Manuscript accepted on :03-06-2022
Published online on: 15-06-2022
Plagiarism Check: Yes
Reviewed by: Dr. Debarshi Kar Mahapatra
Second Review by: Dr. T. Karthikeyan
Final Approval by: Dr. Ian James Martin
Muzna Said Rashed Al-Mamari1 , Shadia Al-Sinawi2
, Shadia Al-Sinawi2 , Fathiya Salim AL-Rahbi2 and Mohamed Mabruk1*
, Fathiya Salim AL-Rahbi2 and Mohamed Mabruk1*
1Department of Allied Health Sciences, College of Medicine and Health Sciences, Sultan Qaboos University, Oman
2Department of Pathology, College of Medicine and Health Sciences, Sultan Qaboos University, Muscat, Oman
Corresponding Author E-mail: mabruk@squ.edu.om
DOI : https://dx.doi.org/10.13005/bpj/2418
Abstract
Epstein Barr virus (EBV) has been incriminated in the pathogenesis of Hodgkin’s lymphoma (HL) and non-Hodgkin’s lymphoma (NHL). The present study aimed to investigate the prevalence and the pattern of the expression of Epstein-Barr in HL and NHL tissue samples obtained from Omani patients attending Sultan Qaboos University Hospital (SQUH). Besides, to compare the sensitivity and specificity of immunohistochemistry(IHC) and in situ hybridization (ISH) for the detection of EBV in HL and NHL and finally to have more understanding of the pathogenesis of EBV in HL and NHL among patients in Oman. Formalin-fixed paraffin-embedded tissue samples consisting of 26 Hodgkin and 34 non-Hodgkin lymphomas were assessed for the presence of EBV by IHC to detect Latent membrane protein (LMP), expression and by using ISH to detect Epstein -Barr encoded RNAs (EBERs). The expression of LMP and EBERs were detected respectively in 46.2% and 57.7% of Hodgkin’s lymphoma cases and were detected in 11.8% and 14.7% respectively of non-Hodgkin’s lymphoma cases. The intensity of LMP-1 and EBER expression was significantly high in mixed cellularity compared to other subtypes. The expression of EBV was detected in transformed cells in both HL & NHL. The expression of EBV in transformed cells in both HL and NHL indicates that EBV may play a pro vital role in the pathogenesis of HL and NHL among patients in Oman. Moreover, this study indicates that IHC is to some degree compatible in terms of sensitivity and specificity to ISH in the detection of EBV in HL and NHL.
Keywords
Epstein Barr virus; Hodgkin’s Lymphoma; In Situ hybridization; Immunohistochemistry; non-Hodgkin’s lymphoma; Omani patients
Download this article as:| Copy the following to cite this article: Al-Mamari M. S. R, Al-Sinawi S, AL-Rahbi F. S, Mabruk M. The Prevalence and the Patterns of the Expression of Latent Epstein-Barr Virus in Hodgkin’s and Non-Hodgkin’s Lymphomas Among Patients in Oman: Immunohistochemistry Versus n Situ Hybridization. Biomed Pharmacol J 2022;15(2). |
| Copy the following to cite this URL: Al-Mamari M. S. R, Al-Sinawi S, AL-Rahbi F. S, Mabruk M. The Prevalence and the Patterns of the Expression of Latent Epstein-Barr Virus in Hodgkin’s and Non-Hodgkin’s Lymphomas Among Patients in Oman: Immunohistochemistry Versus n Situ Hybridization. Biomed Pharmacol J 2022;15(2). Available from: https://bit.ly/39shMcU |
Introduction
Lymphomas consist of various groups of clonal (malignant) lymphoproliferative disorders, classified by WHO based on lymphocytic origin1. The determination of whether the lymphoma is classified as Hodgkin’s lymphoma or non-Hodgkin’s lymphoma is based entirely on the type of the abnormal cells identified in tumor tissues 2.
Hodgkin’s lymphoma is a haematological malignancy characterized by a proliferation of large atypical cells called Hodgkin’s Reed–Sternberg cells (HRCs)2. The WHO classified Hodgkin’s lymphoma (HL) into a classical Hodgkin’s lymphoma and nodular lymphocyte-predominant Hodgkin’s lymphoma 1,2. Classical Hodgkin’s lymphoma (HL) is subdivided into nodular sclerosis, lymphocyte rich, mixed cellularity and lymphocyte depleted HL 1,2. The pathological effects of the virus are controlled by the immune system in healthy individuals while in immunosuppressed individuals, like acquired immune deficiency syndrome (AIDS) patients and transplant recipients, EBV can cause abnormal growth of infected B-cells and this may help in transforming normal lymphocytes 3-5. Many evidence link EBV to Hodgkin lymphoma including detection of EBV nucleic acid in the HRCs, and increased antibody titers to EBV viral capsid antigen 6-11.
