-19 virus variant gamma contains 10 alterations in its spiky glycoprotein, including N501Y and E484K, and open reading frames (ORF1a, ORF1b, and ORF8) gene. The descendant P.1, P.2, P.3, and P.4 of the coronavirus descendant B.1.1.28 have been identified as (VOC). The descendant P.2 (B.1.1.28.2, Zeta variation), discovered in Brazil in October 2020, has only one mutation with P.1: The E484K. The remaining P.2 mutations are unimportant and are only sometimes encountered in other forms. E484K in S-gene, A119S in N-gene, 5’UTR C100U, plus L3468V and synC11824U in ORF1ab-gene are the five P.2-specific mutations. 3’UTR C29754U, F120F (synC28253U) in ORF8, M234I in the N-gene, plus L3930F and synA12964G in ORF1ab are all prevalent mutations in P.2. In Philippines, descendant P.3 (Theta variation) was identified during the second month of 2021. The descendant P.4 is the only extant B.1.1.28 virus. Although its exact origin is unknown. It has the L452R mutation in the spiky glycoprotein, which has also been found in descendant B.1.617 (Delta and Kappa variants) from India and Epsilon variation (descendant B.1.427 and B.1.429) from California. The P.4.1 (VUI-NP13L) branch of this descendant, emerged in Goiás, Brazil, Japan, Netherlands, and England during the year 2020, in spiky glycoprotein, P.4.1 possesses mutations V1176F and D614G.57-60
Delta (B.1.617.2)
The Delta (δ) variant of Covid-19 virus (B.1.617.2), is the major ill-famed variant amongst Covid-19 virus variants. The δ species was identified in December 2020 in India and named during May 2021. The δ variety also exhibits a high rate of transmissibility in comparison with other Covid-19 virus variants that transmitted to more than 175 countries by November 2021. The Public Health England (PHE) mentioned a 10.8 percent secondary firing rate in infected persons of non-travel or unidentified cases for δ in comparison with the 10.2 % α variant; the death rate those persons were 386,835 with δ cases 0.3 %. In addition, 46 % of cases for unvaccinated and 6 % of deaths being below 50 years of age humans. Immunity from prior recoveries or Covid-19 immunizations is required. Therefore, it is regarded to be among the highly contagious respiratory viruses. As a result, the esteemed organizations like WHO and CDC specify this variant as VOC and became the prevalent strain globally.61,62
The δ version was recognized in the beginning in India, as a result as “Indian Variant.” The δ pedigree B.1.617 has been identified as the three variations in India. After that, WHO issued the tag of δ to descendant B.1.617.2 on May 2021. Three sub descendants come under descendant B.1.617. Among them, B.1.617.1 sub-descendant are named as Kappa. After some time, another VUI was identified as both sub-variants, such as B.1.617.2 and B.1.617.3. In addition, B.1.617.1 and B.1.617.3 variants don’t have T478K. At the same time, ECDC released a concise maintaining of the three sub-descendant B.1.617 regarding VOI. Additionally, the δ versions further subcategorized in the variants (AY.1 – AY.28) in August 2021. However, these variants (AY) have been found in all over the world. Among them AY.4 to AY.11 variants were the largest in the United Kingdom, whereas in USA and Mexico, various types of variants also present such as AY.2, AY.3, AY.13, AY.14, AY.25, AY.20.63-64
The δ variant had mutated the gene expressing the spiky glycoprotein of Covid-19 virus, resulting in the replacements T478K, P681R, and L452R, which have been reported to impact the viral infection. Mutations also specify the changes in genes expressing its spiky glycoprotein resulting in the alterations of D614G, T478K, P681R, and L452R. The descendant of B.1.617.2 genome of δ has thirteen alterations that cause mutation in the peptide chain of proteins it encodes.65-66
These spiky glycoproteins of the δ variant of the virus have all codes where among them, four are especially concern; In the D614G, aspartic acid (Asp)-to-glycine (Gly) replacement at site 614 is conserved through other highly pathogenic variants such as α, Beta (β), and Gamma (ϒ). In the second mutation T478K, the transition occurred at point 478 from threonine (Tyr) to lysine (Lys). Furthermore, at position 452 of L452R mutations, the alteration from leucine-to arginine increases the spiky glycoprotein affinity towards the ACE-2 receptor and lowers immune system recognition capabilities. In the mutation P681R, the substitution at site 681 from proline-to-arginine may upgrade the variant’s cell-level transmissibility by splitting S precursor protein to the active S1/S2 configuration. Finally, another mutation of E484Q does not exist in the genome of B.1.617.2.67-69
