Manuscript accepted on :05-11-2021
Published online on: 24-11-2021
Plagiarism Check: Yes
Reviewed by: Dr. Ismail Ismail Abdullahi
 , Dr. Sherif Ramzy
, Dr. Sherif Ramzy
Second Review by: Dr. Hanefi ÖZBEK

Final Approval by: Dr. Fai Poon
Dalia Zaafar1 , Toka Elemary2, Yara Abdel Hady2 and Aya Essawy3
, Toka Elemary2, Yara Abdel Hady2 and Aya Essawy3
1Department of Pharmacology and Toxicology, Faculty of Pharmacy, MTI University, Cairo, Egypt.
2Department of Clinical Pharmacy, Faculty of Pharmacy, Cairo University, Cairo, Egypt.
3Department of Clinical Pharmacy, Faculty of pharmacy, MTI University, Cairo, Egypt.
Corresponding Author E-mail: dalia.zaffar@pharm.mti.edu.eg
DOI : https://dx.doi.org/10.13005/bpj/2277
Abstract
The term "non-druggable" refers to a protein that cannot be targeted pharmacologically; recently, significant efforts have been made to convert these proteins into targets that are reachable or "druggable." Pharmacologically targeting these difficult proteins has emerged as a major challenge in modern drug development, necessitating the innovation and development of new technologies. The idea of using RNA-targeting therapeutics as a platform to reach unreachable targets is very appealing. Antisense oligonucleotides, nucleic acid or aptamers, RNA interference therapeutics, microRNA, and synthetic RNA are examples of RNA-targeting therapeutics. Many of these agents were FDA-approved for the treatment of rare or genetic diseases, as well as molecular markers for disease diagnosis. As a promising type of therapeutic, many studies are being conducted in order for more and more of them to be approved and used in different disease treatments and to shift them from treating rare diseases only to being used as more specific targeting agents in the treatment of various common diseases. This article will look at some of the most recent technological and pharmaceutical advances that have contributed to the erosion of the concept of undruggability.
Keywords
Antisense Oligonucleotides; Aptamers; Interference Therapeutics; Microrna; Synthetic RNA
Download this article as:| Copy the following to cite this article: Zaafar D, Elemary T, Hady Y. A, Essawy A. RNA-targeting Therapy: A Promising Approach to Reach Non-Druggable Targets. Biomed Pharmacol J 2021;14(4). |
| Copy the following to cite this URL: Zaafar D, Elemary T, Hady Y. A, Essawy A. RNA-targeting Therapy: A Promising Approach to Reach Non-Druggable Targets. Biomed Pharmacol J 2021;14(4). Available from: https://bit.ly/3xgMD3I |
Introduction
Recently using RNA-targeting therapeutics as drugs to control disease-relevant gene expression as a treatment is very promising. These RNA targeting drugs include antisense oligonucleotides (ASOs), nucleic acid or peptide aptamers, RNA interference therapeutics (RNAi), microRNA therapeutics, and synthetic mRNAs. These drugs can engage several “difficult to drug” targets, including enzymes, ion channels, receptors, kinases, or transporters, giving hope for treating uncontrollable diseases. Some of these therapeutics are already in clinical practice and approved by the FDA, such as Nusinersen 1, which is splicing switching ASOs used to treat spinal muscular atrophy. Patisiran is an RNAi drug approved by the FDA for treating hereditary transthyretin amyloidosis 2. A number of RNA targeting drug candidates can be directed towards liver conjugation, GalNac, an amino sugar that can be added to siRNA, and ASO drugs to help better cellular uptake of the liver due to its unusually high expression on the hepatocyte’s surface. Inclisiran is a GalNac-conjugated siRNA drug that decreases LDL cholesterol expression through the downregulation of PCSK9 3. This drug is now being studied in different studies in order to treat several cardiovascular patients with elevated LDL levels. If successful, RNA-targeting therapies can be used in the treatment of diseases that affect a large proportion of the population beyond rare diseases. Additional RNA therapeutics had to be explored, approved and used in life-saving applications from various rare, aggressive diseases which did not yet have an effective treatment. Currently, hundreds of RNA-targeted research drugs for different indications of diseases are under clinical development, including neurodegeneration, metabolism, cardiovascular, and various cancers.
RNA-targeting therapeutics may be promising in treating several central nervous system diseases, especially genetic ones. It has been a challenge to deliver such drugs into their proper target tissues. Several experimental trials used different methods of delivery. Dahlman and colleagues tried to use nanoparticles for RNA targeting drug delivery with promising results 4. Sugo and colleagues revealed that siRNAs conjugation with antibodies could ease recognizing different specific surface receptors and consequently direct the drugs to muscle tissue 5 or cancer cells, as reported by Bäumer and colleagues 6. In addition, Kamerkar and colleagues reported that siRNAs could be delivered against KRAS in their pancreatic cancer experimental model using exosomes 6. In this review, the authors discussed and reviewed some of these promising drugs, their effect, expected mechanism of action, and adverse reactions. Table.1 lists different studies of RNA targeting therapies on the human, cell line, or animal and their type and role (Table.1.)