On the other hand, non-Hodgkin’s lymphomas are the hematologic malignancy with the highest prevalence worldwide and all types of lymphoma other than Hodgkin’s are categorized under NHL 1,2. Non-Hodgkin’s lymphoma (NHL) is classified into indolent (low grade) NHL and highly aggressive (intermediate and high grade) NHL based on WHO classification 1,2. Indolent (low grade) NHL is subdivided into follicular lymphoma, small lymphocytic lymphoma, marginal zone lymphoma, lymphoplasmacytic lymphoma, mantle cell lymphoma, and mycosis fungoid lymphoma (T-cell) 2. Highly aggressive (intermediate and high grade) NHL is sub-classified into diffuse large B cell lymphoma, Burkitt lymphoma, primary CNS lymphoma, lymphoblastic lymphoma(most are T cell), anaplastic large cell lymphoma (T cells) and Sezary syndrome (T cells). 12 Many studies have shown a consistent association between EBV and HL worldwide 12,13 but no study was shown an association between EBV and HL and NHL in Oman so far.
EBV was detected for the first time in a Burkitt lymphoma cell line in the year 1964. EBV has been implicated in the pathogenesis of Hodgkin’s and non-Hodgkin’s lymphoma 14,15. EBV exhibit three different latency programs, these programs describe the EBV gene expression patterns in different cell lines 14-16. Latent EBV genomes can multiply in dividing memory cells (type I), induce B-cell differentiation (type II), or activate naïve B cells (type III) by using different transcription programs 17. EBNA1(Epstein-Barr nuclear antigen-1), EBER1, and EBER2 are only expressed in the type I latency program, which is observed in Burkitt’s lymphoma 13,16. LMP1/2A, EBER, and EBNA1are expressed in the type II latency program, which is seen in Hodgkin’s and lymphoma 4,13,16. The entire latency gene products are expressed in the type III latency program which is often detected during acute infectious mononucleosis 4,6,8,17
Latent membrane proteins (LMP 1,2) are the two latent proteins produced by the EBV genome. LMP1 is considered the major transforming protein of EBV which is involved in the activation and transformation of human B-lymphocytes 3,4,6. The ultimate outcome of the expression of LMP1 expression in the cell is the induction of adhesion molecules on cell surface and up-regulation of anti-apoptotic proteins 3,12.
Furthermore, LMP-1 protein can block apoptosis by the up-regulation of several anti-apoptotic proteins, including Bcl-2, A20 and p53-mediated apoptosis 10 LMP2A and LMP2B are the two distinct proteins yielded by the encoding of the LMP2 gene 17. LMP2A and LMP2B structures are similar and neither LMP2A nor LMP2B is essential for B-cell 3,17
Up to 11 viral genes are expressed during latency of the Epstein-Barr virus (EBV) which encodes up to nine proteins, two of these genes, EBER-1 and EBER-2 18. The viral genes EBER-1 and EBER-2 are transcribed by polymerase III in every latently Epstein-Barr virus-infected cell; they are the most abundant transcripts in latently EBV-infected cells 18,19.
EBER1,2 have served as excellent targets to detect EBV in tumors 18. Several studies have reported the localization of the EBERs, these transcripts were found either in the nuclear membrane in the cytoplasm or the nucleus 20.
Associations between EBV and tumors were made based on serologic and/or epidemiologic findings. 21 However, other specific assays can detect latent EBV infection within the tumor20. Among these methods used for the detection of EBV includes immunohistochemistry (IHC) for latent membrane protein (LMP-1) and in situ hybridization (ISH) for EBER RNAs. These two techniques have become increasingly popular in the laboratory because they can localize virus to the tumor cell, they are easy to apply and can be used on archived pathology specimens 4,21-24.
Immunohistochemistry and in situ hybridization have proved to be effective in detecting EBV in paraffin-embedded tissues 22-24. Histochemical assays are commonly used for localizing nucleic acids and proteins of EBV in malignant/ RS cells 20. However, in situ hybridization(ISH) was shown to be sufficiently sensitive to detect a low copy number of EBV 22-25, and has been recommended as the best technical approach for localizing and detecting EBER in tissue sections 20, It is a gold standard technique but it requires a long time of processing 22-24. Therefore, several studies have detected LMP-EBV and EBER-EBV in HL and NHL by using Immunohistochemistry and in situ hybridization 25.
The objective of the present study is to determines the rate and the patterns of the expression of EBERs and LMP proteins of Epstein –Barr virus (EBV) in formalin-fixed paraffin-embedded tissue Hodgkin’s and non-Hodgkin’s lymphomas tissue samples obtained from 60 Omani patients. The outcome of this study may help to understand the pathogenesis of EBV in Hodgkin’s and non-Hodgkin’s diseases among patients in Oman.
Materials and methods
Specimens
A total of sixty Omani patients who had been histologically diagnosed with Hodgkin and non- Hodgkin lymphomas in the period from 2011 to 2016 were included in the present study. Clinicopathologic data for each of the 60 patients were collected from the Sultan Qaboos University hospital (SQUH) record. The collected data included age, sex, histological subtype and stages. The ethical approval (MREC# 1118) for this research was obtained from the Research Ethics Committee at the College of Medicine and Health Sciences, Sultan Qaboos University.
