Manuscript accepted on :30 Dec 2020
Published online on: --
Plagiarism Check: Yes
Anita Khanna1 , Narendra Digambar Londhe1*
, Narendra Digambar Londhe1* and Shubhrata Gupta1
and Shubhrata Gupta1
1Electrical Engineering Department, National Institute of Technology Raipur, Raipur, India.
Corresponding Author E-mail : nlondhe.ele@nitrr.ac.in
DOI : https://dx.doi.org/10.13005/bpj/2042
Abstract
Bronchial airway structure and morphology identification is very useful for analysis of many lung diseases. Since, the human tracheo-bronchial tree is a dyadic non-symmetric branching network which is very complex and its manual tracing is quite tedious and unwieldy. Moreover, automatic detection techniques for airway are quite challenging. This is due to its complexity and fading off the airway intensity because of the smaller asynchronous branching and noise in the image reconstruction. In this paper, an unsupervised approach for segmentation of localized airway has been proposed after segmenting the lung region. Firstly, airways are segmented out by using 3D region growing techniques with intensity constrained to prevent leakages. This results in limited segmentation of airways due to partial volume effect and leakage risk. Further, deeper bronchial branches are segmented by applying adaptive morphological techniques on 3D segmented lungs. Then, these two results are combined followed by 3D region growing to get complete segmentation of airway. The proposed technique is tested on Exact’09 20 test cases and evaluated by Exact’09 team. The performance of the proposed approach is quite reliable in segmenting distal branches with reasonable leakages. The advantage of this scheme is that it is easy to implement, fully automated, and time efficient.
Keywords
Airway; Computed Tomography (CT); Medical Image Processing; Reconstruction; Region Growing; Segmentation
Download this article as:| Copy the following to cite this article: Khanna A, Londhe N. D, Gupta S. Automated Localized Approach for Airway Segmentation in 3D Chest CT Volume. Biomed Pharmacol J 2020;13(4). |
| Copy the following to cite this URL: Khanna A, Londhe N. D, Gupta S. Automated Localized Approach for Airway Segmentation in 3D Chest CT Volume. Biomed Pharmacol J 2020;13(4). Available from: https://bit.ly/38PPCo8 |
Introduction
Lung airway diseases namely cystic fibrosis, Bronchiectasis, asthma and Chronic Obstructive Pulmonary Disease (COPD) are quite important to be treated soon as they cause the narrowing and blockage of airway tubes which results in breathlessness1. Most of the people suffers from both forms of COPD namely, chronic bronchitis and emphysema which ultimately results in damage of the lung. These diseases are often related to morphological changes. Analysis of chest with Computed Tomography (CT) gives an indication of pathological changes and information about airway related diseases. But manual analysis of 3D CT volume having 300-500 slices in each volume one by one is quite cumbersome and a very slow process. However, the tracing of these structures can be helpful in planning as well as guidance of various surgeries like bronchoscopy, laparoscopy and neurosurgery.
Visual appraisal of tracing and marking a number of airways discerned by the human eye is quite tedious. Further, it is also not possible to be segmented by a simple algorithm. The main reasons for the mentioned problems are noise in the image, limited intensity distribution and diseases like bronchiolitis, mucus and other anomalies which result in non-connected airway segments. Moreover, there is no gold standard for identifying airways. Its judgement is done by generation number (i.e. number of splits in the tree), branch number and total tree length2. As image noise/artifacts have high impact on small airways, the problem of leakage causes fusion of airways with lung tissues having deeper branches of airways. Both semi-automated and automated methods of computerized airway segmentation may relatively reliable and serve a great value.
Segmentation is also very helpful for volume estimation and visualization of structure’s surface region. The most common method of segmentation involves region growing which uses large variation in contrast between tracheas and surrounding tissues3-6 in direction to initialize the location of the seed point. After initialization of seed point, either normal or adaptive 3D region growing approach is applied as general method to segment out large airways from CT dataset. This method is fast and efficient only if intensity threshold criteria is met. Region growing fails in case of small airways because of noise in the images and artefacts that causes broken edges. This results in airway segmentation leaking into lung parenchyma. This also happens due to similar voxel intensities of these two structures in deeper region of lungs.