Table 1: Shows different studies of RNA-targeted therapeutics, types of studies, types of RNA agent used and its role concluded from the study.
| Serial | Author | Year | Type of Study | subject | RNA | role |
| 1 | Gragoudas et al (1) | 2004 | prospective | human | VEGF 165 aptamer (pegaptanib) | Inhibit angiogenesis |
| 2 | Gragoudas (2) | 2004 | prospective | human | VEGF 165 aptamer (pegaptanib) | Inhibit angiogenesis |
| 3 | Tong et al (3) | 2010 | prospective | cell line | Aptamer | conjugation with antiproliferative drugs like Taxol |
| 4 | Dhar et al (4) | 2008 | prospective | cell line | Aptamer | deliver cisplatin nanoparticles to prostate tumor cells |
| 5 | Hagenacker et al (5) | 2020 | prospective | Human | Antisense(Nusinersen) | safe and effective in treating adults with 5q spinal muscular |
| 6 | Dasgupta & Benson (6) | 2019 | prospective | human | Antisense(Inotersen) | supress progression of transhyretin cardiomypathy |
| 7 | Ackermann et al (7) | 2016 | prospective | human | Antisense(Inotersen) | suppress the progression of transhyretin cardiomyopathy |
| 8 | Charlestone et al (8) | 2018 | prospective | human | Antisense ( Eteplirsen) | Therapy for Duchenne muscular dystrophy (DMD) |
| 9 | McCaffrey et al (9) | 2002 | prospective | mice | RNAi | destruction sequence of hepatitis C virus in mice |
| 10 | Song et al (10) | 2003 | prospective | human | RNAi(CCR5,P25) | Supress HIV replication in macrophages |
| 11 | Davis et al (11) | 2010 | prospective | human | RNAi | Therapy for skin cancer melanoma |
| 12 | Yuen et al (12) | 2020 | prospective | RNAi (ARC520) | reduction in HBsAg and HBeAg positive | |
| 13 | Thi et al (13) | 2019 | prospective | cell line | RNAi (ARB-1740) | degredation of HBV viral replication in hepatocytes |
| 14 | Adams et al
(14) |
2017 | prospective | human | RNAi (Patisiran) | supress progression of transhyretin cardiomypathy |
| 15 | Zimmermann et al (15) | 2017 | prospective | human | RNAi (Revusiran) | supress progression of transhyretin cardiomypathy |
| 16 | Varghese et al (16) | 2020 | prospective | human | RNAi (siG12D-LODER) | in combination with gemcitabine for better efficacy in pancreatic cancer |
| 17 | Pipe et al (17) | 2019 | prospective | human | RNAi (Fituiran) | Antithrombin in treating hemophilia A or B |
| 18 | Wang et al (18) | 2020 | prospective | human | miRNA(miR-93) | Molecular marker in prostate cancer |
| 19 | Li et al (19) | 2015 | prospective | human | miRNA(miR-93) | Molecular marker in Head and neck squamous cell carcinoma |
| 20 | Smith et al (20) | 2012 | prospective | cell line | miRNA(miR-93) | Molecular marker in breast cancer |
| 21 | Fang et al (21) | 2012 | prospective | cell line | miRNA(miR-93) | Molecular marker in lung cancer |
| 22 | Fang et al (22) | 2011 | prospective | cell line | miRNA(miR-93) | Molecular marker in glioblastoma |
| 23 | Li et al (23) | 2020 | prospective | human | miRNA(hsa-miR-155-5p and has-miR-532-5p ) | potential pharmacogenomic predictors of ICS response in asthma |
| 24 | Wei et al (24) | 2020 | prospective | human | miRNA(hsa-miR-149, hsa-miR-221, hsa-miR-628-3p and has-miR-654-5p ) | Play a significant role in development and regulation of atrophic bone non-union |
| 25 | Xu et al (25) | 2017 | prospective | cell line | miRNA (miR-30a-5p) | increase baclitaxel sensitivity in lung cancer |
| 26 | Liu et al (26) | 2016 | prospective | cell line | miRNA (miR-30a-5p) | role in cisplatin resistance in ovarian cancer |
Nucleic acid or peptide aptamers
The word ‘Aptamer’ is the Latin word ‘aptare’ for ‘to fit or join,’ and the Greek word ‘mero’ means ‘to part.’ A nucleic acid Aptamer is a short chain of single-stranded DNA or artificial oligonucleotides RNA with nucleotide bases that vary between 40 and 100 nucleotide bases 7. Aptamers are small single-stranded DNAs or RNAs that link with high affinity and selectivity their specific targets. Aptamers can also function as an intracellular supply vehicle. Aptamers may also be crosslinked to small iRNA or miRNA to improve virulence, drug resistance, or pathogensis through their drug delivery 8. They have a unique, flexible 3D structure, high affinity, and high specificity, strongly related to their ligand sequence. These critical features allow them to distinguish between objectives. In addition, aptamers can bind to their cognate target protein’s functional domains, such as allosteric sites 9, 10. Small ions, large proteins, whole cells, viruses, and tissues can be their targets [10]. Various medicines, such as ampicillin aptamer conjugate, can easily be linked to aptamers and offer a better effect 11. One aptamer conjugates for photodynamic therapy (PDT) is (Apt@Au NRs), an aptamer-functionalized gold nanorods. Aptamers are produced by systematic ligand evolution by the exponential method of enrichment (SELEX) or other modified SELEX strategies12,10. Without prior knowledge of the objective, SELEX has the advantage of selecting aptamers. Lately, aptamers have been considered for a wide range of human disorders in several clinical evaluations. They are flexible and efficient instruments for therapeutic objectives 13, 14. Aptamers offer several benefits, making them preferable to the traditional drug used containing antibodies, since aptamers are not immunogenic and have a high cell/tissue selectivity, penetration, and many potential objectives. Aptamers are not immunogenic. They are thermally stable, have fewer lots of batch variability, short production time, and low expense.