Immunohistochemistry and In situ hybridization
Tissue processing
The Hodgkin and non-Hodgkin formalin-fixed paraffin-embedded tissue samples were cut into sequential 4 µm sections by the microtome. One section from each tissue sample was used for hematoxylin and eosin (H&E) staining and the parallel four sections were bound covalently to the glass slides, two sections were used for immunohistochemistry (IHC) experiment and two sections for in situ hybridization experiment. For each experiment, positive and negative controls were included., one section was used as a test section and the parallel section was used as a negative control in which the primary antibodies/EBERs probe was omitted. After sectioning, the slides were incubated in a 60 0C oven for one hour.
Immunohistochemistry
Immunohistochemistry was carried out on HL and NHL tissue samples using the Envision Flex+ High pH (Ref K8002, Dako. Briefly, sections were deparaffinized through two changes of xylene for 5 minutes each. After the deparaffinization, sections were hydrated in ethanol. followed by heat-induced epitope retrieval using the target high pH solution in the detection kit according to the package insert for the primary antibody. The blocking of endogenous peroxidase activity was carried out by the incubation in EnVision Flex Peroxidase-Blocking Reagent (SM801) for 10 minutes. This was followed by washing in Envision Flex Wash Buffer (TBS-DM831) (Dako, Denmark) for 5 minutes.
Then sections were incubated with a primary antibody consisting of a 1:50 mouse monoclonal antibody against EBV-encoded LMP (clone: CS,1-4 and isotype: IgG1, kappa), diluted by Envision Flex Diluent (K8007 -Dako, Denmark). Incubation with the primary antibody was carried out at room temperature for 60 minutes (negative controls were incubated with Tris buffer saline (TBS) during this time). The secondary polyclonal antibody used in the present study ( Dako Envision Flex/HRP SM802) consisted of dextran coupled with peroxidase molecules. Tissue sections were incubated with the secondary antibody at room temperature for 30 minutes. Then, 3,3-diaminobenzidine tetra- hydrochloride (DAB) as a chromogen ( Envision Flex DAB+ chromogen –DM827) was added to each section for 10 minutes . The sections were washed twice with TBS between each step for 5 minutes. Finally, the sections were incubated with Dako Envison Flex Hematoxylin (K8008), as a counterstain for 5 minutes to stain the nuclei, background and non-stained areas, then the sections were dehydrated and cover slipped by using an automatic glass coverslipper.
In situ hybridization
The detection of EBER1 and EBER 2 by in situ hybridization, was carried out by using the Inform EBER Probes (800-2842, Ventana Medical Systems, Roche Diagnostics GmbH, Mannheim Germany). The positive signals for EBER1,2 were detected by Ventana ISH/View Blue detection Kit (Ref: 853-2193) . In situ hybridization was performed by using Ventana Bench Mark Ultra, Medical Systems Inc, Tucson, AZ, USA), following manufacturer instructions.
The Ventana ISH/VIEW Blue detection kit (Ventana Medical Systems, Roche Diagnostics GmbH, Mannheim Germany REF: 853-2193) is composed of a primary mouse anti-fluorescein antibody that when followed by an indirect biotin-streptavidin system allows the detection of INFORM EBER (Epstein Barr Virus Early RNA) hybridized Probe (REF:800-2842) on paraffin-embedded tissue on Ventana BenchMark ULTRA automated slide stainer.
According to the manufacturer protocol, a barcode label with the corresponded protocol was loaded on the autostainer to perform the stain. Prior to the initial use of the INFORM EBER probe and the Ventana ISH/VIEW Blue detection kit, the tissue sections were treated with Ultra Cell conditioning-2 (Ventana. REF:950-223 ) and ISH Protease-2 (REF:780-4148 ). The Ultra CC2 pH 6 solution breaks the covalent bonds formed by formalin fixation and this is done in conjunction with the heating system in Ventana BenchMark ULTRA automated slide stainer. Removing these bonds would aid in the unmasking of target RNA for hybridization to occur. In addition, ISH Protease-2 (REF: 780-4148) was used to the permeable cell membrane and to remove protein that surrounds target RNA sequences. The target nucleic acid sequences were denatured by heating the tissue section and probe solution. Following the denaturing step, the reaction was cooled allowing the labelled nucleic acid probe to hybridize to the complementary nucleic acid sequence in the tissue section. After hybridization of the EBER probe, 10X SSC (REF: 950-110) was used as a hybridization buffer to control stringency for washing steps.
The detection kit contains mouse anti-fluorescein primary antibody that detects Epstein Barr Virus Early RNA sequences which links and bind indirectly to a biotinylated secondary anti body consisting of goat anti mouse IgG. This step is followed by streptavidin alkaline phosphatase enzyme to bind conjugate to biotin present. The complex is visualized with 5-Bromo-4-chloro-3-indolyl phosphate (BCIP) and nitro blue tetrazolium (NBT) chromogens and counterstained by Red counterstain (Ventana Medical Systems, Ref: 760-501).
Histopathological analysis
All immunohistochemistry stained slides were analyzed by an independent experienced pathologist. The histochemical evaluations were carried out using the guidelines published by Gulley, M., et al. 20.
Statistical analysis
Statistical analysis was conducted by using the SPSS program (Statistical Package for Social Sciences) version 23. The retrospectively collected clinicopathological and histopathological data have projected in percentage and frequency.