Delta plus (δ+) variant having K417N mutation that was initially matched to AY.1 and AY.2, then linked with descendant AY.3. Since about October 2021, this AY.3 variations has had an accumulative frequency of around 5% in Americans and 2% globally. The AY.4.2 version maybe 10-15% higher transmittable than the classic Delta type. AY.4.2 increases at a rate of around 15% quicker every week.70
Omicron (B.1.1.529)
Around the end of year 2021, Omicron was transmitted in varied countries, after drastic escalation in Covid-19 cases in Gauteng province of South African region confirming the novel variant’s detection. The first Omicron confirmed case was reported on 9 November 2021, by the end of 2021 it was transmitted and identified at 22 countries including Asian, African, European, American continents. Omicron has many mutations, some of which are alarming. The mutations include T91 in the envelope, P13L, E31del, R32del, S33del, R203K, G204R in the nucleocapsid protein, D3G, Q19E, A63T in the matrix, N211DEL/L212I, Y145DEL, Y144DEL, Y143DEL, G142D, T95I, V70DEL, H69DEL, A67V in the N-terminal domain of the spike, Y505H, N501Y, Q498R, G496S, Q493R, E484A, T478K, S477N, G446S, N440K, K417N, S375F, S373P, S371L, G339D in the receptor-binding domain of the spike, D796Y in the fusion peptide of the spike, L981F, N969K, Q954H in the heptad repeat 1 of the spike as well as multiple other mutations in the non-structural proteins and spiky glycoprotein.71-73
The South African epidemiologists suggested an increased risk of reinfection with Omicron, in comparison to other VOC. The mutations were so promising that many scientists raised the concern of partial resistance of this strain to approved Covid-19 vaccines, although the investigations have been in process. It is still unclear whether the transmission rate of Omicron is more compared to other variants of this virus, or whether infection resulted from Omicron is caused more severe illness. In one of the published reports of WHO on 7 December 2021 it was mentioned that 57 countries globally have identified Omicron cases. However, the actual transmission figure of its spread might be in excess due to retarded declaration and inadequate reconnaissance in some countries.74-76
The delta variant remains the most predominant and is responsible for 99.8% of Covid-19 positive cases globally in relation with previously identified variants. Omicron has enormous mutations in its genomic structure, more than 30 mutations are in the spiky glycoprotein which Covid-19 virus uses to gain entry to our cells, allowing it to multiply and transmit.77-80
New Challenges on Public Health due to New Variants
The countries around the globe are facing the disgraceful public health crisis than ever due to Covid-19 virus. However, in a short period of time since this pandemic started several vaccines have been approved my health authorities but since this virus changes its properties very quickly, so some of the vaccines have been found ineffective against the new variants.1,12-30
It has been reported that humans administered with single and double dose of Covid-19 vaccine have 75-94% less chances to get hospitalized compared with those have not taken any of the vaccine doses. It is well known fact that, vaccination helps boosting a prolonged defense feedback and the severity of Covid-19 can only be reduced by increasing the vaccination programme everywhere in the world. Additionally, it is also important to provide the booster dose of vaccines to reduce morbidity and mortality rate worldwide.55-60
Conclusion
The replication of Covid-19 virus genome leading to emergence of genetic variants, most of these variants influence virus transmission, extremity of disease including the vaccine induced immune response in global population. The variants of Covid-19 virus resulted to increased transmission rates, morbidity, and mortality. Additionally, variants escape detection through diagnostics kits, resulting to decreased susceptibility for treatment by antiviral agents, monoclonal antibodies, and plasma therapy.
The clinical outcomes of infection caused by Covid-19 variants should be explored meticulously in children, pregnant women, and older people. The studies should explore the impact of variants on immune response of the people and response to therapeutics also need to be assessed. The framework of modern techniques of genome sequencing are needed to support large scale surveillance and assessment. This framework will help to identify, track and estimate new variants globally.
