Manuscript accepted on :15-April-2019
Published online on: 09-05-2019
Plagiarism Check: Yes
Reviewed by: Hendry Irawan
Second Review by: Sadegh Rajabi
Final Approval by: Dr. Ayush Dogra
Ramya Premanath*1, Sarika Suresh1, Prathiksha P. Alva1 and Akash S. K2
1Infectious Diseases, Nitte University Centre for Science Education and Research, Paneer Campus, Deralakatte, Mangaluru-575018, India.
2Kasturba Medical College, Hampankatta, Mangaluru-575001, India.
Corresponding Author E-mail: ramya@nitte.edu.in
DOI : https://dx.doi.org/10.13005/bpj/1687
Abstract
Diabetes mellitus, a chronic metabolic disease is increasing worldwide. Diabetic foot infections are one of the most feared and bothersome complications of diabetes caused by different genera of bacteria. There is an increasing evidence which demonstrates the presence of biofilm former's in chronic diabetic foot ulcers which contribute to the development of antibiotic-resistant strains and treatment failure. The present study aimed at isolating bacteria from diabetic wounds, to check for its antibiotic susceptibility and biofilm forming ability. From the diabetic wounds, isolates belonging to the genera of Staphylococcus, Pseudomonas, Klebsiella, Esherichia, Vibrio, Acinetobacter and Citrobacter were recovered. To the best of our knowledge, Vibrio parahaemolyticus was isolated for the first time from diabetic ulcer. Antibiotic sensitivity profile of the organisms infers the presence of multidrug-resistant strains. Majority of bacteria isolated were found to be biofilm formers. High biofilm former's were observed in strains of P. aeruginosa, S. aureus and Klebsiella spp. There was significant association between incubation time and intensity of biofilm formation in P. aeruginosa [ᵡ2 (p< 0.05) = 0.001)], Staphylococcus spp. [ᵡ2 (p< 0.05) = 0.023)] and Acinetobacter spp. [ᵡ2 (p< 0.05) = 0.018)]. The presence of biofilm forming multidrug-resistant bacteria infers the chronic nature of diabetic wounds.
Keywords
Biofilm; Diabetic Wound; Multidrug-Resistant Bacteria; Poly Microbial Infection
Download this article as:| Copy the following to cite this article: Premanath R, Suresh S, Alva P. P, Akash S. K. Biofilm Forming Abilities of Microorganisms Associated with Diabetic Wound Infection: A Study from A Tertiary Care Hospital. Biomed Pharmacol J 2019;12(2). |
| Copy the following to cite this URL: Premanath R, Suresh S, Alva P. P, Akash S. K. Biofilm Forming Abilities of Microorganisms Associated with Diabetic Wound Infection: A Study from A Tertiary Care Hospital. Biomed Pharmacol J 2019;12(2). Available from: https://bit.ly/2Haf3DZ |
Introduction
Diabetes mellitus (DM), a metabolic disorder is increasing at an alarming rate all over the world. India has nearly 33 million diabetic subjects today and tops the list of countries with the highest number of diabetics and considered the “diabetic capital of the world” (1). The number of diabetics in India is expected to rise to a whopping 79.4 million by 2030 (2). Diabetics are more susceptible to infections due to increased glucose levels and suppressed immune response as well as decreased blood flow to extremities that lead to slow healing wounds (3). Diabetic foot infections caused by different genera of bacteria are the most feared complications of diabetes associated with high morbidity which can end up with gangrene and amputation. High incidence of diabetic foot infection in India can be attributed to practices such as bare foot walking, inadequate facilities for diabetic care, illiteracy and low socioeconomic status (4). As bacteria that cause diabetic foot infection have become resistant to number of available antibiotics, the most successful strategies to manage infection is the frequent debridement of foot ulcer (5). Many of the organisms that cause infection have acquired resistance to the available antibiotics making treatment regimen complicated.
The polymicrobial community that cause infection can produce extracellular polymeric substances called biofilms. Biofilm formed performs a dual function in acting as a physical barrier for biological and antimicrobial substances and also facilitate adhesion to surfaces (6). In recent years, biofilms have gained as an important means of survival of microorganisms in hostile environment. Bacteria in biofilm exchange genetic material, communicate with each other, which often result in altered phenotype of bacteria which influences the wound healing process (7). Chronic diabetic foot infection due to biofilm formers contribute to the development of antibiotic resistant strains and treatment failure. Though there are many studies worldwide on this topic, hardly few studies have been conducted in Mangaluru region focusing on the biofilm forming abilities of the organisms isolated from foot wounds. Against this background, our study focused on isolating bacteria from diabetic wound infection, checking for its antibiotic susceptibility and also biofilm forming abilities of these pathogens.