For most variables, two categories were analyzed in pairs as EBV positive versus EBV negative. We analyzed categorized variables using Pearson’s Chi-square and Fisher’s exact tests. Correlations between variables and EBV were assessed using Spearman rank linear test. P-values <0. 5 were considered to be significant.
Results
Latent EBV expression
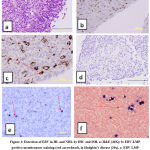
The prevalence of EBV in HL and NHL obtained from Omani patients is summarized in Table 1. EBV expression was considered positive if any Hodgkin’s Reed-Stenberg (HRS) cells were positive by either method immunohistochemistry (IHC) or in situ hybridization (ISH). EBV was detected in the HRS cells in 12 (46.2%) cases out of the 26 HL cases examined by IHC and by ISH, EBV was detected in HRS cells in 15 (57.7%) cases out of the 26 HL cases examined (p-value 0.000049) [Table1]. Three out of the 26 HL cases (11.5%), which were LMP negative by IHC showed EBER positivity by ISH and no HL cases which were EBER negative by ISH showed LMP positivity by IHC. [Table 1; Figure.1].
On the other hand, and in the 34 NHL cases examined, EBV was detected in the atypical cells of NHL cases by ISH and IHC are 5 (14.7%) and 4 (11.8%), respectively (p-value 0.000022). One out of 34 (2.9%) NHL cases which were LMP negative by IHC showed EBER positive by ISH and no NHL cases which were EBER negative by ISH showed LMP positive by IHC. [Table 1]
Positive tissue sections for LMP showed brown diffuse membranous stain (Figure 1b,c) and positive tissue sections for EBER showed blue nuclear stain (Figure.1, e,f) of HRS cells of Hodgkin’s disease.
Table 1: The expression of EBER and LMP in HL and NHL in Omani patients
| number of cases | LMP-EBV positivity by IHC(%) | EBER-EBV positivity by ISH(%) | |
| HL | 26(43.3%) | 12 (46.2%) | 15(57.7%) |
| NHL | 34 (56.7%) | 4 (11.8%) | 5 (14.7%) |
| total | 60 (100%) | 16 (26.7%) | 20 (33.3%) |
The pattern of LMP-EBV expression in HL and NHL
Four out of nine (44.44%) EBV-LMP positive mixed cellularity HL cases showed cytoplasmic staining while five out of nine (55.55%) EBV-LMP positive mixed cellularity HL cases showed membranous staining (Figure. 1). However, one out of three (33.33%) EBV-LMP positive nodular sclerosis HL cases showed cytoplasmic staining while two out of three (66.67%) EBV-LMP positive nodular sclerosis HL cases showed membranous staining (p-value 0.735) which indicates that there was no statistically significant correlation between the pattern of expression and HL histological subtype.
On the other hand, one out of two (50%) EBV-LMP positive diffuse large B cell lymphoma of NHL cases showed cytoplasmic staining while one out of two (50%) EBV-LMP positive diffuse large B cell lymphoma of NHL cases showed membranous staining. All EBV-LMP positive follicular lymphoma of NHL cases showed cytoplasmic staining. All EBV-LMP positive T cell lymphoma NHL cases showed membranous staining. The (p-value= 0.368), indicates that there was no statistically significant correlation between the pattern of expression and NHL histological subtype. LMP protein was detected in atypical B cells (CD20 positive) in all the NHL subtypes except T cell lymphoma which is detected in atypical T cells. [Table 2].
Table 2: The expression of LMP in NHL in atypical B and T cells
| NHL histological subtypes | Stage of lymphocyte | marker |
| Burkett lymphoma | B cell | CD 20 positive |
| diffuse large B cell lymphoma | B cell | CD 20 positive |
| Follicular lymphoma | B cell | CD 20 positive |
| chronic lymphocytic leukaemia | B cell | CD 20 positive |
| T cell lymphoma | T cell | CD 3 positive |
Latent EBV expression in correlation to age and sex
Hodgkin’s lymphoma
The median age of the patients was 34 years (ranged from 14 to 80 years). The expression of LMP between the age of 14-34 years was 5 LMP positive out of 13 (38.5%) and age of 35-80 years was 7 LMP positive out of 13 (53.8%) [p-value= 0.695] (Table.3), while the expression of EBER between the age of 14-34 years was 6 EBER positive out of 13 (46.2%) and age of 35-80 years was 9 EBER positive out of 13 (69.2%) (p-value 0.428) [Table 3].
The expression of LMP between males patients was 8 LMP positive out of 17(47.1%) and females patients were 4 LMP positive out of 9( 44.4%) [p-value= 1.000] (Table 3), while the expression of EBER between males was 9 out of 17 (52.9 % ) and females were 6 out of 9 (66.7%) [p-value= 0.683].
Therefore, the expression of EBV (EBER&LMP) in HL was NOT statistical significantly correlated to age and sex. The frequency of EBV positive cases in Hodgkin’s disease (2011-2016) in correlation to sex and age are summarized in Table 3.
