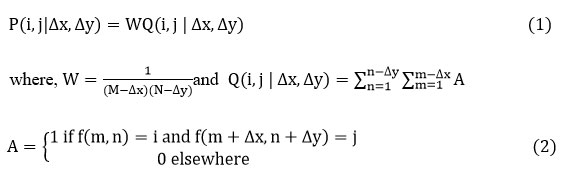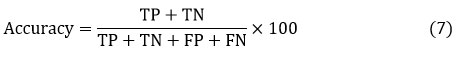Manuscript accepted on :06-04-2022
Published online on: 06-05-2022
Plagiarism Check: Yes
Reviewed by: Dr. Narayanappa Krishnappa
Second Review by: Dr. Mandar Malawade
Final Approval by: Dr. H Fai poon
Swaroopa H N* , Basavaraj N Jagadale
, Basavaraj N Jagadale , Priya B S
, Priya B S , Omar Abdullah Murshed Farhan Alnaggar
, Omar Abdullah Murshed Farhan Alnaggar  and Abhisheka T E
and Abhisheka T E
Department of PG studies and research in Electronics, Kuvempu University, Shankaraghatta, Shimoga, India.
Corresponding Author E-mail: swaroopampl@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/2414
Abstract
In recent years, bio-medical image segmentation is established itself as base for image analysis. This article proposes a new method in developing a robust wavelet based medical image fusion technique for image segmentation. A GLCM (Gray Level Co-occurrence Matrix) based statistical method is used to extracts the texture features of the image decomposed at single level and the image is segmented based on region growing method. The combination of these two along with fusion technique gives a new segmented image. The results indicate the efficiency of the proposed method in segmenting the both normal cell images as well as darker cell images.
Keywords
Cell image; GLCM; Image analysis; Region growing; Segmentation; Wavelet transform
Download this article as:| Copy the following to cite this article: Swaroopa H. N, Jagadale B. N, Priya B. S, Alnaggar O. A. M. F, Abhisheka T. E. Bio-Medical Image Segmentation using Wavelet Based Fusion Technique .Biomed Pharmacol J 2022;15(2). |
| Copy the following to cite this URL: Swaroopa H. N, Jagadale B. N, Priya B. S, Alnaggar O. A. M. F, Abhisheka T. E. Bio-Medical Image Segmentation using Wavelet Based Fusion Technique .Biomed Pharmacol J 2022;15(2). Available from: https://bit.ly/38aibQL |
Introduction
The advances in digital technology as well as in the light microscopy field in recent years, resulted in growing importance of digital cellular imaging and computerized image analysis in cell biology. The use of medical image processing techniques, increase the speed of analysis and can supersede some limitations inherent to the human visual system. They also provide better and early solutions for medical emergencies, including, detection of abnormalities in critical medical conditions. The cell image analysis is an important step in biological applications to understand cell behavior and its response to different cell culturing conditions etc. The use of image segmentation algorithms increases the accuracy of data analysis which forms the basis for further analysis of the image. There are several image segmentation techniques used for these kind of analysis. Image segmentation uses features extracted from image using mathematical morphological operations 1, 2, based on erosion and dilation of the image. The watershed segmentation is obtained by partitioning an image into primitive regions using gradient magnitude and markers 3, 4. The segmentation by indexing pixel clustering technique is very useful in gray level as well as in color image segmentation 5. The K-means algorithm is an unsupervised technique, which separates the clusters based on user requirement and helps in finding specified number of clusters (K), which are denoted by their center point6. FCM (fuzzy C-means clustering) algorithm is a form of image segmentation technique, in which each point is assigned based on probability criteria corresponding to that cluster, with varying degree of membership 7-14. The GLCM (Gray Level Co-occurrence Matrix) is mainly used to drawing out texture features of image, and is more useful in segmentation of remote sensing images, medical images and texture based images 15-23. Region growing method is one of the efficient region based segmentation method, which gives the segmented image by performing region based operations 24-27. This method is mainly used in detecting tumor in brain image, cancer cell detection in liver images 28, 29.
Discrete wavelet transform (DWT)
In DWT, the digital image is divided into four sub-images and named as A1 (approximation coefficient or A1 band) and D1 (detailed coefficients or H1, V1, D1 bands). However D1 contains edge/texture details of the image, whereas A1 represents most of the visual details of the image 1-3, 6. A1 sub-image is subjected to the further preprocessing steps as it contains most of the image information. The A1 sub-image is subjected to second level of decomposition and so on16, 18, 19, 30-33.
