Manuscript accepted on :12-June-2019
Published online on: 26-06-2019
Plagiarism Check: Yes
Reviewed by: Mr. Guy-Armel Bounda
Second Review by: Sumit Kushwaha
S. M. Vijaya1 and K. Suresh2
1RRCE, Bengaluru, 560074 - India.
2College of Engineering and Technology, Bengaluru, 560049 - India.
Corresponding Author E-mail: vijaya@rrce.org
DOI : https://dx.doi.org/10.13005/bpj/1730
Abstract
Robotic guided medical system requires efficient mechanism of compression of Region of Diagnostics Interest (RODI) in medical images to overcome the tradeoff among efficiency and time which is a computationally challenging task. This task involves the requirement of suitable noise filtering, segmentation, critical feature selection especially at corners of RODI and encoding process. This paper proposes a framework namely ICRODI to evaluate a hybrid approach of compression for region of diagnostic interest in Brain MRI as well as for rest of the region. The approaches used are median filter, thresholding as pre-processing and fuzzy c-mean clustering, Harris corner detection, s-shape fuzzy for segmentation and feature point selection optimization. Further alpha hull of the convex hull is used for getting the volume of the mass and finally the wavelet co-efficient based compression is applied. The effectiveness of the proposed ICRODI is validated by evaluating MSE and PSNR for both RODI and Non-ROSI. The average value of the PSRN for RODI is found approximately 49 % higher as compared to the non-RODI and MSE of the RODI is reduced by approximately 33% as compared to the non-RODI after simulating the process on a numerical simulation platform. The achieved results are quite promising and could be optimized for the VLSI implementation in future.
Keywords
Brain MRI; Image Coding; Medical Image Processing; Medical Robotics; Region of Interest Compression
Download this article as:| Copy the following to cite this article: Vijaya S. M. Suresh K. ICRODI: Image Compression of Region of Diagnostics Interest (RODI) using Layer Segmentation and Wavelet. Biomed Pharmacol J 2019;12(2). |
| Copy the following to cite this URL: Vijaya S. M. Suresh K. ICRODI: Image Compression of Region of Diagnostics Interest (RODI) using Layer Segmentation and Wavelet. Biomed Pharmacol J 2019;12(2). Available from: https://bit.ly/2J5NLOT |
Introduction
In the world of ubiquitous computing many advanced medical systems have been visualized on the basis technologies such as robotics, artificial intelligence, advanced communication and Network system etc., along with digital image processing.1,2 There exist limitations of bandwidth to transfer a high dimension data of real-time medical images. Therefore, an efficient technique to be developed to transfer only region of diagnostic interest (RODI) through the communication channel without losing its essential features. The first requirement towards achieving this goal is to adopt a suitable noise reduction method for the Brain-MRI which improvises the signal to noise (SNR) by keeping both spatial and contrast resolution optimal. The suitable noise reduction method ultimately improvises the significance of critical diagnostics.3 The work of Majumdar et al8 have developed different models of noises for realization of the filtering as well as extraction of the some textural along with geometric feature of the image. The noise reduction further could be enhanced for the better vision. The automatic segmentation is an open research problem for the brain MRI as it a quite challenging task due to heterogeneity into the geometric contour of tumor tissues as well as the overlapping similarity among even normal tissues and tumor.4,5 In past many semi-automatic and automatic method for the segmentations are proposed where mostly a technique of global thresholding is used which exhibit potential error.6,7 To overcome these error the methods of active contour (AC) is used.9,10,11 In order to extract the critical mass RODI, these methods exploit the primary contours, whereas the accurate positioning of these contour is of high importance due to non-alignment tendency of the energy release. To overcome this the contour is localized to the approximation of the RODI and avoid the wrong edge which might be arises due to various artifacts as well as the noises. The proposed ICRODI, in this paper is motivated by the work by Jiyo et al (2016),12 for the automatic segmentation mainly for bone critically from the CT-images used in the diagnostic of spine disorders. Like,12 ICRODI extract features from the Brain-MRI to build the initial level of contour which is further converted into fuzzy corner metric (FCM) on the basis of the intensity of the Brain MRI input image data. The ICRODI uses s-Shape convex hull as Alpha hull for the corners detection optimization and finally wavelet based image coding is done to get optimal PSNR and MSE for the RODI as compared to the non-RODI region as these regions are not very critical from the view point of diagnosis.