To extract higher generation airways, authors have used morphological, knowledge/template based, vesselness filter, shape analysis, fuzzy logic and recently convolution neural network techniques. Morphology based reconstruction techniques were used to detect airways and connect them7-9. This method is quite efficient but performance deteriorates when the slices are not perpendicular. In knowledge or template-based methods, prior knowledge of anatomical structure10, set of predefined masks11-12 or a tree structure13 are used to facilitate the search of airways in lung section. These techniques are time taking and fails to detect very small airways. Other methods include enhancement of the tubular structures based on vesselness filter14-16. Method proposed in16 further improved the basic method14 by considering the ratio of Hessian eigenvalues to remove the dependency on the structure contrast. This method suffers drawback of poor response at bifurcations and small structures. Analysis of shape was carried out by identifying elliptical structure of airways and using a graph search technique to combine the detected tubes17. To avoid leakages and minimize the possibility of falsely detected airways, fuzzy rules were proposed in18-20. Further, graph refinement approach as a probabilistic model is shared in21.
In comparison to hand crafted traditional approaches in7-21, Convolutional Neural Network (CNN) based self-learning techniques are becoming quite popular. Image classification task proposed by22-24 was outperformed by25-27 which uses weight-sharing techniques to train the filter banks. Classification based leakage detection and removal was proposed by28 to minimize the leakage problem. A 3D U-Net based architecture was proposed in29-31 for extraction of airways from volumetric image dataset. High quality segmented airways from incomplete labelled training datasets method was also recommended32. Voxel-by-voxel airway segmentation using 2.5D CNN which is trained in supervised way is featured in33 and CNN with Unet architecture is proposed by34. CNN methods require lot of data preparation and performance deteriorates for small airways especially in low-attenuated areas.
In this paper, an automated and unsupervised method for airway segmentation involving reconstruction morphology and two stages of region growing is proposed. The first stage of region growing is performed to detect trachea and part of airways which are basically large sized. The reconstruction morphological technique is employed to detect and segment distal airways in the localized areas of segmented lung. Then, the results of these two approaches are joined and further followed by second stage of region growing for refining the segmentation result by removing non-airway sections. The proposed approach is applied on 20 sets of test datasets provided by Exact’09 team35 during the airway segmentation challenge.
The rest of the paper is prepared as follows. In Section 2, airway anatomy with branch details are given. Section 3 describes the materials and methodology used for airway segmentation. The results and discussion part are shown in Section 4. Further, brief summary and conclusion of proposed work with future scope are presented in Section 5.
Airway Anatomy
The respiratory system which helps in exchange of air has three major parts: airways, lungs and respiratory muscles. Nose, pharynx, larynx and trachea comprise of conducting zone which are outside the lungs whereas bronchi and bronchioles forms respiratory zones which are inside the lungs. The length of the trachea is about 4.5 inch and diameter is 1 inch. Trachea extends into two main bronchi which are located inside the lungs. Right bronchus is longer than left one. Each bronchus partitions into secondary bronchi which again branches off to tertiary bronchi. These are further subdivided into terminal bronchioles. There are around 30,000 bronchioles which lead to alveoli via alveoli ducts. There are 23 generations from trachea to last order of bronchioles1. Conducting zone represent first 16 generations while 17-23 generations are represented by respiratory zone. The airway architecture is illustrated in Fig 1. The estimation of number of branches in every generation is indicated in Table 1.