Gray level co-occurrence matrix
There are various algorithms available for extracting texture features of the images. Texture feature is nothing but pattern of data, involving structure of image and its statistical information 15, 16. The GLCM approach is a popular method based on texture feature of gray level consociation probability 17, 18. It measures the second-order statistical properties of the digital images and brings out the spatial relationship of specific pair of pixel values and it occurs in neighboring pixels in an image19, 20. The GLCM properties allows faster recognition of various types of cell images with high accuracy, consumes less time for execution and provides good sensitivity dependent features after preprocessing stages 21-23.
A GLCM is a statistical method, where, the each rows and columns is equal to the number of gray levels, G, in the image. The array element shows frequency ratio between the two pixels, which are isolated by a pixel spacing , and occur within a given vicinity, one having intensity and another with intensity . Moreover the array element contains the second order statistical probability values for variations between gray levels and at a certain shift in spacing ‘s’ and at a certain angle (θ) 19, 22, 23. Given a neighborhood of an input image containing G gray levels from 0 to , let is the intensity at selected m, line n of the neighborhood. The relative frequency matrix is given by,
Region growing
Region growing is a form of image segmentation method, which selects the particular region to segment the interested area of the image. It groups the pixels according the pre-defined criteria 24,25. The algorithm grows the seed regions until all image pixels have been incorporated and finally, method identifies the individual regions to be segmented 26. This method provides clear edge properties and separates regions in a simple way, by choosing multiple criteria at the same time 27. It performs well with respect to noise and it has less algorithm complexity compare to FCM based methods.
The region growing method works on the following rules. Here represents the ith region and predicts the region of interest in a logical way and ∅ is the null set.
All the interested pixels must include in a region to finish the segmentation process.
The regions must be disjoint.
Region of interest must be connected with often predefined criteria.
In the proposed method, the combination of both Region growing and GLCM segmented images are fused to obtain more meaningful segmented results. Sample images are taken from cellaVision and CellAtlas, and the experimental studies are focused mainly, on blood cells like proerythroblast, basophilic erythroblast which comes under erythropoiesis family. The attributes of these cells are large, round and centrally placed nucleus with fine chromatin structure and narrow dark blue cytoplasma edges. This paper aims to identify and extract this proerythroblast, basophilic erythroblast nucleus.
This paper is divided into four sections, the Section 1, carries introduction, Section 2, covers the proposed method, section 3, portraits the flowchart and algorithm of the proposed work. The results of the proposed method are presented in section 4, and conclusions are furnished in section 5.
Proposed method
The proposed segmentation method is carried out on cell images. In this method, first, the cell image is subjected to median filter to denoise the image without losing edge details. A statistical GLCM method is used to extract the textural information from the filtered image. Then, this median filtered image is segmented using region growing method to separate the regions based on some pre-defined criteria. In this work, single seed region growing techniques is employed, where the difference between the mean of region and pixel intensity value are used as the measure of similarity.
Image fusion method adds the two or more processed images of the same object type into a solo image 12. The endower of image fusion, especially in bio-medical imaging, is to create new images that are more meaningful for human visual perception 27. The segmentation of cell images is mainly based on abrupt changes in image features, like color information, intensity, boundary/texture features and statistical information’s. In this work, the average fusion technique of the GLCM segmented image and region growing segmented images are taken in order to reach desired and meaningful result, which leads a new segmented image. The proposed work is very useful to extract the required area of the cell image.
Flow chart
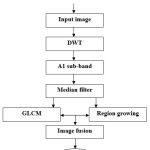 |
Figure 1: Schematic flowchart of the proposed method |
Algorithm steps
Input: Cell image to be segmented
Output: Segmented image
A DWT (discrete wavelet transform) decomposes the cell image into scale 1.
Sub-image (A1) is denoised by using median filter.
GLCM texture feature classifier is used to extract the essential information from A1 and A1 is segmented simultaneously using region growing method.
Final image obtained by fusing of two segmented outcomes.
Results and discussions
Performance matrices
Accuracy
It specifies the pixels of the image which are perfectly classified. Perfection of the segmentation technique [34] is given by
Pixels which are perfectly incorporated to the given class are represented by TP and TN represents the pixels which are not belonging to the given class. FN is wrongly predicted pixels, which are not belonging to the given class.