It has been reported that new strains of Covid-19 virus are around 50% more contagious than the first strain of Covid-19. The approved vaccines are likely to be less effective against the new strains and warrant further studies to confirm the efficacy. The disease control should continue to be prioritized to minimize the risk of mortality. It is highly recommended that vaccination technology must be updated regularly to increase the immunity of humans against different variants of Covid-19 virus and vaccination programmes for booster dose should be implemented at massive scale in all countries. The global synchronized regulatory processes are required to increase fast development, assessment of reformed vaccines targeting the variants.
Acknowledgment
The work on this review was supported by University of Sharjah, United Arab Emirates and Meerut Institute of Engineering and Technology, Meerut, Uttar Pradesh, India.
Conflict of interest
The authors declare no conflict of interest.
Funding Sources
There is no funding Source.
References
- Chaudhary S, El-Shorbagi AN, Gupta RK, Kumar A. The recent updates on approaches and clinical trials status of covid-19 vaccines developed globally. Biomed. Pharmacol. J., 2021; 14(3): 1109-1124.
CrossRef - Arvin AM, Fink K, Schmid MA, Cathcart A, Spreafico R, Havenar-Daughton C, Lanzavecchia A, Corti D, Virgin HW. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature, 2020; 584: 353-363.
CrossRef - Jiang S. Don’t rush to deploy COVID-19 vaccines and drugs without sufficient safety guarantees. Nature, 2020; 579: 321.
CrossRef - Khamsi R. If a coronavirus vaccine arrives, can the world make enough? Nature., 2020; 580: 578-580.
CrossRef - Callaway E. The underdog coronavirus vaccines that the world will need if front runners stumble. Nature, 2020; 585: 332-333.
CrossRef - Dror AA, Eisenbach N, Talber S, Morozov NG, Mizrachi M, Zigron A, Srouji S, Sela E. Vaccine hesitancy: the next challenge in the fight against COVID-19. Eur. J. Epidemiol., 2020; 35: 775-779.
CrossRef - Woo PC, Huang Y, Lau SK, Yuen K. Coronavirus genomics and bioinformatics analysis. Viruses., 2010; 2(8): 1804-1820.
CrossRef - Zhang L, Wang W, Wang S. Effect of vaccine administration modality on immunogenicity and efficacy. Expert Rev. Vaccines., 2015; 14(11): 1509-1523.
CrossRef - Zheng J. SARS-CoV-2: An emerging coronavirus that causes a global threat. Int. J. Biol. Sci., 2020; 16(10): 1678-1685.
CrossRef - Le TT, Andreadakis Z, Kumar A et al. The COVID-19 vaccine development landscape. Nature, 2020; 19: 306.
CrossRef - Hossain MK, Hassanzadeganroudsari M, Apostolopoulos V. The emergence of new strains of SARS-CoV-2. What does it mean for COVID-19 vaccines? Expert Rev. Vaccines., 2021; 20(6): 635-638.
CrossRef - Ball P. The lightning-fast quest for COVID vaccines-and what it means for other diseases. Nature., 2021; 589(7840): 16-18.
CrossRef - Callaway E. The coronavirus is mutating-does it matter? Nature., 2020; 585(7824): 174-177.
CrossRef - Mahase E. Covid-19: what new variants are emerging and how are they being investigated? BMJ., 2021; 372: n158.
CrossRef - Zhang L, Shen FM, Chen F, Lin Z. Origin and evolution of the 2019 novel coronavirus. Clin. Infect. Dis., 2020; 71(15): 882-883.
CrossRef - Zhang YZ, Holmes EC. A genomic perspective on the origin and emergence of SARS-CoV-2., 2020; 81(2): 223-227.
CrossRef - Zhang J, Jia W, Zhu J, Li B, Xing J, Liao M, Qi W. Insights into the cross-species evolution of 2019 novel coronavirus. Infect., 2020; 80(6): 671-693.
CrossRef - Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol., 2019; 17: 181-192.
CrossRef - Sanjuan R, Domingo-Calap P. Mechanisms of viral mutation. Mol. Life Sci., 2016; 73(23): 4433-4448.
CrossRef - Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Rev. Microbiol., 1997; 51: 151-178.