![Figure 1: Schematic diagram of airway layout [36]](https://biomedpharmajournal.org/wp-content/uploads/2021/01/Vol13_No4_Aut_Ani_Fig1-150x150.jpg) |
Figure 1: Schematic diagram of airway layout [36] |
Table 1: Number of airway branches with respect to generation [34]
| S. No. | Generation Number | Number of Branches |
| 1 | 0 th | 1 |
| 2 | 1 st | 2 |
| 3 | 2 nd | 4 |
| 4 | 3 rd | 8 |
| 5 | 4 th | 16 |
| 6 | 5 th | 32 |
| 7 | 6 th | 64 |
| 8 | 7 th | 128 |
| 9 | 8 th | 256 |
| 10 | 9 th | 512 |
| 11 | 10 th | 1024 |
| 12 | 11 th | 2048 |
| 13 | 12 th | 4096 |
| 14 | 13 th | 8192 |
| 15 | 14 th | 16384 |
| 16 | 15 th | 32768 |
| 17 | 16 th | 65536 |
| 18 | 17 th | 131072 |
| 19 | 18 th | 262144 |
| 20 | 19 th | 524288 |
| 21 | 20 th | 1048576 |
| 22 | 21 st | 2097152 |
| 23 | 22 nd | 4194304 |
| 24 | 23 rd | 8388608 |
Materials and Methods
In an input volumetric 3D chest scan, let I denote a voxel (x, y, z) and I(x,y,z) denotes the intensity value of the voxel. The chest scan I consist of stack of isotropic 2-D transverse-plane sections denoted by Δx and Δy, which is equal to 0.5mm. The section spacing is denoted by Δz and ranges from 0.6mm to 1.25mm. The proposed scheme commences with preprocessing of images to minimize noise artifacts. In general, greyscale range in CT images range from -1024 to 700 Hounsfield units (HU). Lung tissues filled with air and airway lumen are in range from -1024 HU and -850 HU 6. The high-density tissues like airway walls and blood vessels are in range between -300 to -50 HU and harder ones like bones are above 300 HU. The dataset used in proposed work consists 20 volumetric 3D chest scan of test cases from Exact’09 challenge35.
The flow diagram of the proposed scheme is illustrated in Fig. 2. The 3D chest scan through CT generates volumetric images in the form of contiguous slices which serves as input data in the proposed scheme. The images reconstructed with high-spatial frequency gives well defined sharp edges of airway outline but mostly noise which is a common factor in imaging. At starting, data is preprocessed to minimize unwanted noise. In the proposed scheme, input data is passed through two stages individually. The first stage comprises of thresholding and 3D region growing approaches in which trachea are detected and conducting zones branches are segmented from the input data. Whereas in second stage, the airway region present inside the lung area is segmented out after segmenting the region of interest (i.e. lung). The output of these two stages are combined. For refining the segmentation result, 3D region growing method is applied once again in the combined output of the two stages.
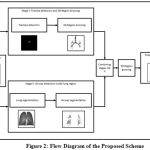 |
Figure 2: Flow Diagram of the Proposed Scheme |
(i) Trachea detection and 3D Region Growing (Stage I)
In this stage, first few images of volume CT are used as first slice may not start with trachea. Using Otsu’s thresholding method, all intensity values in image is set as 1 if it is greater than globally determined threshold value or as 0 otherwise. Then, the border is cleared to remove outside region. Now, only trachea is present with some small region having intensity value of 1. After removing these small regions, we trace trachea whose center coordinates can be found out which acts as input seed point for region growing method.
The basic 3D region growing with trachea location as seed point gives trachea, main bronchus, secondary bronchioles and some branches going inside the lungs. The 3D region growing is a very simple and efficient technique to separate out regions having constraint intensities. The main idea starts with seed point extracted from trachea location. If intensity of voxel Iv(x,y,z) < T, then voxel is added with the seed else rejected. This condition is checked for six-connected path of voxels. Threshold T is a constraint value which is decided by following two constraints6 :
- Current voxel grey level should be within 5% the classified airway lumen voxel’s mean grey level value.
- Six connected neighbors of working voxel should have grey level values within 5% of mean airway lumen grey level value.
If the range of threshold T is kept large, detection of airway lumen can slip into lung parenchyma area. T is obtained experimentally by observing the range of distribution from manual selection and studying the pattern of pixel intensity distribution inside airway lumen. Some of the output of stage I is shown in Fig 3.
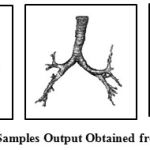 |
Figure 3: Samples Output Obtained from Stage I |
(ii) Airway segmentation inside lung region (Stage II)
In this stage, the two lungs are segmented out initially for achieving localized detection of airway pixel inside the lung area which includes distal airways also. For this, optimal thresholding is implemented on slices. The selection of threshold is adaptively done based on pixel range at the background and foreground. Optimal thresholding method leads to enhancement of the image. Then, the morphological open and close operations are applied which results in segmentation of entire two lung regions from trachea, main bronchi, outer area and background. The label matrix of different areas is obtained by connected component analysis. This helps in removing unwanted regions and acquiring the right and left lungs. The sample results of segmented lung in 3D view are shown in Fig. 4.