Materials and methods
Sample collection
Clinical samples (wound swab) from patients with a history of diabetes was collected from Justice K.S Hegde hospital, Deralakatte, Mangaluru, by taking clearance from the institutional ethics committee (INST.EC/2017-18/003) before commencement of this work. Patient consent was taken before sample collection and was anonymized. Sample collection was carried for a period of 3 months between January and March, 2018. Collected swabs were enriched in brain heart infusion broth for the period of 8 hours.
Selective Isolation
Culture from the enrichment media was streaked onto nutrient agar and colonies that developed were inoculated onto different selective media viz. cetrimide agar, mannitol salt agar, MacConkey agar, Leeds Acinetobacter agar and thiosulphate citrate bile salt sucrose agar.
Identification of bacteria
Phenotypic identification was done by performing Gram staining and an array of biochemical tests. Genotypic identification was carried out by polymerase chain reaction using genus and species specific primers. The list of primers used in the study is given in Table 1.
Table 1: Name and sequence of primers used for identification of different bacteria.
| Bacteria | Primer | Oligonucleotide sequence (5’–3’) | Annealing temperature | Amplicon size (bp) |
| Staphylococcus8 | ST1
ST2 |
GGC CGT GTT GAA CGT GGT CAA ATC A
TTA CCA TTT CAG TAC CTT CTG GTA A |
55 | 370 |
| Staphylococcus aureus 9 | SA1
SA2 |
AAT CTT TGT CGG TAC ACG ATA TTC TTC ACG
CGT AAT GAG ATT TCA GTA GAT AAT ACA ACA |
57 | 104 |
| Staphylococcus epidermidis 9 | SE1
SE2 |
ATC AAA AAG TTG GCG AAC CTT TTC A
CAA AAG AGC GTG GAG AAA AGT ATC A |
56 | 128 |
| Vibrio 10 | rpoA1
rpoA2 |
CGT AGC TAG AGG CAA AGA TGA
AAG CTG GAA CAT AAC CAC GAA |
55 | 197 |
| Acinetobacter 11 | RpoB1
RpoB2 |
CCT TCA TGA CCT GGA ACG GAT A
TCC AGG ATC TGA CCG ACG TTC AT |
59 | 940 |
| Citrobacter freundii 12 | Cfa 1
Cfa 2 |
TTG GCG TCC AGC GCA TTC A
AAT TCC AGC CTT CGG CAA ACG |
57 | 100 |
| Escherichia coli 13 | UIDA1
UIDA2 |
AAA ACG GCA AGA AA AGC AG
ACG CGT GGT TAC AGT CTT GCG |
63 |
147
|
| Klebsiella spp. 14 | KpNM1
KpNM2 |
ATT TGA AGA GGT TGC AAA CGA T
TTC ACT CTG AAG TTT TCT TGT GTT C |
55 | 130 |
| Vibrio parahaemolyticus 15 | Tlh1
Tlh2 |
AAAGCGATTATGCAGAAGCACTG
GCTACTTTCTAGCATTTTCTCTGC |
55 | 450 |
| Pseudomonas aeruginosa 16 | OprL1
OprL2 |
ATG GAA ATG CTG AAA TTC GGC
CTT CTT CAG CTC GAC GCG ACG |
55 | 504 |
Antibiotic susceptibility testing
All the confirmed isolates were subjected to antibiotic susceptibility test by employing Kirby Bauer disc diffusion method (17). Antibiotics norfloxacin, imipenem, tetracycline, gentamicin, amoxyclav, ampicillin, amoxicillin, ciprofloxacin, cefoxitin, cefotaxime were used for Gram negative bacteria, vancomycin, penicillin G, amoxyclav, azithromycin, tetracycline, trimethoprim, and oxacillin were used for Gram positive bacteria and cefoperazone, piperacillin, levofloxacin, gentamicin, amikacin, imipenem, aztreonam, cefoperazone/sulbactam, piperacillin/tazobactam, ceftazidime, netillin, ciprofloxacin, tobramycin were used for P. aeruginosa. The zones of inhibition (mm) that developed after an incubation period of 24 h were measured.
