Manuscript accepted on :18-12-2021
Published online on: 28-12-2021
Plagiarism Check: Yes
Reviewed by: Dr. Bhavana Gundavarapu
Second Review by: Dr. Victor Oti
Final Approval by: Dr. Ayush Dogra
Kamilia Qudsiani , Sutriyo*
, Sutriyo* and Ratika Rahmasari
and Ratika Rahmasari
Faculty of Pharmacy, Universitas Indonesia, Depok, Indonesia, 16424
Corresponding Author E-mail: sutriyo@farmasi.ui.ac.id
DOI : https://dx.doi.org/10.13005/bpj/2304
Abstract
Nucleoside analogue antiviral remdesivir works by inhibiting the RNA-dependent RNA polymerase enzyme and terminating the viral replication. Currently, remdesivir is under a clinical trial for its activity against SARS-CoV-2. In the blood, remdesivir will undergo an enzymatic reaction to become monophosphate analogue form which is difficult to penetrate into the cell membrane. PAMAM (polyamidoamine) dendrimer is a good carrier to encapsulate remdesivir as a water-insoluble drug (0,339 mg/mL). Entrapment of remdesivir in the PAMAM cavity avoided remdesivir molecules to not undergo the enzymatic reactions. This study aimed to synthesize, characterize and evaluate cellular uptake of PAMAM-Remdesivir conjugate. PAMAM-Remdesivir was prepared with various stirring times (3, 6, 12, 24, and 48 hours). The conjugates were characterized to observe the size and particle distribution using Particle Size Analyzer, encapsulating efficiency using UV-Vis Spectroscopy, interaction between PAMAM and remdesivir particle using Fourier Transform Infrared Spectroscopy and cellular uptake of PAMAM-RDV using Fluorescence Microscope. The optimized stirring time of PAMAM-Remdesivir conjugate was 24 hours wich resulted the particles charge of + 23,07 mV of zeta potential, 1008 nm of particle size, 0,730 of PDI, and 69% entrapment efficiency. In addition, the FTIR analysis showed that remdesivir molecules successfully conjugated to PAMAM. Thus, through strring optimization time, the remdesivir molecules were successfully entrapped to PAMAM cavity. The cellular uptake in Vero Cell of PAMAM-RDV conjugated fluorescein isothiocyanate was observed with fluorescence microscope and had a stronger intensity than remdesivir only solution.
Keywords
Conjugate; Fluorescein; Isothiocyanate; Polyamidoamine; Remdesivir
Download this article as:| Copy the following to cite this article: Qudsiani K, Sutriyo, Rahmasari R. Polyamidoamine-Remdesivir Conjugate: Physical Stability and Cellular Uptake Enhancement. Biomed Pharmacol J 2021;14(4). |
| Copy the following to cite this URL: Qudsiani K, Sutriyo, Rahmasari R. Polyamidoamine-Remdesivir Conjugate: Physical Stability and Cellular Uptake Enhancement. Biomed Pharmacol J 2021;14(4). Available from: https://bit.ly/3z8flEP |
Introduction
Nucleoside analogue antiviral remdesivir is effectively in inhibiting Ebola and Marburg virus infections 1. This antiviral works by inhibiting the RNA-dependent RNA polymerase enzyme, thus terminate the viral replication 1,2. Currently, remdesivir is under a clinical trials of its activity against SARS-CoV-2. Previously, the antiviral activity of remdesivir against SARS-CoV-2 has been evaluated in vitro and in vivo. As result, remdesivir has better activity against SARS-CoV-2 than other antiviral 3. It indicates that remdesivir has potential as an antiviral for SARS-CoV-2.
The nucleoside triphosphate analogue from remdesivir can form a complex with RNA-dependent RNA-polymerase (RdRp) virus to inhibit the synthesis of viral RNA in cells. However, the result of in vivo pharmacokinetic test of remdesivir shows that the antiviral will undergo an enzymatic reaction become metabolite alanine, and rapidly become a monophosphate analogue after administrated intravenously 5 6 .The monophosphate analogue of remdesivir is negatively charged and polar which is difficult to penetrate into the cell membrane, while antiviral activity is highly dependent on the accumulation of drug molecules in the cells to cause an effect 4, 5. Therefore, remdesivir requires a drug delivery system that protects it from the enzymatic reactions berfot penetrating into the cells.