The proposed framework for the ICRODI takes the advantages of median filtering for the noise reduction and uses thresholding technique for the contrast enhancement. The segmentation and feature selection is optimized by exploiting the optimality of the fuzzy c-mean clustering for layered segmentation which is further optimized for the corner point feature selection using Harris corner detection followed by the s-shaped fuzzy corner optimization to get the critical mass of the RODI by getting alpha hull of convex hull and finally the compression process of image encoding is adopted using wavelet. The section 2.0 provides details of the related work in the same filed, section 3.0 illustrates the process model of ICRODI, section 4.0 describes the algorithms used in ICRODI, section 5.0 provides the results and analysis of the ICRODI for a given dataset to compute performance metrics of the model such as PSNR and MSE, followed by section 6.0 a conclusion, which also discuss the future scope of the work.
Related Work
The effective mechanism of compression of the region of interest with critical diagnosis of the particular region in different medical images are a critical topic of research to have advantages for the optimal uses of the disk storage, transmission of the image data through a constraints bandwidth channels as well as for many advance medical systems [Cite Madam Previous work if nay else cite a paper which talks importance of the work].Few recent work on the compression of region of interest is takes here to under the current line of research in the related domain. The approach of ROI localization and then compression approach is studied by Liaghati et al (2016) for hyperspectral image to meet the challenges pose by the communication channels constraints. They have proposed lossless compression mechanism for the ROI by adopting bi- level block maping to get most probable blocks using optimized Huffman coding technique. Their method promises significant improvement over JBIG2 and JPEG 2000 when applied on ROI maps.13 Apart from another image the study has been carried out on the medical image. The work of Dong et al(2016) claims to have a GUI based tool where the ROI can be extracted from the slide images and provision lossless compression to the extracted ROI as well claims 40 times higher compression ratio. The model was validated by block matching approach for detecting malaria cells from the whole slide images and exhibits better result as compared to Bzip2, Shape- Adaptive Jpeg 2000 (SAJPEG2) which provides new vision for the medical images to intact the complex geometry.14 Yee et al (2017) have focused on the compression requirement of growing amount of medical images from the storage view point manageability. They have proposed a new format called “Better Portable Graphics (BGP)”. They divide the input image into ROI and Non-ROI then apply lossless BPG to ROI then by combining they create a final image and found improvement approx. 10 to 25% in the compression as compared to the traditional method for medical image compression.15 The work on especially Brain-MRI tumor detection then Tumor region compression along lack in the literatures, whereas few significant primary work is found into work of Vilas et al (2016)16 and many other details is found towards tumor analysis in brain MRI in the work of Bauer et al (2013).17 The section 3.0 explain the process flow of the prosed model of ICRODI.
Process Model of ICRODI
The methodology adopted for the proposed image compression process for the region of diagnostic interest is shown in the fig x in as block by block operations performed.
 |
Figure 1: Block wise operation of ICRODI.
|
The component wise processing blocks of the ICRODI is shown in the fig.2 below, which include the operations of non-linear digital filtering of the input Brain-MRI images using median filter for the noise reduction followed by the contrast enhancement using thresholding.
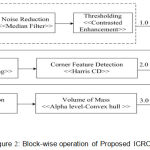 |
Figure 2: Block-wise operation of Proposed ICRODI.
|
For the dimension reduction an approach of layer segmentation is adopted with customized function development. The most critical aspect of handling fuzzy ness and principal components of the features on the corner a Harris corner point detection mechanism is used followed by the optimization using Fuzzy Corner Point is used to consider the important points which are not taken into previous steps of feature extraction/selection and finally the Alpha Hull Based Active Contour System optimization is performed. Finally, compression and reconstruction are done using wavelet. Section 5.0 illustrates the values of PSNR and MSE for the test Brain MRI images.
Algorithms used in ICRODI
This section describes overall implementation algorithm of the proposed ICRODI system from Brain-MRI input stage till image compression and reconstruction. The 2-D median filtering uses the eq. (1), and the value of PSNR is computed using eq. (2).
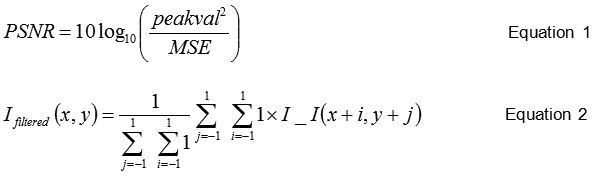
The S -Shaped membership function while optimization of the feature points is a mapping on vector I. The parameter x and y locate the extreme of the sloped portion of the curve. The eq. (3) is used for the same.
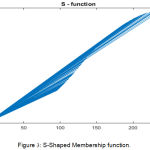 |
Figure 3: S-Shaped Membership function.
|
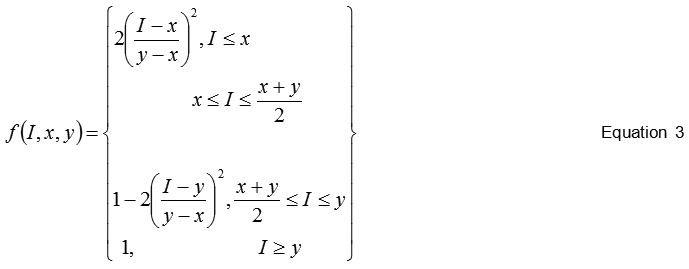
The optimization of the active contour method is motivated by the work of,12 which is to minimize the following energy in eq. (4).