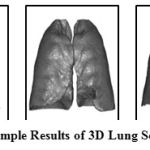 |
Figure 4: Sample Results of 3D Lung Segmentation |
The CT slices are smoothened with 4-connected low pass filter to reduce noise and followed by gray-scale reconstruction7-8. Reconstruction of image is often referred as geodesic operator of morphology operation. This concept can be applied in both binary and grayscale images.
(a) Binary reconstruction: In binary reconstruction, successive geodesic dilations are performed inside a mask X using a set Y and non-empty connected components of X are progressively filled with interaction with Y37. The reconstruction of X with Y X is obtained by Eq. (1):
where;
![]()
Marker (Y) is a subset of mask (X), B is the structuring element, Marker Y is the localized area where dilation is performed, X is the image operated on, is dilation operator and denotes intersection.
(b) Grayscale reconstruction: In grayscale image, reconstruction is useful for image filtering, segmentation as well as in feature extraction tasks. The concept of binary reconstruction can be extended to grayscale. Here, it takes the successive thresholds Tk(I) of image I into consideration for decomposition of image. With this, the peaks of the mask marked by the marker image are extracted by reconstruction. Alternately, this transformation can be introduced by the use of geodesic dilation.
Airways and vessels pictorially represent grayscale valleys and peaks in lung images. Using grayscale reconstruction technique, the local minima in lung images is detected to locate airway pixels. As diameter of airways vary from few centimeters to millimeters7, when it goes down in the lung Then, repeated closing operations are implemented with multiscale structure element Bn represented by Eq. (2) to achieve marker image.
![]()
where; B is 4-connected binary structure, which is n times dilated. Here, n ranges from 1 to 8. The local valleys are filled by morphological closing to get marker image which is expressed as Eq. (3).
![]()
where; X is original CT image, i is slice number, ⊕& Θ are dilation and erosion operators respectively. The image with highlighted airway locations is formed by computing maximum value of gray-level from pixel-to-pixel represented by Eq. (4).
![]()
To get potential airway locations, grayscale difference image is computed by using Eq. (5).
![]()
The difference image D shows the bright clusters of pixels according to their depth and locations in original image. But not all of the bright clusters are airways, some of them are non-airways also. To achieve probable airways, difference image D is thresholded with Th. The threshold value (Th) is found out by using Eq. (6); in which f is set at 0.3.
Th=f*(DPixelmaxvlaue-DPixelminvlaue ) (6)
The output of this step gives not only airways but few non-airway regions too as the thresholding can’t give this demarcation strictly. Some of the sample output of segmented airways present inside the lungs are shown in Fig 5.
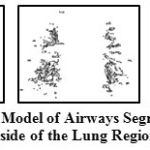 |
Figure 5: 3D Model of Airways Segmented from Inside of the Lung Region |
(iii) Final 3D Airway segmentation
The airways segmented from stage I gives preliminary results with trachea, main bronchi and secondary bronchi. The stage II is performed to get much deeper airway branches imbibed in both the lung regions. The results of these two stages are combined to get airway tree. The combined airway in this stage contains many false positive cases which were detected in stage II. Therefore, 3D region growing approach is performed once again to exclude those false segmented regions to get the refined segmentation result. The sample of final segmentation result of 3D airway volume are shown in Fig. 6.
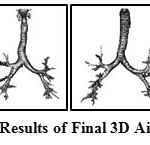 |
Figure 6: Sample Results of Final 3D Airway Segmentation |
Results and Discussion
The proposed approach has been employed for automatic segmentation of 3D airways. This approach is tested on publicly available twenty test cases of Exact’09 challenge. For evaluation, seven measures namely tree length, tree length detected, branch count, branch detected, leakage count, leakage volume and false positive rate are used to assess the airway segmentation scheme. In the branch length and count assessment, trachea is excluded. For leakage measures, both main bronchi and trachea are excluded. The result obtained in each stages of proposed scheme for Case 36 are shown in Fig. 7. This can be observed that after combining the results of two approaches of stages I & II, we get airway with many false cases as shown in Fig 7(c). Most of these false cases are removed by performing region growing once again. The final result is shown in Fig 7(d). The result of 3D airway segmentation obtained from proposed scheme is evaluated by Exact’09 team in terms of above-mentioned measures and tabulated in Table 2. Fig. 8 shows tree branches detected by proposed approach along with true positive and false positive rendering.