PAMAM is a with polyamide branch dendrimer with tertiary amine as the nucleus. It also has a cavity to encapsulate hydrophobic drug molecules. Therefore, PAMAM is a good carrier to encapsulate remdesivir as a water-insoluble drug (0,339 mg/mL). Commercially, PAMAM is available from the 1st to the 10th generations. The 4th generation is less toxic than the 5th or the 6th, and has a greater capacity of drug encapsulation than the previous generation 6, 7. Therefore, in this study PAMAM G4 was used. In physiological pH, encapsulated remdesivir in the PAMAM cavity will be protected from the environment, thusthe remdesivir molecules do not undergo the enzymatic reactions. 8, 9.
Currently, the synthesize of encapsulated remdesivir in PAMAM has not been reported yet. Thus, this study aimed to synthesize the PAMAM-Remdesivir (PAMAM-RDV) conjugate with has better stability than remdesivir without PAMAM. The conjugate will be characterized by examine the size and particle distribution using Particle Size Analyzer (PSA), encapsulating efficiency (EE) using UV-Vis spectroscopy, and interaction between PAMAM and remdesivir particle using Fourier Transform Infrared (FTIR) spectroscopy, and its cellular uptake at Vero cell by Flourescence Microscope.
Material and Methods
Reagents and Instruments
Remdesivir (Glentham Life Sciences), PAMAM G4 10% in methanol (Merck), dimethyl sulfoxide (Merck), water for injection (PT. Ikapharmindo Putramas), potassium bromide (Merck), Dulbecco’s Modified Eagle’s Medium (DMEM) (Merck), formaldehide (Merck). Spectrophotometer UV-Vis V-530 (Jasco, Japan), NANO-ZS series Malvern Zetasizer (Malvern Instruments, Malvern, UK), FTIR spectrophotometer 8400S (Shimadzu, Japan), fluorescence microscope opthiphot 2 (Nikon, Japan)
Methods
Synthesis of PAMAM-RDV conjugate
The stock solution of remdesivir was prepared by dissolving the remdesivir powder into 1 mL of DMSO. The mixture was stirred slowly with a spin bar on a magnetic stirrer until it dissolved. Then, 9 mL of water for injection was added to the mixture to obtain a concentration of 332 µM stock solution of remdesivir. The encapsulation of remdesivir into the PAMAM cavity was made using the molarity ratio (10:20). The conjugate was made by stirring remdesivir in 10% DMSO for 10 minutes at room temperature. The solution was added by 10% PAMAM and stirred for 3, 6, 12, 24, and 48 hours. The conjugate was purified using a centrifugation ultrafiltration tube (MWCO of 10 kDa) at 5000 rpm for 90 minutes, the pure conjugate on the top of the tube was stored at 4°C and kept away from light.
Characterization of PAMAM-RDV conjugate
Encapsulation efficiency
Preparation of a standard solution for the calibration curve was carried out by making remdesivir stock solution (332 µM) by dissolving 5 mg of remdesivir into 10 mL of 10% DMSO. The solution was diluted to nine concentrations; 7,5 µM; 10 µM; 12,5 µM; 15 µM; 17,5 µM; 20 µM; 22,5 µM; 25 µM; 27,5 µM; 30 µM. Each solution was analyzed using a UV-Vis spectrophotometer at a wavelength of 246 nm. Therefore, the calibration curve with the relationship between absorbance (x) and concentration (y) was made and a linear regression equation is obtained.
The encapsulation efficiency of PAMAM-RDV conjugate was aimed to determine the % value of remdesivir encapsulation efficiency in the PAMAM. The remdesivir particle that did not encapsulate to the PAMAM cavity from the filtrate of the centrifugation ultrafiltration tube (MWCO: 10 kDa) were measured using UV-Vis spectrophotometer at the 246 nm wavelength.
The % value of encapsulation efficiency is calculated by the equation:
% Encapsulation efficiency =
Where:
RDV1: The concentration of remdesivir in PAMAM-RDV conjugate
RDV2: The concentration of remdesivir in the filtrate of the centrifugation ultrafiltration tube
Particle size and zeta potential measurement
Particle size, distribution, and polydispersity index value were determined by Particle Size Analyzer NANO-ZS series Malvern Zetasizer (Malvern Instruments, Malvern, UK), using the photon correlation spectroscopy method. The conjugate solution was put into the analysis cuvette at a temperature of 25°C. The zeta potential of PAMAM-RDV conjugate was analyzed to determine the particle charge of the conjugate, the analysis was carried out using Particle Size Analyzer (Delsa Nano C type) with electrophoretic light scattering method. The sample was placed in a cuvette and analyzed at room temperature (25°C).