Table3: The frequency of EBV positive cases in Hodgkin’s disease (2011-2016) in correlation to sex and age.
| Hodgkin’s disease | LMP-EBV positivity by IHC(%) | EBER-EBV positivity by ISH(%) | |
| sex
male female |
17 (65.4%) 9 (34.6%) |
8 (47.1%) 4 (44.4%) |
9(52.9%) 6 (66.7%) |
| Age(years)
14-34 35-80 |
13 (50.0%) 13 (50%) |
5 (38.5%) 7 (53.8%) |
6 (46.2%) 9 (69.2%) |
| total | 26 (76.5%) | 12 (46.2%) | 15 (57.7%) |
Non- Hodgkin’s lymphoma
The median age of the NHL patients was 56 years (ranged from 4 to 91 years). The expression of LMP between the age of 4-55 years old patients was 1 LMP positive out of 17(5.9% ) and age of 56-91 years was 4 LMP positive out of 17 ( 23.5% ) [p-value= 0.319], while the expression of EBER between the age of 4-55 years was 2 EBER positive out of 17 (11.8% ) and age of 56-91 years was 4 EBER positive out of 17 ( 23.5%) [p-value 0.641].
The expression of LMP between males was 4 LMP positive out of 15 (26.7 %) and females was 1 LMP positive out of 19 (5.2%) [p-value= 0.146], while the expression of EBER between males was 4 EBER positive out of 15 (26.7%) and females was 2 EBER positive out of 19 ( 10.5%)
[p-value= 0.370) [Table 4].Therefore, the expression of EBV (EBER & LMP) in NHL was NOT statistical significantly correlated to age and sex.
Table 4: The frequency of EBV positive cases in Non-Hodgkin’s disease (2011-2016) in correlation to sex and age.
| Non-Hodgkin’s disease | LMP-EBV positivity by IHC(%) | EBER-EBV positivity by ISH(%) | |
| sex
male female |
15 (44.1%) 19 (55.9%) |
4 (26.7%) 1 (5.2%) |
4(26.7%) 2(10.5%) |
| Age(years)
4-55 56-91 |
17 (50.0%) 17 (50.0%) |
1 (5.9%) 4 (23.5%) |
2 (11.8%) 4(23.5%) |
| total | 34 (56.7%) | 5 (14.7%) | 6(17.6%) |
Latent EBV expression in correlation to histological subtype
Hodgkin’s lymphoma
Nine patients out of 26 (34.6%) were classified as Nodular sclerosis (NS), ten out of 26 (38.5%) were of mixed cellularity (MC), while only seven out of 26 (26.9%) were of the Lymphocyte Predominant (NLPHL) subtype. The frequency of the expression of latent EBV in HL in correlation with histological types is summarised in Table 5.
Table 5: The frequency of EBV positive cases of HL in correlation to histological subtype.
| Hodgkin’s disease | LMP-EBV positivity by IHC(%) | EBER-EBV positivity by ISH(%) | |
| Nodular sclerosis | 9 (34.6%) | 3 (33.3%) | 4 (44.4%) |
| Mixed cellularity | 10 (38.5%) | 9 (90.0%) | 10 (100%) |
| Lymphocyte Predominant | 7 (26.9%) | 0 (0%) | 1 (14.3%) |
| total | 26 (43.3%) | 12 (46.2%) | 15 (57.7%) |
Non- Hodgkin’s lymphoma
Sixteen patients out of 34 (47.1%) were classified as diffuse large B lymphoma, ten out of 34 (29.4%) were of Follicular lymphoma, four out of 34 (11.8%) were of Burkett lymphoma, two out of 34 (5.9%) were of chronic lymphocytic leukaemia/small lymphocytic lymphoma (CLL/SLL), and also two out of 34 (5.9%) were of the T cell lymphoma (MC) subtype. The frequency of the expression of latent EBV in NHL in correlation with histological types is summarised in Table 6.
Table 6: The frequency of EBV positive cases of NHL in correlation to histological subtype.
| Non-Hodgkin’s disease | LMP-EBV positivity by IHC(%) | EBER-EBV positivity by ISH(%) | |
| Burkett lymphoma | 4 (11.8%) | 1 (25%) | 1 (25%) |
| diffuse large B cell lymphoma | 16 (47.1%) | 3 (18.8%) | 4 (25.0%) |
| Follicular lymphoma | 10 (29.4%) | 1 (10.0%) | 1 (10.0%) |
| chronic lymphocytic leukaemia/small lymphocytic lymphoma (CLL/SLL). | 2(5.9%) | 0 (0%) | 0 (0%) |
| T cell lymphoma | 2 (5.9%) | 1 (50%) | 1 (50%) |
| total | 34 (56.7%) | 6 (17.6%) | 7 (20.6%) |
Discussion
In the present study, the age of most HL and NHL cases was between the age of 35-80 years. This was in agreement with the results obtained in previous North American study 26, which showed that most of the Hodgkin lymphoma (HL) cases were between the age of 50-74 years. This is, however, in contrast to other studies conducted in other Arab countries like Kuwait, Jordan and Egypt in which the disease occurred earlier 25,27,28. This may indicate that in Oman the age distribution of HL followed a similar pattern to the other developing countries. The immunity of the host is known to decrease with increasing age and these findings suggest that EBV positivity in Reed–Sternberg cells in HL in the present study may be correlated with relative immunity impairment in elderly. The immunity impairment could contribute to defective control of EBV infection, resulting to a higher risk of cell transformation and the development of malignancies 10.