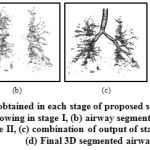 |
Figure 7: Results obtained in each stage of proposed scheme for Case 36:(a) simple region growing in stage I, (b) airway segmented from lung region in stage II, (c) combination of output of stage I & II, and (d) Final 3D segmented airway |
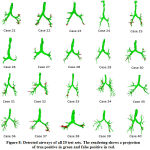 |
Figure 8: Detected airways of all 20 test sets. The rendering shows a projection of true positive in green and false positive in red. |
Table 2: Result obtained from proposed scheme for twenty cases in the test set of Exact’09 datasets
| Case No. | Branch count | Branch detected (%) | Tree length (cm) | Tree length detected (%) | Leakage count | Leakage volume (mm3) | False positive rate (%) |
| Case 21 | 113 | 56.8 | 56.0 | 50.7 | 40 | 1209.4 | 14.45 |
| Case 22 | 54 | 14.0 | 37.7 | 11.4 | 34 | 2172.6 | 22.29 |
| Case 23 | 33 | 11.6 | 25.7 | 9.9 | 16 | 319.7 | 4.85 |
| Case 24 | 48 | 25.8 | 46.4 | 28.5 | 5 | 41.5 | 0.37 |
| Case 25 | 110 | 47.0 | 83.0 | 32.9 | 48 | 4956.7 | 21.58 |
| Case 26 | 20 | 25.0 | 14.0 | 21.3 | 2 | 3.0 | 0.19 |
| Case 27 | 48 | 47.5 | 33.8 | 41.7 | 14 | 66.3 | 1.25 |
| Case 28 | 65 | 52.8 | 45.0 | 41.0 | 13 | 719.4 | 9.96 |
| Case 29 | 86 | 46.7 | 56.5 | 40.9 | 20 | 310.1 | 3.87 |
| Case 30 | 94 | 48.2 | 63.3 | 41.4 | 22 | 1102.3 | 13.86 |
| Case 31 | 112 | 52.3 | 76.0 | 43.3 | 29 | 1210.3 | 9.88 |
| Case 32 | 104 | 44.6 | 85.1 | 39.1 | 40 | 2622.7 | 16.73 |
| Case 33 | 78 | 46.4 | 56.6 | 38.5 | 27 | 1020.0 | 14.36 |
| Case 34 | 250 | 54.6 | 180.2 | 50.4 | 154 | 2310.4 | 9.12 |
| Case 35 | 140 | 40.7 | 104.2 | 33.7 | 54 | 677.3 | 5.02 |
| Case 36 | 120 | 33.0 | 111.3 | 27.0 | 50 | 1330.7 | 11.62 |
| Case 37 | 54 | 29.2 | 42.0 | 23.6 | 17 | 2376.2 | 20.99 |
| Case 38 | 36 | 36.7 | 27.8 | 41.8 | 14 | 2478.6 | 31.35 |
| Case 39 | 111 | 21.3 | 94.3 | 23.0 | 51 | 605.5 | 5.82 |
| Case 40 | 110 | 28.3 | 97.0 | 25.1 | 48 | 1879.5 | 11.00 |
| Mean | 89.3 | 38.1 | 66.8 | 33.3 | 34.9 | 1370.6 | 11.43 |
| Std. dev. | 51.0 | 13.8 | 38.6 | 11.7 | 32.4 | 1208.7 | 8.26 |
| Min | 20 | 11.6 | 14.0 | 9.9 | 2 | 3.0 | 0.19 |
| 1st quartile | 48 | 25.8 | 37.7 | 23.6 | 14 | 319.7 | 4.85 |
| Median | 90 | 42.7 | 56.6 | 36.1 | 28 | 1155.9 | 10.48 |
| 3rd quartile | 113 | 52.3 | 97.0 | 41.8 | 50 | 2376.2 | 20.99 |
| Max | 250 | 56.8 | 180.2 | 50.7 | 154 | 4956.7 | 31.35 |
While comparing the performance of proposed scheme with reference standard presented by Exact’09 team35, it is found that the tree length detected (%) and branch detected (%) of Cases 21, 27, 35, 37, and 38, is quite comparable. When considering Case 38, the result obtained from proposed scheme are same as reference standard in terms of branch detected (%) and tree length detected (%). From Fig. 8, it can be observed that Case 24, 26, and 27 have less false positive. Therefore, the false positive rate and leakage volume is found to be small. Hence, the leakage data are acceptable in such cases but somewhat found to be on higher side in other cases. This is primarily due to the lower dose (noisy) and different severity level abnormalities in Exact’09 datasets. Moreover, segmentation of very small bronchioles is indeed a difficult task as the quality of image is affected by radiation dose, slice thickness and reconstruction kernel. The diminishing size of small bronchioles and gradual reduction in intensity variation between airway lumen and airway walls in deeper section of the lungs leads to blurring effect.