CrossRef - Taha BA, Al-Jubouri Q, Al Mashhadany Y, Zan MSDB, Bakar AAA, Fadhel MM, Arsad N. Photonics enabled intelligence system to identify SARS-CoV 2 mutations. Microbiol. Biotechnol., 2022; 29: 1-16.
CrossRef - Noh JY, Jeong HW, Shin EC. SARS-CoV-2 mutations, vaccines, and immunity: implication of variants of concern. Signal Transduct. Target Ther., 2021; 6(1): 203.
CrossRef - Jia Z, Gong W. Will mutations in the spike protein of SARS-CoV-2 lead to the failure of COVID-19 vaccines? Korean Med. Sci., 2021; 36(18): 1-11.
CrossRef - Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science., 2020; 367(6483):1260-1263.
CrossRef - Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front. Immunol., 2020; 11: 576622.
CrossRef - Koyama T, Weeraratne D, Snowdon JL, Parida L. Emergence of drift variants that may affect COVID-19 vaccine development and antibody treatment. Pathogens., 2020; 9(5): 324.
CrossRef - Liu Z, VanBlargan LA, Bloyet LM, Rothlauf PW, Chen RE, Stumpf S, et al. Identification of SARSCoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 2021;29(3):477-488.
CrossRef - Chan KK, Dorosky D, Sharma P, Abbasi SA, Dye JM, Kranz DM, Herbert AS, Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science., 2020; 369: 1261-1265.
CrossRef - Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science., 2020; 369: 1014-1018.
CrossRef - Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science., 2020; 369: 643-650.
CrossRef - Kumar N, Quadri S, AlAwadhi AI, AlQahtani M. COVID-19 recovery patterns across Alpha (B.1.1.7) and Delta (B.1.617.2) variants of SARS-CoV-2. Immunol., 2022; 13: 812606.
CrossRef - Meyer M, Holfter A, Ruebsteck E, Gruell H, Dewald F, Koerner RW, Klein F, Lehmann C, Huenseler C, Weber LT. The alpha variant (B.1.1.7) of SARS-CoV-2 in children: first experience from 3544 nucleic acid amplification tests in a cohort of children in Germany. Viruses., 2021; 13(8): 1600.
CrossRef - Brookman S, Cook J, Zucherman M. Effect of the new SARS-CoV-2-variant B.1.1.7 on children and young people. Lancet Child. Adolesc. Health., 2021; 5(4): e9-e10.
CrossRef - Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir. Med., 2021; 9(2): e20-e21.
CrossRef - Khan A, Gui J, Ahmad W, Haq I, Shahid M, Khan AA, Shah A, Khan A, Ali L, Anwar Z, Safdar M, Abubaker J, Uddin NN, Cao L, Wei DQ, Mohammad A. The SARS-CoV-2 B.1.618 variant slightly alters the spike RBD-ACE2 binding affinity and is an antibody escaping variant: a computational structural perspective. RSC Adv., 2021;11(48): 30132-30147.
CrossRef - van Oosterhout C, Hall N, Ly H, Tyler KM. COVID-19 evolution during the pandemic- Implications of new SARS-CoV-2 variants on disease control and public health policies., 2021; 12(1): 507-508.
CrossRef - Solen K,Delphine P, Sandrine I, Isabelle S, Julien P, Nicolas R, Julien R, Timothée B, Thomas V, Olivier S, Laurent B, Hélène P, David V. Transmission of SARS-CoV-2 Alpha Variant (B.1.1.7) From a BNT162b2-Vaccinated Individual. Open Forum Infect Dis., 2021; 8(8): 1-4.
CrossRef - Foresta C, Rocca MS, Di Nisio A.Gender susceptibility to COVID-19: A review of the putative role of sex hormones and X chromosome. J. Endocrinol. Invest., 2021; 44: 951-956.
CrossRef - Santos JC, Passos GA. The high infectivity of SARS-CoV-2 B.1.1.7 is associated with increased interaction force between Spike-ACE2 caused by the viral N501Y mutation. bioRxiv 2021:2020.12.29.424708.
CrossRef - Abu-Raddad LJ, Chemaitelly H, Butt AA. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N. Engl. J. Med., 2021; 385: 187-189.
CrossRef - Thiagarajan K. Why is India having a covid-19 surge? BMJ., 2021; 373: n1124.