Sensitivity
Sensitivity is calculated through positive predictive and negative predicted values, which is the ratio of true outcome of all segmented results 34.
Dice co-efficient
Dice-coefficient examines the identical things in between segmented image and the ground truth image, which is retrieved from manual segmentation. Dice coefficient of segmented image 35 is given by
where, |M| and |N| cardinalities of two sets.
Jaccard co-efficient
Jaccard co-efficientis as same as the dice coefficient, which calculates the commonness in-between ground truth data with different segmentation methods employed in this paper and 35 is given by
Experimental results
 |
Figure 2: a. Input cell image1, b. Groundtruth image, c. FCM thresholding segmented image, d. FCM clustering segmented image, f. Proposed method segmented image. |
 |
Figure 3: a. Input image, b. Groundtruth image, c. FCM thresholding segmented image, d. FCM clustering segmented imge, f. Proposed method segmented image. |
 |
Figure 4: a. Input image, b. Groundtruth image, c. FCM thresholding segmented image, d.FCM clustering segmented imge, e.Proposed method segmented image. |
Table 1: Comparison of segmentation results.
| Image | accuracy | sensitivity | Dice co-efficient | Jaccard co-efficient | ||||||||
| FCMC | FCMT | Our method | FCMC | FCMT | Our
method |
FCMC | FCMT | Our method | FCMC | FCMT | Our method | |
| Cell image 1 | 92.8% | 97.3% | 97.5% | 96.5% | 97.9% | 1 | 96.2% | 98.6% | 98.7% | 95.6% | 97.2% | 97.4% |
| Cell image 2 | 89.3% | 96.7% | 96.8% | 97.3% | 97.3% | 99.9% | 94.3% | 98.2% | 98.3% | 89.3% | 96.4% | 96.6% |
| Cell image 3 | 81.0% | 89.3% | 90.7% | 96.9% | 89.6% | 99.9% | 89.4% | 93.3% | 94.7% | 80.9% | 87.4% | 90.0% |
Table 2: Execution time comparison of segmentation methods
| Image | Time in sec | ||
| FCMC | FCMT | Proposed
method |
|
| Cell image 1 | 9.731 | 54.628 | 11.340 |
| Cell image 2 | 13.680 | 39.714 | 11.894 |
| Cell image 3 | 13.190 | 29.834 | 14.031 |
Conclusion
This paper presents a new algorithm called wavelet based fusion technique, combines GLCM based statistical method and region growing methods efficiently via image fusion technique to produce a better segmented image. However, the utilization of median filter followed by level 1 wavelet decomposition is one of the key feature to achieve the effective segmentation.
The results of this method are compared with that of wavelet based FCM thresholding and FCM clustering methods and found that segmentation using wavelet based fusion technique increases the efficiency of the segmentation, significantly, in terms of correctness, sensitivity, dice co-efficient and jaccard co-efficient. The algorithm is computationally fast and efficient. The results show that the proposed method performs better even for the darker cell images. It is found that sensitivity parameter in performance metrics increases approximately 5% and this improve in sensitivity and accuracy are the major contribution of this work and is evident from the results obtained from the developed algorithm.
Acknowledgement
Author wish to acknowledge cellaVision and CellAtlas, morphology experts for the sample datasets (cell images) used in this work (https://www.cellavision.com/en/cellavision-cellatlas).
Conflict of Interest
There is no conflict of interest.
Funding Sources
There is no funding source.
References
- Wavelet transform and morphology image segmentation algorism for blood cell department of computer science, hunan college of humanities science and technology, 417000; 2.college of software, hunan university,410081; 978-1-4244-2800-7/09/$25.00 ©2009 ieee.
- Anantha Sivaprakasam, E.R. Naganathan, V. Saravanakumar, Wavelet based cervical image segmentation using morphological and statistical operations, jour of adv research in dynamical & control systems, vol. 10, 03-special issue, 2018.
- Xiaoqiang Ji, Yang Li, Jiezhang Cheng, Yuanhua Yu*, Meijiao Wang, Cell image segmentation based on an improved watershed algorithm, 2015 8th international congress on image and signal processing (cisp 2015), 978-1-4673-9098-9/15/$31.00 ©2015 ieee.