The longer lengths of branches have been segmented from proposed scheme when compared with what retrieved from simple region growing method in addition to greater number of branch count. The side-by-side visual comparison of airway segmentation result for all twenty test cases with single region growing method and proposed scheme is depicted in Fig 9.
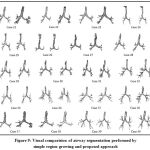 |
Figure 9: Visual comparision of airway segmentation performed by simple region growing and proposed approach |
Nevertheless, the results of the proposed scheme from unsupervised approach serves as a riveting datum for further improvements that can be done with more elaborative approaches. However, this usually requires more program coding (with its maintenance costs), physical models, parameter changes (with its overfitting difficulties) and run-time. The charm of the proposed algorithm strengthens from the fact that it’s fully automatic, simple in implementation, quick runtime and requires no training process. CNN techniques28,31-34 requires lot of data preparation and are computationally expensive. Further, no parameter tuning has been done as in8. We worked on online provided data of Exact’09 which consists of all types of data achieved from different sites as compared to9,8,15.
Airway tree segmentation is a difficult task from volumetric CT images of clinical-quality or low-dose. The anatomy-concerned reasons are airway obstructions related and movements induced by heartbeat. Airway wall’s grey level values changes substantially as moving gradually deep through airway generations. The walls of the airways are mostly in the range of 0 to −200 HU in the trachea part. But further down the tree, the gray values in the 5th or 6th generation (for example) are found to be in the range of −300 to −800 HU. This variation occurs due to the limited volume effect affected by the X-ray beam collimation and also because the airway walls get thinner in the deeper-generation airways which gets more sensitive during the CT scanner’s sampling process.
Conclusion
A simple, robust, time efficient and unsupervised approach for airway segmentation based on localized search of airway regions inside segmented lung region is proposed in the paper. A 3D region growing algorithm provides the primary branches of airways which is followed by airways segmentation inside the two lung lobes. Then, these results are combined and again 3D region growing is applied in order to reject non-airways detected in the process. The performance of the proposed work is tested on Exact’09 test cases. The proposed approach includes automatic algorithm for achieving precise seed location in trachea. One of the difficulties in a generalized segmentation algorithm is that the chest CT scans are acquired from different sites using different CT scanners, scanning protocols, and reconstruction parameters. CT scans also suffer from noise and low contrast, particularly at peripheral branches. This creates a challenge for automatic methods to balance the extraction of deeper airway branches and avoid the leakage of airway region into the surrounding lung parenchyma. The evaluations on Exact’09 platform is used for comparative analysis only as there is no gold standard for airway segmentation till date. Though the proposed approach is found to be quite promising, stable and time efficient across scanning parameter variations, the absence of small parenchymal leakages is still not guaranteed. The proposed scheme can be extended for distal and peripheral branches identification for further scope of research. This may lead to improve the existing results in future by integrating with the proposed algorithm.
Acknowledgments
The authors would like to thank the EXACT’09 team for providing the quantitative results obtained from proposed approach, which is listed in Table 2.
Conflict of Interest
None declared
Funding Source
The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- Athanazio R. Airway disease: similarities and differences between asthma, COPD and bronchiectasis. Clinics 67(11):1335-43 (2012).