CrossRef - Suvvari TK, P C, Kuppili S, Kandi V, Kutikuppala LVS, Kandula VDK, Mishra S, Sarangi AK, Mohapatra RK, Dhama K. Consecutive hits of COVID-19 in India: The mystery of plummeting cases and current scenario. Razi. Inst., 2021; 76(5): 1165-1174.
- Prathipati KK, Mishra M, Rathod B, Tripathy JP, B H S, Bidkar V, Dabhekar S, Shete V, Deepa G. Symptomatology and relationship between symptoms and duration among COVID-19 patients in a COVID-19 care hospital in central India. , 2022; 14(1): e21541.
CrossRef - Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. Virol., 2010; 84, 3134-3146.
CrossRef - Kistler KE, Bedford T. Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229E. ,2021;10: e64509.
CrossRef - Day T, Gandon S, Lion S, Otto SP. On the evolutionary epidemiology of SARS-CoV-2. Biol., 2020;30: R849-R857.
CrossRef - Karim SSA, de Oliveira T. New SARS-CoV-2 variants -Clinical, public health, and vaccine implications. Engl. J. Med., 2021; 384: 1866-1868.
CrossRef - V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Rev. Microbiol., 2020;19: 155-170.
CrossRef - Yadav PD, Sapkal GN, Ella R, Sahay RR, Nyayanit DA, Patil DY, Deshpande G, Shete AM, Gupta N, Mohan VK, Abraham P, Panda S, Bhargava B. Neutralization of Beta and Delta variant with sera of COVID-19 recovered cases and vaccines of inactivated COVID-19 vaccine BBV152/Covaxin. J. Travel Med., 2021; 28(7): 1-3.
CrossRef - Le TT, Cramer JP, Chen R, Mayhew S. Evolution of the COVID-19 vaccine development landscape. Nat. Rev. Drug Discov., 2020;19: 667-668.
CrossRef - Offit PA. Covid-19 boosters-Where from here? Engl. J. Med., 2022; 386:1661-1662.
CrossRef - Giovanetti M, Benedetti F, Campisi G, Ciccozzi A, Fabris S, Ceccarelli G, Tambone V, Caruso A, Angeletti S, Zella D, Ciccozzi M. Evolution patterns of SARS-CoV-2: Snapshot on its genome variants. Biophys. Res. Commun., 2021; 538: 88-91.
CrossRef - Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, Zhang Z, Fan P, Dong Y, Yang Y, Chen Z, Guo Y, Zhang J, Li Y, Song X, Chen Y, Xia L, Fu L, Hou L, Xu J, Yu C, Li J, Zhou Q, Chen W. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. , 2020; 369(6504): 650-655.
CrossRef - Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, von Gottberg A, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao NY, Korsman S, Davies MA, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LCJ, Kosakovsky Pond SL, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C, de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. , 2021; 592(7854): 438-443.
CrossRef - Chakraborty D, Agrawal A, Maiti S. Rapid identification and tracking of SARS-CoV-2 variants of concern. The Lancet., 2021; 397(10282): 1346-1347.
CrossRef - Medema G, Been F, Heijnen L, Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: Opportunities and challenges. Curr. Opin. Environ. Sci. Health., 2020, 17, 49-71.
CrossRef - Duong D. Alpha, Beta, Delta, Gamma: What’s important to know about SARS-CoV-2 variants of concern? CMAJ., 2021; 193(27): E1059-E1060.
CrossRef - Yépez Y, Marcano-Ruiz M, Bezerra RS, Fam B, Ximenez JP, Silva WA Jr, Bortolini MC. Evolutionary history of the SARS-CoV-2 Gamma variant of concern (P.1): a perfect storm. Genet. Mol. Biol., 2022; 45(1): e20210309.
CrossRef - Cheng MH, Krieger JM, Banerjee A, Xiang Y, Kaynak B, Shi Y, Arditi M, Bahar I. Impact of new variants on SARS-CoV-2 infectivity and neutralization: A molecular assessment of the alterations in the spike-host protein interactions. iScience., 2022; 25(3):103939.
CrossRef - Perez-Gomez R. The Development of SARS-CoV-2 Variants: The Gene Makes the Disease. J Dev Biol., 2021; 9(4): 58.
CrossRef - Yu F, Lau LT, Fok M, Yiu-Nam Lau J,ZhangCOVID-19 Delta variants- Current status and implications as of August 2021. Precis. clin. med., 2021; 4(4): 287-292.