In the present study, the slight increase of the EBV expression in the male patients in comparison to female in both HL and NHL was in agreement with other studies conducted in other Arab countries like Kuwait 25, Egypt 28 and worldwide 29, however, this differed from a previously published study carried out by Al-Safi in 2007 in Iraq and showed the equal incidence of HL in both females and males 30, this finding could be explained by the smaller number of samples used in Al-Safi study.
In the current study, LMP-1 expression was positive in 46.2% of HL cases. This expression was low in comparison to previously published studies with a percentage of 75% for Iraqi patients30 and 63% for Egyptian patients 28, 60% for Nigerian patients 6, 82% for Indian 31, and 93% for Iranian patients 32, whereas it was similar to developed countries, with percentages of 20-50% for North American patients 26,33.
The high expression of LMP-1 in mixed cellularity HL that was seen in the present study was in agreement with other studies carried out in Jordan 27, China 34 and Rio de Janeiro 35.
In the present study, the LMP-1 expression was positive in 11.8% of NHL and to some extents, EBV expression was low when compared to other studies conducted in developing countries like Iran in which LMP -1 was found in 30% of non-Hodgkin lymphoma cases 36. In the present study, the Burkitt’s lymphoma among NHL cases was most commonly associated with EBV infection in which LMP and EBERs were expressed in 25% of Omani Burkitt’s lymphoma patients. This is to some degree comparable to previous studies, in which EBV associated with Burkitt’s lymphoma documented in 29 % of NHL USA patients 37, while in Brazil, 87% of Burkitt’s lymphomas patients were EBV positive 38.
Furthermore, in the current study, LMP protein expression of EBV was detected in 18.8% of diffuse large B cell lymphoma, 10% of follicular lymphoma and 50% of T cell lymphoma. A higher incidence was detected in a previous study conducted in Pakistan39 and showed that 44.4% of diffuse large B cell lymphoma and 22.2% of follicular lymphoma were positive for LMP protein expression by immunohistochemistry. However, they detected a lower incidence in the LMP expression in T cell lymphoma of 11.1% 39, in comparison to 50% detected in the present study.
The detection of LMP expression in both HL and NHL cases in the present study, with a higher rate of LMP expression in HL and since LMP is considered as the major EBV oncogene and is essential for B-cell immortalization. thus, we may conclude that the presence of EBV in HL may indicate that EBV plays an important role in the pathogenesis of the HL disease in Omani patients.
In the current study, there was a significant correlation between LMP-1 and EBER expression in HL and NHL by both ISH and IHC. Therefore, we can conclude that IHC was equivalent in terms of sensitivity and specificity to ISH for the detection of EBV in formalin-fixed paraffin-embedded tissue samples. This may provide a cheaper and technically simple approach in the detection of EBV in formalin-fixed paraffin-embedded tissue samples. This finding was in agreement with the results of Van Gorp et al 40and Zong-Li 41.
Conclusion
The present study provides evidence of an association between EBV and Hodgkin’s and non-Hodgkin’s lymphomas among patients in Oman and shows that this association is more frequent in mixed cellularity subtype. Moreover, the detection of LMP and EBER in HRS cells suggests that EBV may be involved in the pathogenesis of Hodgkin’s and non-Hodgkin’s lymphomas among patients in Oman. It also demonstrates that IHC is similar to some degrees in terms of sensitivity and specificity to ISH in the detection of EBV in HL and NHL. Due to the high cost of in situ hybridization, the present study encourages Pathologists to replace ISH with a cheaper and technically simpler IHC approach. Furthermore, the high prevalence of EBV in HL encourages conducting future studies on the effect of anti-herpes virus drug on the treatment of EBV positive HL and NHL cases.
Acknowledgement
The authors would like to thank the head and the staff of the Department of Pathology at Sultan Qaboos University for allowing us to use their laboratory facilities.
Conflict of Interest
The authors declare no conflict of interest.
Funding Source
There are no funding for this project.
References
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J . WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. WHO publication, 4th Edition, Volume 2; 2017
- de LevalL, Elaine S. Lymphoma Classification. The Can J; 26 ( 3):176-185 (2020).
CrossRef - Sandvej K, Hamilton-dutoit S. and Pallesen, G. Influence of Epstein-Barr virus-encoded latent membrane protein 1 on the expression of CD23 antigen, ICAM-1 and LFA-3 in Hodgkin and Reed-Sternberg Cells. A Morphometric Analysis.Leukemia & Lymphoma; 9(1-2): 95-101. (1993).
CrossRef - Niedobitek G. Immunohistochemical detection of the Epstein-Barr Virus–encoded latent membrane protein 2A in Hodgkin’s Disease and Infectious Mononucleosis. Blood; 90(4): 1664-1672 (1997).
CrossRef - Ben Dhiab M, Ziadi S, Hanene S, Teheni L, Mounir T. Prognostic significance of Epstein-Barr virus (EBV) infection in Hodgkin lymphoma patients. J Infect Chemother; 23(3):121-130 (2017).