Qualitative and quantitative assay for biofilm
Congo red Method
Qualitative detection of biofilm formation was carried out by Congo red method (18) Biofilm formers formed black colonies with a dry crystalline consistency.
Microtitre Plate Method
Biofilm quantification was carried out according to the method of O’Toole and Kolter with slight modification. In a microtitre plate, 100 µl of the diluted culture was taken and incubated for 24 h at 37ºC. Using PBS of pH 7.4, the adherent cells were washed thrice. 125 µl of 0.1% freshly prepared crystal violet was added to the dried pellet, and incubated for 10 min. 200 µl of 30% acetic acid was added to the stained and washed pellet, and incubated for 15 min for stain solubilisation. To a fresh plate, 100 µl from the well was transferred and optical density was measured at 600 nm in an ELISA reader (Biorad, USA). Reduction in the biofilm formation was measured in terms of per cent inhibition as [(OD of control – OD of treated)/OD of control] x100. The biofilm formed by the confirmed isolates was compared with positive culture of different organisms. The biofilm formers were grouped as weak biofilm formers (OD600 0.071 – 0.142), moderate biofilm formers (OD600 0.142 – 0.284) and high biofilm formers (OD600 ≥ 0.284) (42). Biofilm formation was quantified at different time intervals (24, 48 and 72 h).
Results
Isolation and Identification of Bacteria
Out of 133 colonies found growing on the selective media, 36 developed on Mac Conkey agar, 26 on thiosulphate citrate bile salt sucrose agar, 27 on cetrimide agar, 36 on mannitol salt agar and 8 on Acinetobacter agar. The development of bacteria was predominantly more on mannitol salt agar and cetrimide agar indicating the presence of large number of Staphylococcus spp. and P. aeruginosa respectively. The isolates were identified after performing an array of biochemical tests in addition to molecular confirmation. Staphylococcus spp. and P. aeruginosa were found to be the predominating organisms isolated from diabetic wounds. The number of organisms isolated is given in (Graph.1). The most important observation from the study is the isolation of V. parahaemolyticus from diabetic wounds. To the best of our knowledge, this is the first report in India to encounter V. parahaemolyticus in diabetic wounds.
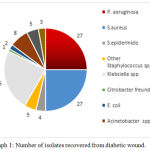 |
Graph 1: Number of isolates recovered from diabetic wound.
|
Antibiotic Susceptibility Test
Antibiotic susceptibility test was performed for the confirmed 107 isolates. Susceptibility of bacterial isolates to the different antibiotics used is shown in (Graph. 2). Among the Gram positive isolates, resistance for oxacillin was significantly high. Bacteria were found to be sensitive for tetracycline followed by vancomycin (Graph.2A). Among the Gram negative isolates other than P. aeruginosa, resistance was significantly more to ampicillin and amoxicillin when compared to other antibiotics used (Graph.2B). Around 60% of the isolates were sensitive to imipenem. In general, more number of isolates was found to be resistant to the antibiotics used. All isolates of E. coli were completely resistant to the antibiotics used other than tetracycline. Klebsiella spp. (94% isolates) showed highest resistance to amoxicillin. Acientobacter spp. were highly sensitive to imipenem and resistant towards ampicillin and amoxyclav. Complete resistance was found to cefocitin/cloxacillin among the Vibrio spp. P. aeruginosa isolates were generally sensitive to all the antibiotics used (Graph.2C). Nearly 40% of the isolates showed resistance to cefperazone/sulbectum combination. Around 85% of isolates showed least resistance to ciprofloxacin.
 |
Graph 2: Antibiotic susceptibility of bacteria isolated from diabetic wounds.
|
2A: Susceptibility of Gram positive bacterial isolates; 2B: Susceptibility of Gram negative bacterial isolates other than P.aeruginosa; 2C: Susceptibility of P. aeruginosa.