FTIR Spectrum of PAMAM-Remdesivir
The FTIR spectrum was analyzed by FTIR spectrophotometer 8400 S (Shimadzu, Japan). Analysis of conjugate PAMAM-RDV was carried out to identify the interaction of molecules, chemical structures, molecular bonds, and functional groups of the PAMAM-RDV conjugate. The samples were ground in the mortar with KBr (250 mg) and inserted into the FTIR analysis ring to observed the spectra. The samples were tested at 4000 to 400 cm-1 wavenumber. The FTIR spectrum of the PAMAM-remdesivir conjugate was compared with the FTIR spectrum of pure remdesivir and PAMAM
Cellular uptake evaluation
Fluorescein isothiocyanate (FITC)-remdesivir and FITC-PAMAM-RDV were prepared (1:1) by dissolved 1 mL of FITC (10 g/mL) in 1 mL of PAMAM-RDV and remdesivir-only solutions. These mixtures were incubated overnight in the dark at room temperature 11, 12. Vero cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) at pH 7.4 at 37°C in 5% CO2. The cells were put into an 8-well chamber (10,000 cells per well) in 500 L of growth medium 11, 12. The Vero cell medium was removed and washed with sterile PBS three times, then added the 4% formaldehyde, and incubated for 1 hour at room temperature. The formaldehyde was removed and the slide chamber was washed by sterile PBS three times. Samples (FITC-remdesivir, FITC-PAMAM-RDV, and FITC) were added to cells and incubated at 37°C with 5% CO2 for 15 minutes. The slide chamber was then washed with sterile PBS three times, dried and observed under a fluorescence microscope using a B-2A light filter to observed the cellular uptake 15.
Results and Discussion
Synthesis of PAMAM-RDV conjugate
In order to maximally dispersed remdesivir, the remdesivir were diluted in 10% DMSO at low stirrer speed (250 rpm) for 10 minutes. The stock solution of remdesivir that has been prepared cannot be stored for more than 24 hours either at room temperature or in the refrigerator (4°C) because it will cause the remdesivir powder that has been dispersed became settle at the bottom of the solution. Thus, the prepared stock solution should be used for conjugate preparation on the same day.
The ratio concentration of remdesivir and PAMAM to synthesis conjugate PAMAM-RDV was 1:2. The conjugate was synthesized by dissolving 301 µL remdesivir from the stock solution in 10 mL of WFI and followed by addition of 35 µL PAMAM. Since PAMAM was not stable and easily degraded by light 10, the mixture of PAMAM and remdesivir was put in erlemeyer flask coated with aluminium foil. We optimeized the stirring time (at 150 rpm) for 3, 6, 12, 24, and 48 hours at room temperature. Optimization of stirring time is important to obtain the conjugate with the highest encapsulation efficiency of remdesivir. The result of encapsulation effiiciency is show in Table 1.
In order to separate the non encapsulated remdesivir in PAMAM, the purification process of conjugate was performed. The purification was carried out using a centrifugation-ultrafiltration tube at molecular weight cut off (MWCO) 10 kDa). Since the molecular weight of remdesivir is less than PAMAM, thus the free remdesivir will pass the filtration tube. Purification carried out for seven cycles each at 5000 rpm 90 minutes until the volume of the filtrate no longer increased, as shown fig 1, resepectively. The PAMAM-RDV conjugate was detained at the top of the filter while the free molecule remdesivir at the bottom of the tube. The obtained PAMAM-RDV conjugate was in clear solution form (Fig 2).
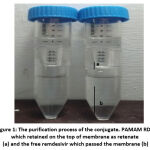 |
Figure 1: The purification process of the conjugate. PAMAM RDV which retained on the top of membrane as retenate (a) and the free remdesivir which passed the membrane (b). |
Characterization of PAMAM-RDV conjugate
Physical stability
In order to evaluate the PAMAM-RDV conjugate physical stability, the solution was kept at room temperature for 24 hours. The conjugate was observed in clear-stated condition however, at remdesivir-only solution, agglomeration was occurred after 24 h as shown at figure 2 (b) This phenomena explained the conjugate PAMAM-RDV maintained the dispersion of remdesivir in the solution as shown at figure 2 (a).