Aptamers are excreted renally with a short half-life in vivo, challenging for therapeutical aptamers development. In furthermore, RNA-based aptamers are susceptible to hydrolytic nuclease breakdowns and several changes have been made to prevent and improve pharmacokinetics 15, including 2-fluoro pyrimidine changes, 2-O-methyl nucleotides and introduction of cholesterol or polyethylene glycol as the anchor group (Figure. 1).
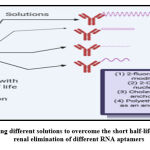 |
Figure 1: showing different solutions to overcome the short half-life and rapid renal elimination of different RNA aptamers |
Macugen®, a VEGF165 aptamer or pegaptanib, is a 2004 FDA-approved aptamer-based drug 16. It strongly impedes angiogenesis by hindering the extracellular form of the endothelial growth factor and by treating adults with age-related wet form macular degeneration. This aptamer-based drug is a 2′fluoropyrimidine aptamer-based modified RNA that contains 2′O-methyl purine modifications to improve its stability against endonucleases, in addition to introducing a 5′-PEG moiety and a 3′dT link to improve its pharmacokinetic profile and protect it from exonuclease17,18.
Aptamers based on RNA are more structurally different form DNA-based Aptamers and are also focused on medical applications. They are used for molecular in vivo imaging 19. Aptamers can be employed as carriers of polymeric NPs, such as polylactides, and antiproliferative conjugating drugs, like taxol 20. Aptamers may also be able to deliver cisplatin to prostate tumor cells from aptamer-functionalized Pt (IV) prodrug-PLGA-PEG nanoparticles 21. It’s of particular interest that aptamers can be used as a specific carrier of therapeutic oligonucleotides for diagnosis, precision medicine, and personalized medicine, such as small-interfering (si)RNAs, miRNAs, short hairpin (sh)RNA’s and antisense oligonucleotides (ASOs) 22.
Antisense Therapeutics
The transcription of Antisense was regarded as a transcriptional noise. This is a generalized phenomenon in both eukaryotic and human transcriptomes, which depends in two ways on the functioning of the antisense RNA in cis or trans. This process can create the long, non-coding RNAs (LncRNAs), one of the most diversified cell transcript groups, that demonstrated multiple function roles, including embryonic pluripotency, differentiation, and growth, in fundamental biological processes23.
The antisense RNA molecule is a unique DNA transcript type with 19-23 nucleotides and an mRNA supplement. In controlling gene expression, antisense RNAs play an essential role in multiple stages including replication, transcription, and translation. Furthermore, the expression of associated genes in host cells can be effectively regulated by artificial antisense RNAs. The research into antisense RNA functions has emerged as a hot study area in the development of antisense RNA24
Cardiomyopathy of TTR is a disease that may be caused by a transthyretin inherited mutation (ATTRm) or by natural transthyretin deposition in older people (ATTRWT). Antisense oligonucleotide (ASO) downregulates both wild and mutant hepatic TTR synthesis25. Inotersen, a transthyretin-specific oligonucleotide, was initially shown to suppress TTR for transgenic mice carrying the human TTR Ile84Ser gene causing decreased immunoglobulin (Ig) and amyloid serum A proteins that restrict or stop the progression of the disease 26.
Dasgupta and Benson reported Inotersen’s tolerability and safety profile. During their research, there were no deaths, and it was well tolerated. The drug-related side effects included minor reactions to the injection site and minor flu-like symptoms post-injection. There was no observation of serious thrombocytopenia or drug-related adverse renal effects. Important negative events included hyperkalemia, heart failure aggravation, urinary retention, atrial fibrillation, bacteremia, lower limb cellulite, anemia, heart pacemaker placement, and decubitus ulcers 25. The progressive degenerative disease, Duchenne muscular dystrophy (DMD), is a recessively linked X-like, evenly lethal neuromuscular disease caused by a lack of dystrophins, resulting in early adolescents losing outpatient ambulation and addiction to wheelchairs27. Eteplirsen is the first FDA-approved DMD therapy which is also prescribed for individuals with genetically confirmed DMD genetic mutation, who are responsive to exon 51 skippings, 28, 29. Contrary to the previously mentioned successful antisense drugs, the addition of apatorsen, which is a non-squamous, non-small-cell lung cancer (NSCLC) anticloveted oligonucleotide targeted at a heat shock protein (Hsp) of 27 mRNA, did not add any value compared to the original doublet protocol as reported in the recent study 30.