- Han sun *, Jingyu yang, Mingwuren. A fast watershed algorithm based on chain code and its application in image segmentation. Pattern Recognition LettersVolume 26 Issue 91 July 2005 pp 1266–1274, https://doi.org/10.1016/j.patrec.2004.11.007 elsevier.com/locate/patrec.
CrossRef - Youguo Li, Haiyan Wu, A clustering method based on k-means algorithm, 2012 international conference on solid state devices and materials science,elsevierphysics procedia 25 ( 2012 ) 1104 – 1109.
CrossRef - Jianwei Liu1, a, Lei Guo1, b *, “An improved k-means algorithm for brain mri image segmentation”, 3rd international conference on mechatronics, robotics and automation (icmra 2015).
- Niladri Shekhar Mishraa, Susmita Ghoshb, Ashish Ghoshc,∗, Fuzzy clustering algorithms incorporating local information for change detection in remotely sensed images, applied soft computing, volume 12, issue 8, august 2012, pages 2683-2692, https://doi.org/10.1016/j.asoc.2012.03.060.
CrossRef - Satya Prakash Sahu*1 , Priyanka Agrawal1 , Narendra D Londhe2 and Shrish Verma3, A new hybrid approach using fuzzy clustering and morphological operations for lung segmentation in thoracic ct images, biomedical & pharmacology journal vol. 10(4), 1949-1961 (2017), doi : https://dx.doi.org/10.13005/bpj/1315.
CrossRef - Priya B S, Basavaraj N Jagadale, Swaroopa H N, Vijayalaxmi Hegde, Multiresolution bio-medical image segmentation using fuzzy c-means clustering, international journal of recent technology and engineering (ijrte), issn: 2277-3878, volume-8 issue-4, november 2019, doi:10.35940/ijrte.d9918.118419.
CrossRef - Chunyuan wan1, Mingquan ye1,*, Chuanwenyao1. Brain mr image segmentation based on gaussian filtering and improved fcm clustering algorithm. changrong wu2.978-1-5386-1937-7/17/$31.00 ©2017 ieee.
- L.N. Murthy1 and B. Anuradha2, Edge enhanced fuzzy c means algorithm for hippocampus segmentation and abnormality identification, biomedical & pharmacology journal vol. 10(4), 1747-1755 (2017), doi : https://dx.doi.org/10.13005/bpj/1288.
CrossRef - Xiangzhi Bai, Chuxiong Sun, Changming Sun, Cell segmentation based on fopso combined with shape information improved intuitionistic fcm, ieee j biomed health inform. 2019 jan;23(1):449-459. doi: 10.1109/jbhi.2018.2803020.
CrossRef - Kamatchi1 and M.Sundararajan2, Diagnosing sinusitis using fractional b-spline wavelet with near infrared spectroscopy, biomedical & pharmacology journal vol. 10(1), 95-103 (2017), doi : https://dx.doi.org/10.13005/bpj/1086.
CrossRef - Gholampour, A.A. Pouyan. The segmentation of fmi image layers based on fcm clustering and otsu thresholding, international journal of scientific & engineering research volume 3, issue 3, march-2012 3 issn 2229-5518.
- Selvathi1 , N.B. Prakash2 *, V. Gomathi3 and G.R. Hemalakshmi3, Fundus image classification using wavelet based features in detection of glaucoma, biomedical & pharmacology journal, june 2018. vol. 11(2), p. 795-805, doi : https://dx.doi.org/10.13005/bpj/1434.
CrossRef - Arivazhagana,*, L. Banesan. Texture segmentation using wavelet transform. 0167-8655/$-see front matter_2003 elsevierb.v. all right reserved.doi:10.1016/j.patrec.2003.08.005.
CrossRef - R Sreeraj1 * and G Raju2, Increasing sensitivity, specificity and ppv for liver tumor segmentation and classification using enhanced glcm, biomedical & pharmacology journal vol. 9(3), 1237-1246 (2016), doi : https://dx.doi.org/10.13005/bpj/1073.
CrossRef - Yuan huang1, Valentin de bortoli2, Fugen zhou1 jérôme gilles3. Review of wavelet-based unsupervised texture segmentation, advantage of adaptive wavelets. iet image process.© the institution of engineering and technology 2018.