 |
Figure 2: The conjugate PAMAM-RDV (a) and Remdesivir-only (b) after 24 hours |
The calibration curve of remdesivir was made from nine series of concentrations; 7,5 µM, 10 µM, 12,5 µM, 15 µM, 17,5 µM, 20 µM, 22,5 µM, 25 µM ,27,5 µM, and 30 µM which were analyzed using a UV-Vis spectrophotometer at 246 nm 12. The obtained calibration curve’s equation was: y= 0.0308x – 0.0222 (Fig 3.) where y is the concentration and x is the absorbance, and the R2 value was 0.993 (Fig 3.). The R2 value which is getting closer to 1 indicates that a high correlation between y and x. In addition, the R2 is also often used to describe data variation, where the desired data is linear 13.
After calculation, the variation coefficient is 1,5%, indicated the data variation is less and calibration curve is acceptable. The remdesivir calibration curve and calibration spectrum can be seen in Fig 3 and 4, respectively.
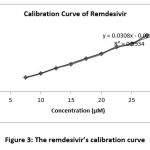 |
Figure 3: The remdesivir’s calibration curve. |
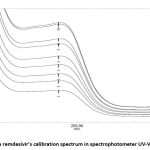 |
Figure 4: The remdesivir’s calibration spectrum in spectrophotometer UV-Vis at 246 nm |
To determine the percentage of encapsu;ated remdesivir molecules in the PAMAM cavity, entrapment efficiency analysis was performed. The indirect method was carried out by measuring the concentration of free remdesivir from the filtrat of purification process as mentioned above. In the synthesize of the PAMAM-RDV conjugate, the conjugate was stirred for 3, 6, 12, 24, and 48 hours using a shaker at room temperature at 150 rpm. The optimization of stirring time awas performed to get the highest entrapment efficiency of remdesivir. The results can be seen in Table 1.
Table 1: The entrapment efficiency of PAMAM-RDV conjugate
| Stirring time (Hr) | EE (%) |
| 3 | 37,969 |
| 6 | 51,101 |
| 12 | 56,500 |
| 24 | 69,082 |
| 48 | 48,435 |
As shown at table 1, 24 hours stirring time showed the highest entrapment efficiency of remdesivir, followed by the 6 hour, and then at the 48, 12, and 3 hours. This indicates that the maximum time required for remdesivir to enter and be entrapped into the PAMAM cavity was 24 hours at 150 rpm. The high entrapment efficiency of remdesivir at 24 h stirring time indicated that the amount of remdesivir encapsulated in the PAMAM cavity was greater than the unencapsulated. PAMAM dendrimers cavities able to encapsulate the hydrophobic drugs by covalent or non-covalent bond 7. However, it was observed that stirring for 48 hours caused the decreasing of entrapment efficiency. This phenomenone occured due to the possibility that PAMAM became unstable at room temperature for too long, thereby reducing its ability to entrap the remdesivir.
Particle size and particle distribution measurement
In this study, particle size (Z-avg) and polydispersity index (PDI) was measured using a Malvern type Particle Size Analyzer (PSA) with the photon correlation spectroscopy (PSC) method. PSC can measure the scattering of light by the Brownian motion of the dispersed particles 12. Similar to the entrapment efficiency test, the samples were PAMAM-RDV conjugates at 3, 6, 12, 24, and 48 hours of stirring. The results can be seen in Table 2, respectively.
Table 2: Particle size (Z-avg) and polydispersity index (PDI) of PAMAM-RDV conjugates
| Stirring time (Hr) | Z-Avg | PDI | Dv90 |
| 3 | 1704,000 | 0,918 | 264,333 |
| 6 | 886,866 | 0,713 | 374,667 |
| 12 | 633,533 | 0,592 | 428,667 |
| 24 | 1008,100 | 0,730 | 399,333 |
| 48 | 7172,667 | 0,559 | 791,567 |
Table 2 shows the largest Z-avg value was obtained at 48 hours and the smallest one was 12 hours of stirring. The smallest Dv90 was the 3 hours of stirring time (264,333 nm) which indicated the 90% of particles was below 264,333 nm. However, the PDI of 3 hours stirring time was 0,918. Polydispersity index or PDI which is also known as heterogeneity index was carried out to see the breadth or narrowness of the particle size distribution. PDI defines as a number on a scale from 0 to 1. A polydispersity index value less than 0.05 indicates a highly monodispersed particle or a narrow particle size distribution, while a broad particle size distribution is greater than 0,714,15, which indicated the 3 hours of stirring time obtained the largest variety of distribution particle.