CrossRef - Gu S., Wang Z., Siegfried J. M., Wilson D., Bigbee W. L., Pu J. Automated lobe-based airway labeling. International Journal of Biomedical Imaging Article ID 382806, 9 (2012).
CrossRef - Tschirren J., Hoffman E. A., McLennan G., Sonka M. Intrathoracic airway trees: segmentation and airway morphology analysis from low-dose CT scans. IEEE transactions on medical imaging 5;24(12):1529-39 (2005).
CrossRef - Lai K., Zhao P., Huang Y., Liu J., Wang C., Feng H., Li C. Automatic 3D segmentation of lung airway tree: a novel adaptive region growing approach. In 2009 3rd International Conference on Bioinformatics and Biomedical Engineering IEEE 11: 1-4 (2009).
CrossRef - Schlathoelter T., Lorenz C., Carlsen I. C., Renisch S., Deschamps T. Simultaneous segmentation and tree reconstruction of the airways for virtual bronchoscopy. In Medical imaging 2002: Image Processing, Proceedings of SPIE 4684: 103-113 (2002).
CrossRef - Fabijańska A. Two-pass region growing algorithm for segmenting airway tree from MDCT chest scans. Computerized Medical Imaging and Graphics. 33 (7): 537-46 (2009).
CrossRef - Aykac D., Hoffman E. A., McLennan G., Reinhardt J. M. Segmentation and analysis of the human airway tree from three-dimensional X-ray CT images. IEEE transactions on medical imaging 22(8): 940-50 (2003).
CrossRef - C. L., Preteux. F., Beigelman-Aubry. C., Grenier. P. Pulmonary airways: 3D reconstruction from multiscale CT and clinical investigation. IEEE transactions on medical imaging 23: 1353-1364 (2004).
CrossRef - Pulagam A. R., Ede V. K., Inampudi R. B. Segmentation of Airways in Lung Region Using Novel Statistical Thresholding and Morphology Methods. Biomedical and Pharmacology Journal 10(4): 2035-43 (2017).
CrossRef - Sonka M., Park W., Hoffman E. A. Rule-based detection of intrathoracic airway trees. IEEE transactions on medical imaging 15(3): 314-26 (1996).
CrossRef - Bartz D., Mayer D., Fischer J., Ley S., del Rio A., Thust S., Heussel C. P., Kauczor H. U., Straßer W. Hybrid segmentation and exploration of the human lungs. In IEEE Visualization 2003, 177-184 (2003).
- Mayer D., Bartz D., Fischer J., Ley S., del Rı́o A., Thust S, Kauczor H. U., Heussel C. P., Hybrid segmentation and virtual bronchoscopy based on CT images1. Academic Radiology. 11(5): 551-65 (2004).
CrossRef - Kaftan J. N., Kiraly A. P., Naidich D. P., Novak CL. A novel multipurpose tree and path matching algorithm with application to airway trees. In Medical Imaging 2006: Physiology, Function, and Structure from Medical Images 6143, p. 61430N (2006).
CrossRef - Frangi A. F., Niessen W. J., Vincken K.L., Viergever M. A. Multiscale vessel enhancement filtering. In International conference on medical image computing and computer-assisted intervention, Springer 130-137. (2006).
CrossRef - Rudyanto R. D., Barrrutia A. M., Diaz A., Ross J., Washko G. R., Solorzano C., Estepar R. S. J. Modelling airway probability. Proceedings in 2013 IEEE International Symposium on Biomedical Imaging, San Francisco, pp 378-381 (2013).
CrossRef - Jerman T., Pernuš F., Likar B., Špiclin Ž. Beyond Frangi: an improved multiscale vesselness filter. In Medical Imaging 2015: Image Processing 2015 9413, p. 94132A.
CrossRef - Graham M. W., Gibbs J. D., Cornish D.C., Higgins W. E. Robust 3-D airway tree segmentation for image-guided peripheral bronchoscopy. IEEE transactions on medical imaging; 29(4): 982-97 (2010).
CrossRef - Park W., Hoffman E. A., Sonka M. Segmentation of intrathoracic airway trees: a fuzzy logic approach. IEEE transactions on medical imaging; 17(4): 489-97 (1998).