![]()
The typical corner detection includes the cornerness evaluation using Hariss corner, where the fuzzy approach is used to optimize the corner matrix and further convex hull is created. This convex hull is optimized for handing sharp converters in the tumor region in Brain MRI.
Algorithm for RODI compression
Input: Brain-MRI Image (I)
Output: Compressed (I), PSNR, MSE
Idenoised ← fmedian(I)
Perform global image thresholding using global Otsu method
Ienhanced ← fmedian(Idenoised)
Layer segmentation
U← ffuzzy-c-mean clustering(Ienhanced), where U is fuzzy partition matrix
Index ← Max(probability) ← Probability ← U-1
[Pixel Label, Index] ← SegmentHarris corner point detection
C ← fcornermatrix(Segment)
If C > 0
(R,G,B) ← Distribute input to output(Segment)
Pick Corner ← fregional maxima(C)
Fuzzy Corner points
fS-shaped membership function()
fc ← multiply image by constant
if fc>0
(R,G,B) ← Distribute input to output(Pick corner)
Apply Alpha Hull as convex hull
[Vertex, Boundary] ← Compute Alpha shape of point set (X, R ), X= coordinate, R= Probe radiusTumor ← segment image into foreground and background using active contour
Compression
Apply true compression of images using wavelets
Reconstruct
Compute PSNR, MSE
Results and Analysis
In order to evaluate the effective of the performance of the ICRODI two performance metrics are taken into consideration 1) Peak Signal to noise ratio (PSNR) and 2) Mean square error (MSE). The average of the PSNR and MSE of both RODI and non-RODI is tabulated in the table 1 and the fig. 2 illustrates the intermediate results of each process.
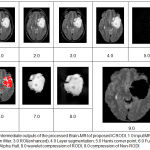 |
Figure 4: Intermediate outputs of the processed Brain-MRI of proposed ICRODI, 1.0 input MRI- image, 2.0 Median filter, 3.0 ROI(enhanced), 4.0 Layer segmentation, 5.0 Harris corner point, 6.0 Fuzzy corner point, 7.0 Alpha Hull, 8.0 wavelet compression of RODI, 9.0 compression of Non-RODI.
|
Table 1: Observation of PSNR and MSE for RODI and Non-RODI region after compression and reconstruction.
| Image Number | RODI | Non-RODI | ||
| PSNR | MSE | PSNR | MSE | |
| 1 | 37.822789 | 3.461563 | 13.499252 | 29.678818 |
| 2 | 43.988636 | 0.819885 | 16.675101 | 23.314758 |
| 3 | 28.891434 | 8.064819 | 14.008082 | 23.167999 |
| 4 | 44.537326 | 0.725189 | 16.542916 | 24.000870 |
The fig 3 and fig 4 shows the comparative graph of both RODI and non-RODI portion of Brain MRI for PSNR and MSE respectively.
It is found that the average of PSNR values of RODI from four different images is 155.24 and of Non-RODI is 25.04, that means the improvement into image quality after compression for RODI is approximately 620% which is quite acceptable as lossless image quality.
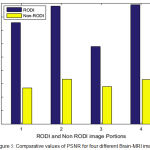 |
Figure 5: Comparative values of PSNR for four different Brain-MRI images.
|
The MSE values of the individual images are 3.4 in place of 29,6, 0.81 in place of 23.31, 8.06 in place of 23.16 and 0,72 in place of 24.0 for image 1, 2, 3 and 4 respectively for RODI and non RODI which is quite consistatnt and the standard deviation is quite low.
The method proposed here can be optimized further to evalaute the bit per pixel to meet the channel contrainst so that without loosing more significant bit teh data can travelk with the limited bandwidth and can truley satisfy the need to RODI image lossless compression. In order to see its realization in real time syncronized application an approach of field programmable gate array(FPGA) approach is planned in the future work as an extension, where the optimization of power and time will be the prime objective function along with the BPP, PSNR and MSE.
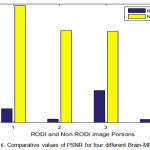 |
Figure 6: Comparative values of PSNR for four different Brain-MRI images.
|
Conclusion
This paper mainly focuses on the effective compression mechanism on the Brain-MRI images especially on the region of interest which are important from the diagnostic view point by exploiting the optimal method noise filtration, segmentation and feature detection. The method is evaluated for the many Brain-MRI images which having tumor into it by computing the value of PSNR and MSE and found that it exhibits better result to be promising as lossless characteristic. In future a very large scale integrated circuit (VLSI) implementation of the method is planned with FPGA realization with an objective of optimal fast computation to make it synchronized with the real-time advance medical operations.