Table 2 shows that the PAMAM-RDV conjugate has the lowest PDI which was stirred for 48 hours (0.559). However, the 48 hours of stirring time obtained the largest Dv90 and Z-avg, 0,559 and 7172,667 nm respectively. The 12 hours of stirring time obtained the good PDI (0,592) and the smallest Z-avg (633,533 nm). Nevertheless, the entrapment efficiency of 12 hours stirring time was 56,5%, smaller than 24 hours of stirring time. The PDI of 24 hours stirring time is 0.730 which indicated the large variety of particle size distribution. However, this conjugate was still optimal because it had the largest number of remdesivir molecules among the others. Thus, conjugates with a stirring time of 24 hours are still acceptable even though the PDI was above 0, 7.
Zeta potential measuremen
The interaction energy between particles in a liquid can be determined by observing the zeta potential value and related to the stability of the particles 16. In this study, the zeta potential value of the PAMAM-RDV conjugate was measured using the Malvern type Particle Size Analyzer (PSA) with the electrophoretic light scattering (ELS) method. The zeta potential of PAMAM-RDV with various strring time shown at table 3 and zeta potential of PAMAM and remdesivir show at table 4 and Fig 5 and 6.
Table 3: Zeta potential of PAMAM-RDV conjugates
| Stirring time (Hr) | ζ Potensial |
| 3 | +1,143 mV |
| 6 | +3,410 mV |
| 12 | +3,063 mV |
| 24 | +23,07 mV |
| 48 | +2,083 mV |
Table 4: Zeta potential of PAMAM-RDV conjugates
| Sample | ζ Potensial |
| PAMAM | +21,7 mV |
| Remdesivir | -26,2 mV |
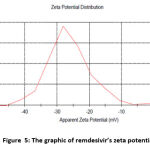 |
Figure 5: The graphic of remdesivir’s zeta potential |
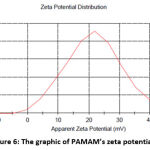 |
Figure 6: The graphic of PAMAM’s zeta potential |
The zeta potential values of the PAMAM-RDV conjugates in Table 3 show that all the conjugates which were stirred for various hours had a positive charge. This due to the positive charge of amine group of PAMAM dendrimer 17. The conjugate stirred for 24 hours had the highest zeta potential value, which was +23.07 mV, while the conjugate stirred for 3 hours had the lowest zeta potential value, which was +1.143. Particles with a zeta potential value greater than +30 mV and less than -30 mV are believed to be strong cationic and anionic. Table 4 shows that remdesivir had a negative charge (-26,2 mV) while PAMAM had a positive charge (+21,7), therefore, it could be concluded that the remdesivir molecules were successfully entrapped in PAMAM because the surface charge of conjugates particles is positive. The presence of positive charge can enhance their binding to the cell surface with the negative charge by electrostatic interactions and enhance the conjugate cellular uptake 20.
Zeta potential can be used as a parameter to predict the stability of a liquid because it is related to the interactions between particles in a colloidal system. High zeta potential values, both negative and positive, are generally stable because the charges between the particles tend to repel each other. On the other hand, a low zeta potential value, both positive and negative, will cause an attraction between the particles and cause particle aggregation in the liquid 16. In this study, the conjugate with the highest zeta potential was obtain from for 24 hours stirring time because it had a particle charge that was closest to +30 mV among other conjugates. Therefore, the best PAMAM-RDV conjugate was stirred for 24 hours. Most cellular membranes have a negative charge, thus particles that have positive zeta potential tend to permeate into the membrane. This will make the PAMAM-RDV conjugate enter the cells easier and increase their cellular uptake 18.