CrossRef - Scudellari M. How the coronavirus infects cells- and why Delta is so dangerous. , 2021; 595: 640-644.
CrossRef - Shahab MS, Imam SS, Jahangir MA. A review on the contemporary status of mutating coronavirus and comparative literature study of current COVID-19 vaccines. Int. J. Pharm. Pharmacol., 2021; 5(1): 1-9.
CrossRef - Roy B, Dhillon JK, Habib N, Pugazhandhi B. Global variants of COVID-19: Current understanding. J. Biomed. Sci., 2021; 8(1): 8-11.
CrossRef - Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, Rakshit P, Singh S, Abraham P, Panda S, Team N. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. 2021; 9(7):1542.
CrossRef - Cyranoski D. Profile of a killer: the complex biology powering the coronavirus pandemic. , 2020; 581(7806): 22-26.
CrossRef - Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020; 98(7): 495-504.
CrossRef - Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. J. Antimicrob. Agents., 2020; 55(3): 105924.
CrossRef - Luo H, Tang QL, Shang YX, Liang SB, Yang M, Robinson N, Liu JP. Can Chinese medicine be used for prevention of corona virus disease 2019 (COVID-19)? A review of historical classics, research evidence and current prevention programs. J. Integr. Med., 2020; 26(4): 243-250.
CrossRef - Maitra A, Sarkar MC, Raheja H, Biswas NK, Chakraborti S, Singh AK, Ghosh S, Sarkar S, Patra S, Mondal RK, Ghosh T, Chatterjee A, Banu H, Majumdar A, Chinnaswamy S, Srinivasan N, Dutta S, Das S. Mutations in SARS-CoV-2 viral RNA identified in Eastern India: possible implications for the ongoing outbreak in India and impact on viral structure and host susceptibility. Biosci., 2020; 45(1): 76.
CrossRef - Ferré VM, Peiffer-Smadja N, Visseaux B, Descamps D, Ghosn J, Charpentier C. Omicron SARS-CoV-2 variant: What we know and what we don’t. Anaesth. Crit. Care Pain Med., 2022; 41(1):100998.
CrossRef - Torjesen I. Covid-19: Peak of viral shedding is later with omicron variant, Japanese data suggest. BMJ., 2022; 376: 89.
CrossRef - Sun Y, Lin W, Dong W, Xu J. Origin and evolutionary analysis of the SARS-CoV-2 Omicron variant. J. Biosaf. Biosecurity., 2022; 4(1): 33-37.
CrossRef - Khandia R, Singhal S, Alqahtani T, Kamal MA, El-Sha NA, Nainu F, Desingu PA, Dhama K. Emergence of SARS-CoV-2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ. Res., 2022; 209: 112816.
CrossRef - Kandeel M, Mohamed MEM, Abd El‐Lateef HM, Enugopala KN, El‐Beltagi HS. Omicron variant genome evolution and phylogenetics. Med. Virol., 2022; 94: 1627-1632.
CrossRef - Kandeel M, Ibrahim A, Fayez M, Al‐Nazawi M. From SARS and MERSCoVs to SARS‐CoV‐2: Moving toward more biased codon usage in viral structural and nonstructural genes. J. Med. Virol., 2020; 92(6): 660‐666.
CrossRef - Kupferschmidt K. Where did ‘weird’ Omicron come from? Science., 2021; 374(6572): 1179.
CrossRef - Abu-Raddad LJ, Chemaitelly H, Butt AA. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N. Engl. J. Med., 2021; 385: 187-189.
CrossRef - Okpeku, M. Possibility of COVID-19 eradication with evolution of a new omicron variant. Dis. Poverty., 2022; 11(30): 1-3.
CrossRef - Wolter N, Jassat W, Walaza S, Welch R, Moultrie H, Groome M, Amoako DG, Everatt J, Bhiman JN, Scheepers C, Tebeila N, Chiwandire N, du Plessis M, Govender N, Ismail A, Glass A, Mlisana K, Stevens W, Treurnicht FK, Makatini Z, Hsiao N, Parboosing R, Wadula J, Hussey H, Davies M-A, Boulle A, von Gottberg A, Cohen C. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet., 2022; 399: 437-446.
CrossRef