Another antisense oligonucleotide is Nusinersen which is capable of modifying the SMN2 gene expression and, subsequently, increase the synthesis of SMN protein and enhance the motor function. The FDA and the European Medicines Agency (EMA) have approved Nusinersen as the first option for 5q spinal musculoskeletal atrophy treatment for patients of different ages and stages. 1, 31.
RNA interference therapy
RNA Interference (RNAi) is a biological process that occurs in several eukaryotic cells; it is a gene silence mechanism after transcription. It is a two-strand RNA (dsRNA) that induces a homologous mRNA sequence-specific degradation. A gradual cleavage of dsRNAs (siRNAs) into 21-23 nucleotides (nt) begins. These native SiRNA duplexes are integrated into a complex of proteins called the RNA-induced silences complex (RISC). The ATP-dependent disassembly of the siRNA duplex creates an active RISC complex that recognizes and cleaves the respective siRNA by guiding the antisense strand of siRNA to protect the genome from retro-transposition by silence or gene expression. In eukaryotes, RNAi is a mechanism where ncRNAs (non-coding RNA) control the post-transcriptional expression of the target gene. This genetic intervention strategy can be clinically implemented using a double-stranded gene RNA (dsRNAs), a small synthetic RNA (siRNA) derived from the genome, and exogenous nucleic acids such as (sh)RNA. siRNAs have also become a robust genetic function study tool 32,33. Silencing any protein with RNAi by selecting the effective target is theoretically applicable. RNAi is also involved in various cellular processes such as viral infection defense, cell transformation, and disease development 34
The detection of the RNAi mechanism and its participation in human diseases allows researchers to develop various forms of RNA molecules and use the RNAi process to target gene expression, as well as to control cellular processes and conditions 34. A regular biomedical research laboratory can manufacture RNAi agents similarly to producing various recombinant/bioengineered DNA and protein macromolecules in living organisms.
In 2002 McCaffrey and RNAi colleagues targeted a hepatitis C virus sequence for destruction in mice5 to demonstrate this therapeutic potential 35. Song and colleagues reported in the following year that RNAi can suppress HIV replication in macrophages 36. The first use of RNAi-media gene silencing in patients with skin cancer melanoma has been reported in phase I clinical trials. The therapy reduced the expression of a gene required for tumor cell multiplication37.
RNAi mediated gene expression suppression and protein production using synthetically prepared siRNAs specifically designed to silence specific genetic sequences of a gene that causes disease. The mechanism is started when a complex consisting of dicer-related helicase 1 (DRH-1) binds the dsRNAs to many SiRNA fragments, which bind the RNA-inducing silencing complex (RISC). The two strands of every synthetic siRNA are separated, allowing the guide strand to match the complementary sequence of the target mRNA. The other strand, called “passenger,” instead, is degenerated and released. Enzymatically activated RISC splits the target mRNA precisely between the complementary nucleotides at positions 10 and 11 in the siRNA guide strand38. The overall result of this process is the degradation of the target mRNA leading to a decline in the target protein level (Fig. 2).
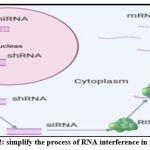 |
Figure 2: simplify the process of RNA interference in mammals. |
ARC-520 is one of the RNAi therapeutics designed to decrease viral and hepatitis B virus (HBV) DNA transcript derived from covalently closed circular DNA. ARC-520 was active both in HBV patients experienced by E-neg (Early Antigen HBeAg negatively) and E-pos (HBeAg positive). The absolute reduction in HBsAg (surface antigen) was moderate, possibly as a result of the HBsAg expression of integrated HBV DNA, indicating the need for RNAi treatments capable of targeting viral transcripts, regardless of origin 39. New therapies are needed to achieve functional cures that can lead to continuous loss of hepatitis B surface antigen (HBsAg) 39. The ARB-1740 is a clinical-stage agent consisting of three Lipid Nanoparticular Nanoparticular siRNAs (LNPs), which protect siRNAs against nuclear plasma degeneration, allowing for the effective intracellular uptake into hepatocytes of the HBV targeting siRNA, a primary site of HBV infection. Once endocytosed, LNPs have been pH-dependently mixed with the endosomal membrane, leading to the release of the encapsulated siRNA into the cytoplasm. It can then bind to the cellular complex (RISC) to detect, cleavage, and subsequently degrade viral transcripts. The function of this agent is to mediate viral transcript degradation, control viral replication, and reduce viral proteins in several HBV cell and animal models38.