Biofilm Assay
Out of 107 isolates checked for their biofilm forming abilities, 80 isolates formed black colonies with a dry crystalline consistency indicating a positive result. Staphylococcus spp. (88%) and P. aeruginosa (88%) were the predominant biofilm formers. In Klebsiella spp. 53% of isolates, in Vibrio spp. 80% of isolates, in Acinetobacter spp. 37% of isolates formed biofilm. Both the E. coli isolates were biofilm formers. In the microtitre plate assay, biofilm formation varied at different time intervals. High biofilm formers were found in Staphylococcus spp., Klebsiella spp. and P. aeruginosa isolates. Majority of the Staphylococcus spp. and E. coli isolates were moderate biofilm formers (92%). The numbers of high biofilm formers (9%) were more in P. aeruginosa when compared to other bacteria. High biofilm formers were not found in E. coli, Vibrio spp. and Acinetobacter spp. Majority of the isolates formed weak and moderate biofilms at 24 h. P. aeruginosa isolates showed moderate biofilm formation at 24 h. But, at 48 h around 27% of them showed high biofilm formation. At 72 h, around 7% of Klebsiella spp. showed high biofilm formation. In P. aeruginosa the number of isolates forming weak biofilm increased at 72 h. In E. coli, Acientobacter spp. and Vibrio spp. the number of isolates forming moderate biofilm increased at 48 and 72 h. There was significant association between incubation time and intensity of biofilm formation in P. aeruginosa [ᵡ2 (p< 0.05) = 0.001)], Staphylococcus spp. [ᵡ2 (p< 0.05) = 0.023)] and Acinetobacter spp. [ᵡ2 (p< 0.05) = 0.018)]. There was no significant association in Klebsiella spp., E. coli and Vibrio spp. The percentage of biofilm formed at different time intervals is shown in (Graph 3).
 |
Graph 3: Biofilm formation in the bacterial isolates at different time intervals (24,48 & 72 h).
|
Discussion
Diabetic foot infections are a major problem worldwide. In India, more than 62 million people have been diagnosed with diabetes. Foot ulcer is the major problem in diabetes which if left untreated, results in limb amputation (19). In the present study, isolation and identification of bacteria causing foot ulcers along their antibiotic susceptibility profile and biofilm forming ability were attempted. As reported from the present study, the percent prevalence of Gram negative bacteria was more than Gram positive bacteria. The result is not in agreement with studies which have exhibited higher proportions of Gram positive bacteria isolated form diabetic wounds (20 , 21). A study from Malaysia has reported Proteus spp. to be the predominating organism in diabetic wound (22). However, Proteus spp. was hardly encountered in this study. S. aureus and P. aeruginosa were the predominant organisms isolated and identified in this study. Contradicting results have been observed in a study which has shown the prevalence of E. coli in diabetic wounds (21). The present study highlights presence of multidrug-resistant bacteria in diabetic wounds as depicted by its resistance to more than one drug used. Gram positive isolates showed resistance to vancomycin in our study. Contradictory results have been seen in a study which has shown 100% sensitivity of S. aureus to vancomycin (22). Gram negative bacteria in the current study have shown significant resistance to amoxicillin+clavulanic acid. The results are in agreement with a study which has shown similar result (21). Resistance to imepenam was around 30%. The results does not correspond with a study has shown 100% sensitivity of Gram negative bacteria towards imepenam (23). Infections with bacteria forming biofilms are difficult to eradicate. These biofilms are not only less susceptible to host cell immune responses but also have a high tolerance to antibiotics than the planktonic cells (24). The resistance of biofilm forming bacteria towards antibiotics is due to obstruction in the permeability of the drug by the polysaccharide matrix (25) and alteration of the drug efficacy in the biofilm environment (3). Not only biofilm effect antimicrobial agents, but also they give protection against host defenses. Biofilms have anti-phagocytic activity and also inactivates complement and antibodies (26). In the present study, 75 per cent of drug resistant bacteria were biofilm formers. The percent of biofilm formers in our study is significantly larger in comparison to a previous study (21) and corresponds to studies by Swarna et al. and James et al. (27, 28). The higher percentage of biofilm formers in diabetic wounds could be due to ineffective debridement procedure or longer duration of ulcer in patients (29). P. aeruginosa was a predominant biofilm former with 89 per cent of the isolates being positive for biofilm formation. This was an expected result as studies have reported biofilm formation by P. aeruginosa more readily in diabetic wound environment (30).
Conclusion
It is clear from the present study that, majority of bacteria isolated from diabetic wounds are multi-drug resistant and moderate-high biofilm formers which resist antibiotic therapy. In order to decrease the undesirable consequences associated with diabetic wounds, it is essential to recognize the biofilm forming abilities of the organism in addition to their antibiotic susceptibility profile. Decline in the morbidity due to diabetic foot ulcers caused by multidrug resistant biofilm producing bacteria is possible by adopting alternative therapies which prevent bacterial attachment, disrupt biofilm and act as quorum sensing inhibitors. Developing new tools to reduce the suffering of diabetic patients with foot ulcers should be taken as a challenging research.