FTIR spectrum of PAMAM-RDV
FTIR spectrum analysis was carried out at wavenumbers 4000 to 400 cm-1 in oder to compare the spectrum of the PAMAM-RDV conjugate with remdesivir and PAMAM. FTIR spectroscopy was used in this analysis because some molecules have strong absorbance in the infrared. The infrared spectrum provides information about molecular structure (peak position and peak intensity). Remdesivir, PAMAM, and PAMAM-RDV was mixed with KBr until it reaches the homogeneous state so the typical spectrum of the sample could be observed. KBr (potassium bromide) pellets are used because they are inert and transparent at wavenumbers 400 cm-1 and above 19
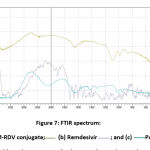 |
Figure 7: FTIR spectrum: (a) PAMAM-RDV conjugate; (b) Remdesivir; and (c) PAMAM |
PAMAM dendrimer has a group of primary amine and secondary amine which are indicated by the presence of bands at wavenumbers 3236 and 3448 cm-1 as shown in Fig. 7 (a), while the presence of bands at wavenumbers 1014 and 1317 cm-1 indicates that the presence of amide and alkane groups, while the wavenumbers at 1668 cm-1 indicated the presence of a carbonyl group. This wavelength indicated the presence of the PAMAM dendrimer. While in the remdesivir (Fig. 7(b)), has a group of alkane which are indicated by the presence of bands at wavenumbers 1600-1680 cm-1, while the presence of bands at wavenumbers 1690-1640 cm-1 indicated the presence of imine group, while the wavenumbers at 1000-1350 cm-1 indicated the presence of amine group, and the wavenumbers 1650-2000 cm-1 indicates the presence of an aromatic group.
The infrared spectrum of the PAMAM-RDV conjugate (Fig. 7(c) has a band at wavenumbers in 1085 cm-1 which indicated the presence of the C-N from the amine group. Amines contained in the conjugate can be derived from remdesivir or PAMAM which has a band in the wavenumber in 1014-1317 cm-1which also comes from the C-N group. In addition, there was broadband of PAMAM-RDV conjugate at a wave number in 3412 cm-1from the N-H group present in both primary and secondary amines. The infrared spectrum of PAMAM and remdesivir also has a band in the area 3138 – 3448 cm-1 which is also derived from the N-H group found in primary or secondary amines.
The infrared spectra of the PAMAM-RDV and remdesivir showed bands at wavenumbers of 1662 and 1647 cm-1, shows that the two samples may have C=N groups of imine compounds. In addition, bands of the C-O or C-O-C groups of the ether compound were also seen at wavenumbers from 1010 to 1292 cm-1 in both the remdesivir sample and the PAMAM-RDV conjugate. Therefore, it could be concluded remdesivir has been encapsulated in PAMAM and formed PAMAM-RDV conjugates because there is a shift in wavenumbers from 3138 to 3431 cm-1 and 3236 to 3448 cm-1 from primary and secondary amine groups in remdesivir and PAMAM to 3412 cm-1 derived from the PAMAM-RDV conjugate. In addition, a wavenumber of 3400 cm-1 from PAMAM-RDV, shows that there is an O-H bond that allows hydrogen bonds to occur between remdesivir and PAMAM molecules in the conjugate.
Cellular uptake evaluation
To evaluate the ability of PAMAM-RDV conjugate to penetrate into the cells, Vero Cells which were normal cells derived from the kidney epithelial cells of African green monkeys was used. A flourescence microscope was used to observe the uptake of PAMAM-RDV in the Vero Cells, with 40 times magnification. The green colour came from the Fluorescein isothiocyanate (FITC), a stain which was conjugated to the samples.
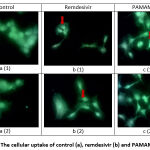 |
Figure 8: The cellular uptake of control (a), remdesivir (b) and PAMAM-RDV (c) |
The Intensity of Vero cell which were treated by FITC only, which was used as control, were showed at Figure 8 a (1) & (b), respectively. At Figures 8 b (1) and b (2), The intensity of FITC which were conjugated by Remdesivir was observed stronger compare to the control. However, the strongest intensity was obtained from FITC which were conjugate by the PAMAM-RDV (Fig. 8 c (1) & (2), respectively).
Generally, the membran cell as a barrier to separate the inside of the cell from the outside and block the particles to enter the cell. The cell membrane has a phospholipid bilayer and other proteins with a negative charge. This causes the membrane to selective permeability with particles. So, the particles that will enter the cell must pass through the cell membrane. The PAMAM-RDV conjugate has a positive charge from the PAMAM. Thus, the conjugate has the ability to bind the negatively charged plasma membrane, then cause pore formation and damage the lipids of the membrane 22, 23.
Fig. 8 a show a weaker intensity of green colour compared to figures B and C in the absence of strong intensity in the centre of the cell. The control was FITC (in 1 mg/mL Dimethyl sulfoxide) which was incubated in cells for 15 minutes. The observed green color at control was due to the presence of Dimethyl sulfoxide which was used as a FITC solvent since it has the ability to penetrate into the cells, as reported previously24. However, the intensity was still weaker than remdesivir and PAMAM-RDV. Thus, it can be concluded from cellular uptake testing that PAMAM-RDV has a stronger intensity than remdesivir.