Patisiran has also been shown to alter the progression of cardiac manifestations of hereditary amyloidosis by FDA-approved RNAi therapy in Phase III trials 2 (Amyloidosis-media-hereditary transthyretin disease is a rare, inherited, life-threatening neurodegenerative (hATTR) disease caused by TTR deposition in the peripheral nervous system, in the heart and gastro-intestinal tract). Revusiran is another medicine that has been tested to treat hATTR. The GalNAc-siRNA conjugate is the first metabolically stabilized to enter clinical trials and targets TTR mRNA targeted like Patisiran 40. siG12D-LODER is a small interfering RNA specifically targeted to the KRAS G12D mRNA mutant, frequent in pancreatic cancers. A phase I clinical trial highlighted the well-tolerated and targeted therapy combination of siG12D-LODER and gemcitabine in patients with locally advanced pancreatic cancer 41. Fituiran is an anti-thrombin-target SiRNA administered once-monthly subcutaneously and has shown a dose-dependent reduction in antithrombin level and increased thrombin production among participants who have no inhibitory effect alloantibodies with A or B hemophilia. Fitusiran has been clinically tested for hemophilia A and B in Phase III 42.
Lumasiran is an RNAi medicine that is used to treat primary hyperoxaluria type 1 of rare genetic disease (PH1). The disease is caused by the overproduction of hepatic oxalate that results in the formation of stones in the kidney that can progress towards kidney failure and systemic oxalosis. This therapeutic agent reduces the production of hepatic oxalate by inhibiting glycolate oxidase. Most lumasiran patients achieved low levels of urinary oxalate excretion. The most common side effect with lumasiran is the injection sitting reaction 43.
Micro RNA Therapeutics
Micro RNA is a small non-coding RNA molecule that induces cleavage or inhibits translation at specific sites with target mRNAs. These miRNAs play an important role in the expression of genes and in a number of other biological processes, including cell death 44. Results showed that miRNAs play an essential role in almost all biochemical processes such as proliferation, differentiation, metabolism and autophagy. In cancer, miRNAs impact many different types of biological processes, including the occurrence of tumors, metastases, invasion, microenvironment, and autophagy 45.
A lot of miRNAs are involved in prostate cancer development and progression. In prostate cancer tissues, the expression of miR-93 shows a significant increase compared to non-cancer prostate tissue 46. miR-93 is highly expressed in T-classification, lymph node metastasis, clinical stage, and poor prognosis in HNSCC squamous cell head and neck patients. MIR-93 may be a key molecular marker for boh the metastasis and prognosis of the lymph node in HNSCC patients 47.
The mechanism of miRNA oncogenicity can be explained in several cancer types, since the miR-93 has been found to regulate cancer metastases by controlling multiple metastasis genes and pathways. In breast cancer, miR-93 participated by inhibiting Smad7 expression and activating the TGF-β signaling pathways in the epithelial-mesenchymal transition (EMT). 48
During lung cancer, miR-93 suppressed LATS2 in vitro. It increased in vivo angiogenesis and lung metastases to promote the development of endothelial cell tubes miR-93 helped endothelial cells to grow, relocate and tube by suppressing the expression of ITGB8, which is the primary receptor in the extracellular matrix proteins and regulates cell-to-ECM adhesion in glioblastomas 50. These research results show that miR-93 contributes to invasion and metastases by controlling several related metastases, including induction of the transition between epithelial and mesenchymal syndrome, stimulation of angiogenesis, and interference with cell-to-ECM adhesion 47.
Jiang and colleagues had a clear association between the long-term responsiveness of ICS (inhaled corticosteroid) and circulating miRNAs. They have shown that miRNAs are potential ICS pharmacogenomic predictors. In particular, 15 miRNAs with a significant change in the ICS treatment effect. Two miRNAs were significantly interlinked with cortisol-dependent NF-kB-ICS (hsa-miR-155-5p and hsa-miR-532-5p). Hsa-miR-155-5p had a good impact on pulmonary function over time, while hsamiR-532-5p had a negative effect on pulmonary function 51.
A recent experimental study found upregulated miRNAs (hsa-miR-149, hsa-miR-221, hsa-miR-628-3p, and hsa-miR-654-5p), by suppressing numerous specific osteogenic genes, may play a significant role in the development and regulation of atrophic bone nonunion 52.
Potential functional target genes for differentially expressed miRNAs have comprised osteogenic and associated regulatory factor genes involved in fracture repair initiations. Multifunctional growth and differentiation factors forming part of the transforming growth factor-beta (TGF-b) superfamily, are bone morphogenetic proteins (BMPs). TGF-b was suggested to play a role in bone remodeling by influencing the differentiation and function of osteoblasts forming from bone and osteoclasts absorbing the bones. BMP-2 is the prominent bone morphogenetic protein used for osteoblast differentiation and function in preclinical and clinical trials. It can also be employed in treating bone abnormalities, nonunion fractures, spinal fusion, osteoporosis, and root canal operations. Liver/bone/kidney alkaline phosphatases and bone gamma-carboxyglutamate proteins indicate mature osteoblasts, and their expression is linked to bone formation and calcification 52.