Acknowledgements
The author is deeply grateful to Hon. A. C. Shanmugam Chairman MCET and Hon. S. Vijayanand Executive Director, and Dr. Balakrishna Principal, Rajarajeswari College of Engineering Bangalore for their kind encouragement and help during the progress of this work.
Conflict of Interest
I hereby disclose of no conflict of interest including honorarium, grants, membership, and employment, ownership of stock or any other interest or non‐financial interest such as personal or professional relation, affiliation and knowledge of the research topic.
References
- C. Hennersperger, B. Fuerst, S. Virga, O. Zettining, B. Frisch, T. Neff, N. Navab. Towards MRI-Based Autonomous Robotic US Acquisitions: A First Feasibility Study. IEEE Transactions on Medical Imaging, 2017; vol. 36, no. 2, pp. 538-548.
- N. Nouri, D. Abraham, J. M. Moureaux, M. Dufaut, J. Hubert and M. Perez. Subjective MPEG2 compressed video quality assessment: Application to Tele-surgery. IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Rotterdam, 2010; pp. 764-767.
- M. A. Ertürk, P. A. Bottomley and A. M. M. El-Sharkawy. Denoising MRI Using Spectral Subtraction. IEEE Transactions on Biomedical Engineering, 2013; vol. 60, no. 6, pp. 1556-1562.
- S. R. Telrandhe, A. Pimpalkar and A. Kendhe. Detection of brain tumor from MRI images by using segmentation & SVM. World Conference on Futuristic Trends in Research and Innovation for Social Welfare (Startup Conclave) Coimbatore, 2016; pp. 1-6.
- N. Behzadfar and H. Soltanian-Zadeh.Automatic segmentation of brain tumors in magnetic resonance images. Proceedings of IEEE-EMBS International Conference on Biomedical and Health Informatics, Hong Kong, 2012; pp. 329-332.
- M. Soltaninejad, G. Yang, T. Lambrou. Automated brain tumour detection and segmentation using super pixel-based extremely randomized trees in FLAIR MRI. International Journal of Computer Assisted Radiology and Surgery, 2017; Springer, Volume 12, Issue 2, pp 183–203.
- M.A. Balafar, A.R.Ramli, M.I.Saripan and S. Mashohor. Review of brain MRI image segmentation methods. Artificial Intelligence Review, 2010; vol. 33(3), pp.261-274.
- V.K. Raj, A. Majumder. Bayesian Regularization-Based Classification for Proposed Textural and Geometrical Features in Brain MRI. Artificial Intelligence Trends in Intelligent Systems 2017; pp 343-353.
- B. F. Zohra, B. Nacéra and T. A. Abdel Malik. Adjustment of active contour parameters in Brain MRI segmentation using evolution strategies.4th International Conference on Electrical Engineering (ICEE), Boumerdes, 2015; pp. 1-7.
- l. Liu. The approach of T1 weighted brain MRI image segmentation. Proceedings of the 33rd Chinese Control Conference, Nanjing, 2014; pp. 4860-4865.
- Freifeld, H. Greenspan and J. Goldberger. Lesion Detection in Noisy Brain Images using Constrained GMM and Active Contours. 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Arlington, 2007; pp. 596-599.
- S. Athertya and G.S. Kumar. Automatic segmentation of vertebral contours from CT images using fuzzy corners. Computers in biology and medicine, 2016; vol. 72, pp.75-89.
- L. Liaghati, H. Shen and W. David Pan. An efficient method for lossless compression of bi-level ROI maps of hyperspectral images. IEEE Aerospace Conference, Big Sky, MT2016; pp. 1-6.
- Dong, H. Shen and W. David Pan. An interactive tool for ROI extraction and compression on whole slide images. IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI), Las Vegas, 2016; pp. 224-227.
- Yee, S. Soltaninejad, D. Hazarika, G. Mbuyi, R. Barnwal and A. Basu. Medical image compression based on region of interest using better portable graphics (BPG). IEEE International Conference on Systems, Man, and Cybernetics (SMC), Banff, AB, 2017; pp. 216-221.
- R. Vilas, S. N. Kulkarni, H. Chiranth and M. Bhille. Segmentation and compression of 2D brain MRI images for efficient tele radiological applications. International Conference on Electrical, Electronics, and Optimization Techniques (ICEEOT), 2016; pp. 1426-1431.
- S. Bauer, R. Wiest, L.P. Nolte and M. Reyes. A survey of MRI-based medical image analysis for brain tumor studies. Physics in medicine and biology, 2013; vol. 58(13), pp.97.