CrossRef - Adelusola K. Epstein Barr virus latent membrane protein-1 in Hodgkin’s lymphoma in Nigerians. Afr Health Sci; 9(3):8-174 (2009)
- TornÓczky T, KelÉnyi G, Pajor L. EBER Oligonucleotide RNA in situ Hybridizationnin EBV Associated Neoplasms. Pathology & Oncology Research; 4(3):201-205 (1998).
CrossRef - Krugmann J, Tzankov L, Gschwendtner A, Fischhofer M, Greil R, Fend F, Dirnhofer S Longer Failure-Free Survival Interval of Epstein-Barr Virus–Associated Classical Hodgkin’S Lymphoma: A Single-Institution Study” Mod Pathol; 16(6):566-573 (2003).
CrossRef - Matsuura H, Kirschner AN, Longnecker R, Jardetzky Crystal Structure of the Epstein-Barr Virus (EBV) glycoprotein H/glycoprotein L (Gh/Gl) Complex. Proceedings of the National Academy of Sciences; 107(52): 2641-2646 (2010).
CrossRef - Murray PG, Young LS. An etiological role for the Epstein-Barr virus in the pathogenesis of classical Hodgkin lymphoma. Blood; 15;134(7):591-596 (2017).
CrossRef - Tanyildiz HG, Yildiz I, Bassullu N, Tuzuner N, Ozkan A, Celkan T, Apak H. The role of Epstein-Barr Virus LMP-1 immunohistochemical staining in childhood hodgkin lymphoma. Iran J Paediat; 25(6):1-5 (2015).
CrossRef - Kanavaros P, Sakalidou A, Tzardi M, Darivianaki K, Delides G, Kazlaris E, Kalmanti M. Frequent Detection of Epstein-Barr Virus (EBV), EBEB Transcripts and Latent Membrane Protein-1 (LMP-1) in Tumor Cells in Hodgkin’s Disease Arising in Childhood.Pathol Res Pract; 90(11): 1026-1030 (1994).
CrossRef - Ben Dhiab M , Sonia Z , Hanene S, Teheni L, Mounir “Prognostic Significance Of Epstein–Barr Virus (EBV) Infection In Hodgkin Lymphoma Patients”. J Infect and Chemoth; 23(3): 121-130 (2017).
CrossRef - Liang, J. Epstein–Barr virus (EBV) DNA in whole blood as a superior prognostic and monitoring factor than EBV-encoded small RNA in situ hybridization in diffuse large B-cell lymphoma.Clin Microbio and Infect; 21(6):596-602 (2015).
CrossRef - Oka K , Nagayama R, Iijima S, Yonekawa N, Hirosawa K, Yatabe Y, Mori N. Epstein-Barr virus-associated lymphoproliferative disorder presenting with classical Hodgkin lymphoma and developing as peripheral T-cell lymphoma 9 years later: A case report of composite lymphoma. Pathol Int; 61(12):752-755 (2011).
CrossRef - Carbone A, Gloghini A, Dotti G. ” EBV-Associated Lymphoproliferative Disorders: Classification and Treatment”. Oncologist; 13(5):577-85 (2008).
CrossRef - Odumade, O, Hogquist K and Balfour H . Progress and problems in understanding and managing Primary Epstein-Barr Virus Infections. Clin Microbiol Rev; 24(1): 193-209 (2011).
CrossRef - Greifenegger N, Jäger M, Kunz-Schughart LA, Wolf H, Schwarzmann F. Epstein-Barr Virus Small RNA (EBER) Genes: Differential Regulation during Lytic Viral Replication. J Virol; 72(11):9323–9328 (1998).
CrossRef - Weinreb M, Day PJ, Niggli F, Powell JE, Raafat F, Hesseling PB, Schneider JW, Mann JR. The role of Epstein-Barr virus in Hodgkin’s disease from different geographical areas. Arch Dis Child; 74(1): 27–31 (1996).
CrossRef - Gulley M, Glaser LS, Craig FE,Borowitz M,Mann RB, Shema SJ, and Ambinder RF . Guidelines for Interpreting EBER in Situ Hybridization and LMP1Immunohistochemical tests for detecting Epstein-Barr Virus in Hodgkin Lymphoma. Am J Clin Pathol; 117(2):259-267 (2002).
CrossRef - Glaser S. Inter- and Intra-Observer Reliability of Epstein – Barr Virus Detection in Hodgkin Lymphoma using Histochemical Procedures.Leukaemia & Lymphoma; 45(3):489-497 (2004).
CrossRef - Mabruk MJ, Flint SR, Coleman DC, Shiels O, Toner M, Atkins GJ.) “A Rapid microwave-in situ hybridization method for the definitive diagnosis of oral hairy leukoplakia: comparison with immunohistochemistry”. J Oral Pathol and Med; 25(4):170-176 (1996).
CrossRef - Yunos AM, Jaafar H, Idris FM, Kaur G, Mabruk M. Detection of Epstein-Barr virus in lower gastrointestinal tract lymphomas: a study in Malaysian patients. Mol Diagn Ther; 10(4):251-260 (2006).
CrossRef - Mabruk MJ. In situ hybridization: detecting viral nucleic acid in formalin-fixed, paraffin-embedded tissue samples. Expert Rev Mol Diagn; 4(5):653-661 (2004).