- Wang min, Zhoushu-dao, Baiheng, Ma ning, Ye song, Sar water image segmentation based on glcm and wavelet textures. institute of mcteorology, pla university of science and technology, 978-1-4244-3709-2/10/$25.00 ©2010 ieee.
- Hammouchea, M. Diafa, J.-G. Postaireb, ∗Auniversitémouloudmammeri. A clustering method based on multidimensional texture Analysis. départementd’automatique, tiziouzou, algeriabuniversité des sciences et technologies de lille—lagis, umrcnrs 8146, 59655 villeneuve d’ascqcedex, 0031-3203/$30.00 _ 2006 pattern recognition society. published by elsevier ltd. all rights reserved.doi:10.1016/j.patcog.2005.11.024.
CrossRef - Abdulkadirsengura, Yanhuiguob. Color texture image segmentation based on neutron sophicsetand wavelet transformation.⇑2011 elsevierinc. all rights reserved.
- Amjadali, Xiaojunjing, Nasirsaleem. Glcm-based fingerprint recognition algorithm. proceedings of ieee ic-bnmt2011.
- Fritz Albregtsenimage processing laboratory. Statistical texture measures, computed from, gray level coocurrence matrices. department of informatics, university of oslo november 5, 2008.
- Frank y. shih*, Shouxiancheng. Automatic seeded region growing for color image segmentation. 0262-8856/$ – see front matter q 2005 elsevierb.v. all rights reserved.doi:10.1016/j.imavis.2005.05.015.
CrossRef - Ammi Reddy Pulagam1 *,Venkata Krishna Rao Ede2 and Ramesh Babu Inampudi3, Segmentation of airways in lung region using novel statistical thresholding and morphology methods, biomedical & pharmacology journal vol. 10(4), 2035-2043 (2017), doi : https://dx.doi.org/10.13005/bpj/1325.
CrossRef - M. Vijaya1 and K. Suresh2, ICRODI: Image compression of region of diagnostics interest (rodi) using layer segmentation and wavelet, biomedical & pharmacology journal, june 2019. vol. 12(2), p. 1015-1021, doi : https://dx.doi.org/10.13005/bpj/1730.
CrossRef - Region based monochrome and ir image fusion using dtcwpt.
- Abd El Kader Isselmou1 *, Guizhi Xu2 and Shuai Zhang3, Improved methods for brain tumor detection and analysis using mr brain images, biomedical & pharmacology journal, december 2019. vol. 12(4), p. 1621-1631, doi : https://dx.doi.org/10.13005/bpj/1793.
CrossRef - Mohammed Lamine Benomar, Amine Chikh, Xavier Descombes, Mourtada Benazzouz, Multi features based approach for white blood cells segmentation and classification in peripheral blood and bone marrow images, international journal of biomedical engineering and technology (ijbet), inderscience, 2021. ffhal-02279352v2.
- Rohit Agrawal, Sachinandan Satapathy, Govind Bagla, Rajakumar K, Detection of white blood cell cancer using image processing, 2019 international conference on vision towards emerging trends in communication and networking (vitecon), 978-1-5386-9353-7/19/$31.00 ©2019 ieee
CrossRef - Sugandha Agarwal1 *, O.P. Singh1 and Deepak Aagaria2, Analysis and comparison of wavelet transforms for denoising mri image, biomedical & pharmacology journal vol. 10(2), 831-836 (2017), doi :https://dx.doi.org/10.13005/bpj/1174.
CrossRef - Jian-jiun ding. The class of time-frequency analysis and wavelet transform, the department of electrical engineering, national taiwan university (ntu), taipei, taiwan, 2007.
- Jian-jiun ding. The class of advanced digital signal processing, the department of electrical engineering, national taiwan university (ntu), taipei, taiwan, 2008.14. w.k. pratt, digital image processing 4th edition, john wiley & sons, inc., losaltos, california, 2007.
- Karlijn, J. van, Stralen1 vianda, S. Stel1 johannes, B. Reitsma2 friedo, W. Dekker13 carminezoccali4 kitty, J.Jager1. Diagnostic methods i: sensitivity, specificity, and other measures of accuracy. https://doi.org/10.1038/ki.2009.92get rights and content, under an elsevier user license.
- Jeroenbertels j.b. and T.E. Optimizing the dice score and jaccard index for medical image segmentation: theory & practice, have contributed equally to this work.11esat, center for processing speech and images, kuleuven, Belgium.