Conclusions
In order to enhance remdesivir physical stability, we successfully synthesized PAMAM-Remdesivir conjugate with the optimum strring time was 24 hours at 150 rpm, followed by ultrafiltration-purification process for 90 min at 5000 rpm. The characterization results showed the PAMAM-Remdesivir conjugate had positive charge of 23,07 mV, 1008 nm of particle size, 0,730 of PDI, and clear-stated solution even after 24 hours incubation. Those results suggested that PAMAM-Remdesivir has better physical stabillity compare to remdesivir only. Furthermore, PAMAM-Remdesivir particle had 69% entrapment efficiency and hydrogen bonds was formed between remdesivir and PAMAM molecules. Thus, indicated the successful remdesivir entrapment into PAMAM. In addition, the PAMAM-Remdesivir was observed had better cellular uptake in Vero Cell than remdesivir only as shown by its higher fluoresensi intensity.
Authors’ Contributions
All authors have contributed according to their duties and responsibilities.
Conflict of Interests
No conflict of interests has been declared by all authors.
Funding Sources
There are no funding Source.
References
- McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res [Internet]. 2020;157(March):104859. Available from: https://doi.org/10.1016/j.phrs.2020.104859
CrossRef - McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res. 2020;157(March).
CrossRef - Vijayvargiya P, Esquer Garrigos Z, Castillo Almeida NE, Gurram PR, Stevens RW, Razonable RR. Treatment Considerations for COVID-19: A Critical Review of the Evidence (or Lack Thereof). Mayo Clin Proc [Internet]. 2020;95(7):1454–66. Available from: https://doi.org/10.1016/j.mayocp.2020.04.027
CrossRef - Frediansyah A, Nainu F, Dhama K, Mudatsir M. Remdesivir and its antiviral activity against COVID-19 : A systematic review. Clin Epidemiol Glob Heal [Internet]. 2020;(July):0–1. Available from: https://doi.org/10.1016/j.cegh.2020.07.011
CrossRef - Yan V, Muller F. Comprehensive Summary Supporting Clinical Investigation of GS-441524 for Covid-19 Treatment. OSF Prepr [Internet]. 2020; Available from: https://osf.io/mnhxu/
CrossRef - Sun D. Commentary Remdesivir for Treatment of COVID-19 : Combination of Pulmonary and IV Administration May Offer Aditional Benefit. 2020;
CrossRef - Lv T, Yu T, Fang Y, Zhang S, Jiang M, Zhang H, et al. Role of generation on folic acid-modified poly(amidoamine) dendrimers for targeted delivery of baicalin to cancer cells. Mater Sci Eng C. 2017;75:182–90.
CrossRef - Choudhary S, Gupta L, Rani S, Dave K, Gupta U. Impact of Dendrimers on Solubility of Hydrophobic Drug Molecules. 2017;8(May):1–23.
CrossRef - Albertazzi L, Serresi M, Albanese A, Beltram F. Dendrimer internalization and intracellular trafficking in living cells. Mol Pharm. 2010;7(3):680–8.
CrossRef - Devarakonda B, Otto DP, Judefeind A, Hill RA, de Villiers MM. Effect of pH on the solubility and release of furosemide from polyamidoamine (PAMAM) dendrimer complexes. Int J Pharm. 2007;345(1–2):142–53.
CrossRef - Oddone N, Zambrana AI, Tassano M, Porcal W, Cabral P, Benech JC. Cell uptake mechanisms of PAMAM G4-FITC dendrimer in human myometrial cells. J Nanoparticle Res. 2013;15(7).
CrossRef - Zhang J, Liu D, Zhang M, Sun Y, Zhang X, Guan G, et al. The cellular uptake mechanism, intracellular transportation, and exocytosis of polyamidoamine dendrimers in multidrug-resistant breast cancer cells. Int J Nanomedicine. 2016;11:3677–90.
CrossRef - Ammerman NC, Beier-sexton M. Growth and maintenance of Vero Cell lines. Curr Protoc Microbiol. 2008.
CrossRef - Jordan P, Alverca E, Andrade M, Dias E, Bento FS, Batore MCC, et al. Toxicon Morphological and ultrastructural effects of microcystin-LR from Microcystis aeruginosa extract on a kidney cell line. 2009;54:283–94.