CrossRef - Makar R, Saji T, Junaid TA. Epstein – Barr virus Expression in Hodgkin’s Lymphoma in Pathol Oncol Res; 9(3): 159-165 (2003).
CrossRef - Diepstra A, van Imhoff GW, Schaapveld M, Karim-Kos H , van den Berg A , Vellenga E, Poppemal S.. Latent Epstein-Barr Virus Infection Of Tumor Cells In Classical Hodgkin’s Lymphoma Predicts Adverse Outcome In Older Adult Patients. J Clin Oncol; 27(23) :3815-3821 (2009).
CrossRef - Almasri N and Khalidi Epstein-Barr virus expression in Hodgkin’s disease in Jordan. Saudi Med J; 25(6): 770-5 (2004).
- Audouin, J, Diebold J, Nathwani B, Ishak E, MacLennan K, Mueller-Hermelink HK, Armitage JO,Weisenburger DD. Epstein–Barr virus and Hodgkin’s lymphoma in Cairo, Egypt. J Hematop; 3(1): 11-18 (2010).
CrossRef - Kapatai, G, and P Murray. Contribution of The Epstein Barr Virus To The Molecular Pathogenesis of Hodgkin Lymphoma. J Clin Pathol; 60(12) :1342-1349 (2006).
CrossRef - Al-Safi RAR.. Immunohistochemical Expression of Epstein -Barr virus Latent Membrane Protein-1 (LMP-1) in Hodgkin’s Lymphoma. MSc Thesis, College of Medicine, Al-Nahrain University, Baghdad. 55-60 (2007).
- Karnik S, Srinivasan B and Nair S.. Hodgkin’s lymphoma: immunohistochemical features and its association with EBV LMP-1. Experience from a South Indian hospital. Pathol; 35: 207-211. (2003.)
CrossRef - Katebi M, Tarhini S, Otrock ZK, Bazarbachi A, Kchour G. Frequency Of Epstein–Barr Virus Expression In Various Histological Subtypes of Hodgkin’S Lymphoma. Histopathol; 52(6) :775-777 (2008).
CrossRef - Chang ET, T, Lennette ET, Weir EG, Borowitz M, Mann R, Spiegelman D, Mueller NE. Heterogeneity of Risk Factors and antibody Profiles in Epstein Barr Virus Genome–Positive and –Negative Hodgkin Lymphoma. The J Infect Dis;18(12): 2271-2281 (2004).
CrossRef - Ping LY, Ding N, Shi YF, Sun L, Zheng W, Xie Y, Wang XP, Tu MF, Lin NJ, Ying ZT, Liu WP, Deng LJ, Zhan C, Tian L, Feng LX, Song YQ, Zhu J. Clinical characteristics and prognosis analysis of patients with LMP-1 positive Hodgkin’s lymphoma after EBV infection. Zhongguo Shi Yan Xue Ye Xue Za Zhi; 22(1):78-84 (2014).
- Loureiro MM, MoraisII JC and Milito CB . Expression of Epstein-Barr virus in patients with Hodgkin’s disease: report of 64 cases from Rio de Janeiro. Brazil J Bras Patol; 40 (1) :37-40 (2004).
CrossRef - Abadi RZM, Sistani NS, Mohtasham N, Hesari KK, Vaezi T, Pazouki M, Shakeri MT. The prevalence of Epstein-Barr virus infection in head and neck non-Hodgkin’s lymphomas in Khorasan, northeast of Iran J Pak Med Assoc; 63(7):882-887 (2013).
- Mbulaiteye SM, Pullarkat ST, Nathwani BN, Weiss LM, Rao N, Emmanuel B, Cozen W . Epstein-Barr virus patterns in US Burkitt lymphoma tumors from the SEER residual tissue repository during 1979-2009. APMIS; 122(1): 5-15 (2014).
CrossRef - Klumb CE, Hassan R, De Oliveira DE, De Resende LMM, Carriço MK, Dobbin J, Pombo-De-Oliveira MS, Bacchi CE, Maia RC. Geographic variation in Epstein–Barr virus associated Burkitt’s lymphoma in children from Brazil. Int J Cancer; 10(8) :66–70 (2004).
CrossRef - Ishtiaq S, Hassan U, Mushtaq S, Akhtar N. Determination of frequency of Epstein-Barr virus in non- Hodgkin lymphomas Using EBV latent membrane protein 1 (EBV-LMP1) immunohistochemical staining. APJCP; 14(6):3963-7 (2013)..
CrossRef - Van Gorp, J, Jacobse KC, Broekhuizen R, Alers J, van den Tweel JG, and de Weger RA. Encoded Latent Membrane Protein 1 Of Epstein-Barr Virus On Follicular Dendritic Cells In Residual Germinal Centres In Hodgkin’s Disease. J Clin Pathol;; 47(1):29-32 (1994).
CrossRef - Zong-Li Qi ,Xi-Qun Han, Jun Hu Guang-Hua, Wang Jin-Wei, Gao Xin Wang, Dao-Yan Liang. Comparison of three methods for the detection of Epstein-Barr virus in Hodgkin’s lymphoma in paraffin-embedded tissues. Molec Med Report; 7(1): 89-92 (2013).
CrossRef