Type 1 spinal muscle atrophy (SMA) is a rare progressive neuromuscular disease due to low motor neuron functional (SMN) protein levels. Risdiplam is an oral medicinal product administered for treating the SMA type 1 infants at a dose of 0.2mg/kg, increasing the functional SMN protein that leads to mRNA shifting to SMN2mRNA exorption 53
MiR-30a over-expression has been shown to increase the sensitivity of paclitaxel by decreasing the expression of the cell apoptosis promoter BCL-2; in the same way, over-expression of BCL-2 increases tolerance of paclitaxel, reducing the expression of MiR-30a. This data shows that miR-30a controls paclitaxel’s vulnerability by BCL-2 downregulation 54.
In cisplatin resistant ovarian cancer patients, MiRNA was found upregulated in drug-resistant cells; homo sapiens (hsa)-miR-30a-5p and hsa-miR-34c-5p. The expression of has- miR- 30a- 5p was highly upregulated in two forms of resistant ovarian cancer cell lines compared to those of chemotherapy-responsive lines. The resistance mechanism can be attributed to the increased miRNA 30a 5p expression, which could enhance the cellular growth and colony development potential and improve both cellular migration and invasion. Therefore, miRNA 30a 5p is projected to become an essential promising target for treatment that will be resistant to ovary cancer 55.
Conclusion
It is clear now that more RNA targeting agents need to be discovered. They are promising agents that target a specific enzyme or protein to treat a particular disease or can be used as a molecular marker for a particular disease. Scientists are now working hard to enable RNA targeting agents for different common cardiovascular and hepatic disorders. More studies needed to be held to investigate other drugs and help for FDA approvals
Conflict of Interest
The authors declare no competing financial interests or personal relationships could have influenced the work reported in this review article.
Funding Sources
There is no funding source.
References
- S. Finkel et al., “Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy,” N. Engl. J. Med., vol. 377, no. 18, pp. 1723–1732, Nov. 2017.
CrossRef - Adams et al., “Trial design and rationale for APOLLO, a Phase 3, placebo-controlled study of patisiran in patients with hereditary ATTR amyloidosis with polyneuropathy,” BMC Neurol., vol. 17, no. 1, pp. 1–12, Sep. 2017.
CrossRef - Ray, “Inclisiran in patients at high cardiovascular risk with elevated LDL cholesterol,” N. Engl. J. Med., vol. 376, pp. 1430–1440, 2017.
CrossRef - E. Dahlman et al., “In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight,” Nat. Nanotechnol., vol. 9, no. 8, pp. 648–655, 2014.
- Sugo et al., “Development of antibody-siRNA conjugate targeted to cardiac and skeletal muscles,” J. Control. Release, vol. 237, pp. 1–13, Sep. 2016.
CrossRef - Bäumer et al., “Antibody-mediated delivery of anti-KRAS-siRNA in vivo overcomes therapy resistance in colon cancer,” Clin. Cancer Res., vol. 21, no. 6, pp. 1383–1394, Mar. 2015.
CrossRef - Kudłak and M. Wieczerzak, “Aptamer based tools for environmental and therapeutic monitoring: A review of developments, applications, future perspectives,” Crit. Rev. Environ. Sci. Technol., vol. 50, no. 8, pp. 816–867, Apr. 2020.
CrossRef - Y. Gong et al., “SiRNA-mediated gene silencing of MexB from the MexA-MexB-OprM efflux pump in Pseudomonas aeruginosa,” BMB Rep., vol. 47, no. 4, pp. 203–208, 2014.
CrossRef - Toh, M. Citartan, S. Gopinath, T. T.-B. and, and undefined 2015, “Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay,” Elsevier.
CrossRef - Vahidi, N. Nafissi-Varcheh, … B. K.-I. journal of, and undefined 2017, “Challenges to design and develop of DNA aptamers for protein targets. II. Development of the aptameric affinity ligands specific to human plasma coagulation,” ncbi.nlm.nih.gov.
- Zhang, B. Soontornworajit, Z. Zhang, N. Chen, and Y. Wang, “Enhanced loading and controlled release of antibiotics using nucleic acids as an antibiotic-binding effector in hydrogels,” Biomacromolecules, vol. 13, no. 7, pp. 2202–2210, Jul. 2012.
CrossRef - Tabarzad and M. Jafari, “Trends in the Design and Development of Specific Aptamers Against Peptides and Proteins,” Protein Journal, vol. 35, no. 2. Springer New York LLC, pp. 81–99, 01-Apr-2016.
CrossRef - Urmann, S. Arshavsky-Graham, J. G. Walter, T. Scheper, and E. Segal, “Whole-cell detection of live: Lactobacillus acidophilus on aptamer-decorated porous silicon biosensors,” Analyst, vol. 141, no. 18, pp. 5432–5440, Sep. 2016.