CrossRef - Rizi F. Y., Ahmadian A., Sahba N., Tavakoli V., Alirezaie J., Fatemizadeh E., Rezaie N. A hybrid fuzzy based algorithm for 3d human airway segmentation. In IEEE 2008 2nd International Conference on Bioinformatics and Biomedical Engineering 16: 2295-2298 (2008).
- Xu Z., Bagci U., Foster B., Mansoor A., Udupa J. K., Mollura D. J. A hybrid method for airway segmentation and automated measurement of bronchial wall thickness on CT. Medical image analysis 24(1): 1-7 (2015).
CrossRef - Selvan R., Welling M., Pedersen J. H., Petersen J., de Bruijne M. Mean field network based graph refinement with application to airway tree extraction. In International Conference on Medical Image Computing and Computer-Assisted Intervention Springer, Cham 750-758 (2018)
CrossRef - Krizhevsky A., Sutskever I., Hinton G. E. Imagenet classification with deep convolutional neural networks. In Advances in neural information processing systems 1097-1105 (2012).
- Sermanet P., Eigen D., Zhang X., Mathieu M., Fergus R., LeCun Y. Overfeat: Integrated recognition, localization and detection using convolutional networks. arXiv preprint arXiv 1312.6229 (2013).
- Szegedy C., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D., Erhan D., Vanhoucke V., Rabinovich A. Going deeper with convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition 1-9 (2015).
CrossRef - LeCun Y., Bottou .L, Bengio Y., Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE 86(11): 2278-324 (1998).
CrossRef - LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 521(7553): 436-44 (2015).
CrossRef - Schmidhuber J. Deep learning in neural networks: An overview. Neural networks 1;61: 85-117 (2015).
CrossRef - Charbonnier J. P., Van Rikxoort E. M., Setio A. A., Schaefer-Prokop C. M., van Ginneken B., Ciompi F. Improving airway segmentation in computed tomography using leak detection with convolutional networks. Medical image analysis 36: 52-60 (2017).
CrossRef - Milletari F., Navab N., Ahmadi S. A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In 2016 IEEE Fourth International Conference on 3D Vision (3DV) 565-571 (2016).
CrossRef - Çiçek Ö., Abdulkadir A., Lienkamp S. S., Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In International conference on medical image computing and computer-assisted intervention Springer, Cham 424-432 (2016).
CrossRef - Meng Q., Roth H. R., Kitasaka T., Oda M., Ueno J., Mori K. Tracking and segmentation of the airways in chest CT using a fully convolutional network. In International Conference on Medical Image Computing and Computer-Assisted Intervention Springer, Cham 198-207 (2017).
CrossRef - Jin D., Xu Z., Harrison A. P., George K., Mollura D. J. 3D convolutional neural networks with graph refinement for airway segmentation using incomplete data labels. In International Workshop on Machine Learning in Medical Imaging Springer Cham 141-149 (2017)
CrossRef - Yun J., Park J., Yu D., Yi J., Lee M., Park H. J., Lee J. G., Seo J. B., Kim N. Improvement of fully automated airway segmentation on volumetric computed tomographic images using a 2.5 dimensional convolutional neural net. Medical image analysis 51:13-20 (2019).
CrossRef - Juarez A. G., Tiddens H. A., de Bruijne M. Automatic airway segmentation in chest CT using convolutional neural networks. In Image Analysis for Moving Organ, Breast, and Thoracic Images Springer, Cham 238-250 (2018).
CrossRef - Lo P., Van Ginneken B., Reinhardt J. M., Yavarna T., De Jong P. A., Irving B., Fetita C., Ortner M., Pinho R., Sijbers J., Feuerstein M. Extraction of airways from CT (EXACT’09). IEEE Transactions on Medical Imaging 31(11): 2093-107 (2012).
CrossRef - Weibel E. R., Cournand A. F., Richards D. W. Morphometry of the human lung. Berlin: Springer (1963).
CrossRef - Vincent L. Morphological grayscale reconstruction in image analysis: applications and efficient algorithms. IEEE transactions on image processing 2(2): 176-201 (1993).
CrossRef