CrossRef - Sun X-Y, Gan Q-Z, Ouyang J-M. Size-dependent cellular uptake mechanism and cytotoxicity toward calcium oxalate on Vero cells. Sci Rep [Internet]. 2017;7(1):1–12. Available from: http://dx.doi.org/10.1038/srep41949
CrossRef - Kulhari H, Pooja D, Prajapati SK, Chauhan AS. Performance evaluation of PAMAM dendrimer based simvastatin formulations. Int J Pharm [Internet]. 2011;405(1–2):203–9. Available from: http://dx.doi.org/10.1016/j.ijpharm.2010.12.002
CrossRef - Sahakijpijarn S, Moon C, Koleng JJ, Christensen DJ, Williams RO. Development of remdesivir as a dry powder for inhalation by thin film freezing. Pharmaceutics. 2020;12(11):1–27.
CrossRef - Prichard L, Barwick V. Preparation of Calibration Curves A Guide to Best Practice Contact Point : Prepared by : Lgc. 2003;(September 2003):1–27.
- Jain R, Savla H, Naik I, Maniar J, Punjabi K, Vaidya S, et al. Novel nanotechnology based delivery systems for chemotherapy and prophylaxis of tuberculosis [Internet]. Handbook of Nanomaterials for Industrial Applications. Elsevier Inc.; 2018. 587–620 p. Available from: http://dx.doi.org/10.1016/B978-0-12-813351-4.00034-1
CrossRef - Mudalige T, Qu H, Van Haute D, Ansar SM, Paredes A, Ingle T. Characterization of Nanomaterials: Tools and Challenges [Internet]. Nanomaterials for Food Applications. Elsevier Inc.; 2018. 313–353 p. Available from: http://dx.doi.org/10.1016/B978-0-12-814130-4.00011-7
CrossRef - Raval N, Maheshwari R, Kalyane D, Youngren-Ortiz SR, Chougule MB, Tekade RK. Importance of physicochemical characterization of nanoparticles in pharmaceutical product development [Internet]. Basic Fundamentals of Drug Delivery. Elsevier Inc.; 2018. 369–400 p. Available from: http://dx.doi.org/10.1016/B978-0-12-817909-3.00010-8
CrossRef - Feng Y, Kilker SR, Lee Y. Surface charge (zeta-potential) of nanoencapsulated food ingredients [Internet]. Characterization of Nanoencapsulated Food Ingredients. Elsevier Inc.; 2020. 213–241 p. Available from: http://dx.doi.org/10.1016/B978-0-12-815667-4.00007-9
CrossRef - Abedi-Gaballu F, Dehghan G, Ghaffari M, Yekta R, Abbaspour-Ravasjani S, Baradaran B, et al. PAMAM dendrimers as efficient drug and gene delivery nanosystems for cancer therapy. Appl Mater Today [Internet]. 2018;12:177–90. Available from: https://doi.org/10.1016/j.apmt.2018.05.002
CrossRef - Leng ZH, Zhuang QF, Li YC, He Z, Chen Z, Huang SP, et al. Polyamidoamine dendrimer conjugated chitosan nanoparticles for the delivery of methotrexate. Carbohydr Polym. 2013;98(1):1173–8.
CrossRef - Rasmussen MK, Pedersen JN, Marie R. Size and surface charge characterization of nanoparticles with a salt gradient. Nat Commun. 2020;11(1):1–8.
CrossRef - Smith, BC. Fundamentals of fourrier transform infrared spectroscopy. Second edition. CRC Press. 2011.
CrossRef - Donahue ND, Acar H, Wilhelm S. Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Adv Drug Deliv Rev [Internet]. 2019;143:68–96. Available from: https://doi.org/10.1016/j.addr.2019.04.008
CrossRef - Kheraldine H, Rachid O, Habib AM, Al Moustafa AE, Benter IF, Akhtar S. Emerging innate biological properties of nano-drug delivery systems: A focus on PAMAM dendrimers and their clinical potential. Adv Drug Deliv Rev [Internet]. 2021;(xxxx):113908. Available from: https://doi.org/10.1016/j.addr.2021.113908
CrossRef - Wang H, Zhong CY, Wu JF, Huang Y Bin, Liu CB. Enhancement of TAT cell membrane penetration efficiency by dimethyl sulphoxide. J Control Release. 2010;143(1):64–70.
CrossRef