CrossRef - Marton, F. Cleto, M. A. Krieger, and J. Cardoso, “Isolation of an aptamer that binds specifically to E. coli,” PLoS One, vol. 11, no. 4, p. e0153637, Apr. 2016.
CrossRef - D. Kovacevic, J. C. Gilbert, and B. Jilma, “Pharmacokinetics, pharmacodynamics and safety of aptamers,” Advanced Drug Delivery Reviews, vol. 134. Elsevier B.V., pp. 36–50, 01-Sep-2018.
CrossRef - W. M. Ng and A. P. Adamis, “Anti-VEGF aptamer (pegaptanib) therapy for ocular vascular diseases,” in Annals of the New York Academy of Sciences, 2006, vol. 1082, pp. 151–171.
CrossRef - S. Gragoudas, A. P. Adamis, E. T. Cunningham, M. Feinsod, and D. R. Guyer, “Pegaptanib for Neovascular Age-Related Macular Degeneration,” N. Engl. J. Med., vol. 351, no. 27, pp. 2805–2816, Dec. 2004.
CrossRef - S. Gragoudas, “VEGF Inhibition Study in Ocular Neovascularization–1 (VISION–1): Efficacy Results From Phase II/III MacugenTM (Pegaptanib Sodium) Clinical Trials | IOVS | ARVO Journals,” Investig. Ophthalmolgy Vis. Sci., vol. 45, no. 13, 2004.
- Bouvier-Müller and F. Duconge, “Application of aptamers for in vivo Molecular Imaging and Theranostics,” Elsevier.
- Tong, L. Yala, T. Fan, J. C.- Biomaterials, and U. 2010, “The formulation of aptamer-coated paclitaxel–polylactide nanoconjugates and their targeting to cancer cells,” Elsevier, vol. 11, no. 31, pp. 3043–3053, 2010.
CrossRef - Dhar, F. X. Gu, R. Langer, O. C. Farokhza, and S. J. Lippard, “Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA – PEG nanoparticles,” Proc. Natl. Acad. Sci. U. S. A., vol. 105, no. 45, pp. 17356–17361, Nov. 2008.
CrossRef - Nuzzo, G. Roscigno, A. Affinito, F. Ingenito, C. Quintavalle, and G. Condorelli, “Potential and challenges of aptamers as specific carriers of therapeutic oligonucleotides for precision medicine in cancer,” Cancers, vol. 11, no. 10. MDPI AG, p. 1521, 01-Oct-2019.
CrossRef - E. Villegas and P. G. Zaphiropoulos, “Neighboring gene regulation by antisense long Non-Coding RNAs,” Int. J. Mol. Sci., vol. 16, no. 2, pp. 3251–3266, 2015.
CrossRef - zhong Xu, J. lan Zhang, and W. guo Zhang, “Antisense RNA: the new favorite in genetic research,” J. Zhejiang Univ. Sci. B, vol. 19, no. 10, pp. 739–749, 2018.
CrossRef - R. Dasgupta and M. D. Benson, “Treatment of ATTR cardiomyopathy with a TTR specific antisense oligonucleotide, inotersen,” Amyloid, vol. 26, no. sup1, pp. 20–21, 2019.
CrossRef - J. Ackermann et al., “Suppressing transthyretin production in mice, monkeys and humans using 2nd-Generation antisense oligonucleotides,” Amyloid, vol. 23, no. 3, pp. 148–157, 2016.
CrossRef - Kole and A. M. Krieg, “Exon skipping therapy for Duchenne muscular dystrophy,” Adv. Drug Deliv. Rev., 2015.
CrossRef - S. Charleston et al., “Eteplirsen treatment for Duchenne muscular dystrophy,” Neurology, vol. 90, no. 24, pp. e2135–e2145, 2018.
CrossRef - Exondys 51 [package insert]., “Dosing Information,” Cambridge, MA: Sarepta Therapeutics, Inc.; 2018., 2018. .
- R. Spigel et al., “A Randomized, Double‐Blinded, Phase II Trial of Carboplatin and Pemetrexed with or without Apatorsen (OGX‐427) in Patients with Previously Untreated Stage IV Non‐Squamous‐Non‐Small‐Cell Lung Cancer: The SPRUCE Trial,” Oncologist, vol. 24, no. 12, 2019.
CrossRef - Hagenacker et al., “Nusinersen in adults with 5q spinal muscular atrophy: a non-interventional, multicentre, observational cohort study,” Lancet Neurol., vol. 19, no. 4, pp. 317–325, Apr. 2020.
CrossRef - Das, S. Musetti, and L. Huang, “RNA Interference-Based Cancer Drugs: The Roadblocks, and the ‘Delivery’ of the Promise,” Nucleic Acid Ther., vol. 29, no. 2, pp. 61–66, 2019.
CrossRef - Gao, M. Cheng, X. Zuo, J. Lin, K. Hoogewijs, and M. P. Murphy, “Active RNA interference in mitochondria,” Cell Res., no. March, pp. 1–10, 2020.
- Yu, C. Jian, A. H. Yu, and M. Tu, “Pharmacology & Therapeutics RNA therapy : Are we using the right molecules ?,” Pharmacol. Ther., vol. 196, pp. 91–104, 2019.
CrossRef - P. McCaffrey, L. Meuse, T. T. T. Pham, D. S. Conklin, G. J. Hannon, and M. A. Kay, “RNA interference in adult mice,” Nature, vol. 418, no. 6893, pp. 38–39, 2002.
CrossRef - Song et al., “Sustained Small Interfering RNA-Mediated HumanImmunodeficiency Virus Type 1 Inhibition in PrimaryMacrophages,” J. Virol., vol. 77, no. 13, pp. 7174–7181, Jul. 2003.
CrossRef - E. Davis et al., “Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles,” Nature, vol. 464, no. 7291, pp. 1067–1070, Apr. 2010.
CrossRef - P. Thi et al., “ARB-1740, a RNA Interference Therapeutic for Chronic Hepatitis B Infection,” ACS Infect. Dis., vol. 5, no. 5, pp. 725–737, May 2019.
CrossRef - F. Yuen et al., “RNA Interference Therapy With ARC-520 Results in Prolonged Hepatitis B Surface Antigen Response in Patients With Chronic Hepatitis B Infection,” Hepatology, vol. 72, no. 1, pp. 19–31, 2020.
CrossRef - S. Zimmermann et al., “Clinical Proof of Concept for a Novel Hepatocyte-Targeting GalNAc-siRNA Conjugate,” Mol. Ther., vol. 25, no. 1, pp. 71–78, Jan. 2017.
CrossRef - M. Varghese et al., “A phase II study of siG12D-LODER in combination with chemotherapy in patients with locally advanced pancreatic cancer (PROTACT).,” J. Clin. Oncol., vol. 38, no. 15_suppl, pp. TPS4672–TPS4672, May 2020.
CrossRef - Pipe et al., “Fitusiran, an RNAi Therapeutic Targeting Antithrombin to Restore Hemostatic Balance in Patients with Hemophilia a or B with or without Inhibitors: Management of Acute Bleeding Events,” Blood, vol. 134, no. Supplement_1, pp. 1138–1138, Nov. 2019.
CrossRef - F. Garrelfs et al., “Lumasiran, an RNAi Therapeutic for Primary Hyperoxaluria Type 1,” N. Engl. J. Med., vol. 384, no. 13, pp. 1216–1226, 2021.
- Issa, “Intracellular microRNA expression patterns influence cell death fates for both necrosis and apoptosis,” Handb. Med. Image Comput. Comput. Assist. Interv., vol. 8, no. 5, p. 55, 2020.
- A. Miska, “How microRNAs control cell division, differentiation and death,” Curr. Opin. Genet. Dev., vol. 15, no. 5 SPEC. ISS., pp. 563–568, 2005.
CrossRef - Wang et al., “Increased expression of microRNA-93 correlates with progression and prognosis of prostate cancer,” Medicine (Baltimore)., vol. 99, no. 22, p. e18432, 2020.
CrossRef - Li et al., “Increased expression of miR-93 is associated with poor prognosis in head and neck squamous cell carcinoma,” Tumor Biol., vol. 36, no. 5, pp. 3949–3956, 2015.
CrossRef - L. Smith et al., “The miR-106b-25 cluster targets Smad7, activates TGF-β signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer,” Oncogene, vol. 31, no. 50, pp. 5162–5171, 2012.
CrossRef - Fang et al., “MiR-93 enhances angiogenesis and metastasis by targeting LATS2,” Cell Cycle, vol. 11, no. 23, pp. 4352–4365, 2012.
CrossRef - Fang et al., “MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8,” Oncogene, vol. 30, no. 7, pp. 806–821, 2011.
CrossRef - Li et al., “Circulating MicroRNAs and Treatment Response in Childhood Asthma,” Am. J. Respir. Crit. Care Med., vol. 202, no. 1, pp. 65–72, 2020.
CrossRef - Wei et al., “Experimental study of expression profile and specific role of human microRNAs in regulating atrophic bone nonunion,” Medicine (Baltimore)., vol. 99, no. 36, p. e21653, 2020.
CrossRef - Baranello et al., “Risdiplam in Type 1 Spinal Muscular Atrophy,” N. Engl. J. Med., vol. 384, no. 10, pp. 915–923, 2021.
CrossRef - Xu et al., “miR-30a-5p enhances paclitaxel sensitivity in non-small cell lung cancer through targeting BCL-2 expression,” J. Mol. Med., vol. 95, no. 8, pp. 861–871, 2017.
CrossRef - Liu et al., “Expression of microRNA-30a-5p in drug-resistant and drug-sensitive ovarian cancer cell lines,” Oncol. Lett., vol. 12, no. 3, pp. 2065–2070, 2016.
CrossRef