Acknowledgements
We thank Nitte (Deemed to be University) for all the facilities provided.
Ethical Approval
The study was approved by the Institutional Ethics Committee of the Nitte University Centre of Science Education & Research (INST. EC/2017-18/003), Mangaluru, India.
References
- Murugan S, Mani K. R and Devi U. Prevalence of Methicillin Resistant Staphylococcus aureus among diabetic patients with foot ulcer and their antibiotic susceptibility patterns. Clin. Diagn. Res., 2008; 2: 979-984.
- Ramachandran A. Epidemiology of diabetes in India-three decades of research. J. A Physicians India., 2005; 53: 34-38
- Trivedi U, Paremeswaran S, Armstrong A, Burgueno-Vega D, Griswold J, Dissanaike S et al. Prevalence of multiple antibiotic resistant infections in diabetic versus non diabetic wounds. Pathog., 2014; ID 173053
- Viswanathan V, Thomas N, Tandon N, Asirvatham A, Rajasekar S, Ramachandran A et al. Profile of diabetic foot complications and its associated complications – A multi centric study from India. J. APhysicians India., 2005; 53: 933-936.
- Dowd SE, Wolcott R.D, Sun Y, Mc Keehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PloS One. 2008; 3: e3326.
- Schierle C.F, De la Garza M, Mustoe T.A, Galiano R. Staphylococcal biofilms impair wound healing by delaying re epithelialization in a murine cutaneous wound model. Wound Repair Regen., 2009; 17: 354-359.
- Murali T.S, Kavitha S, Spoorthi J, Bhat D.V, Prasad A.S, Upton Z et al. Characteristics of microbial drug resistance and its correlates in chronic diabetic foot ulcer infections. Med. Microbiol., 2014; 63: 1377-1385.
- Martineau F, Picard F.J, Ke D, Paradis S, Roy P.H, Ouellette M, et al. Development of a PCR assay for identification of staphylococci at genus and species levels. Clin. Microbiol., 2001; 39:2541-2547.
- Martineau F, Picard F.J, Lansac N, Ménard C, Roy P.H, Ouellette M, et al. Correlation between the Resistance Genotype Determined by Multiplex PCR Assays and the Antibiotic Susceptibility Patterns of Staphylococcus aureus and Staphylococcus epidermidis. Agents. Chemother., 2000; 44:231-238.
- Defoirdt T, Boon N, Sorgeloos P, Verstraete W, Bossier P. Quorum sensing and quorum quenching in Vibrio harveyi: lessons learned from in vivo work. ISME J., 2008; 2:19.
- Wang J, Ruan Z, Feng Y, Fu Y, Jiang Y, Wang H, et al., Species distribution of clinical Acinetobacter isolates revealed by different identification techniques. PLoS One. 2014
- Kaclíková E, Krascsenicsová K, Pangallo D, Kuchta T. Detection and quantification of Citrobacter freundii and braakii by 5′-nuclease polymerase chain reaction. Curr. Microbiol., 2005; 51:229-232.
- Bej A.K, Dicesare J.L, Haff L, Atlas R. Detection of Escherichia coli and Shigella spp. in water by using the polymerase chain reaction and gene probes for uid. Appl. Environ. Microbiol., 1991; 57:1013-1017.
- Lee S.S, Chen Y.S, Tsai H.C, Wann S.R, Lin H.H, Huang C.K, et al., Predictors of septic metastatic infection and mortality among patients with Klebsiella pneumoniae liver abscess. Infect. Dis., 2008; 47: 642-650.
- Bej A.K, Patterson D.P, Brasher C.W, Vickery M.C, Jones D.D, Kaysner C. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tlh, tdh and trh. J. Microbiol. Methods., 1999;36:215-225.
- Salman M, Ali A, Haque A. A novel multiplex PCR for detection of Pseudomonas aeruginosa: A major cause of wound infections. J. Med. Sci., 2013; 4:957
- Bauer A.W, Kirby W.M, Sherris J.C, Turck M. Antibiotic susceptibility testing by a standardized single disk method. J. Clin. Path., 1966;45:493.
- Freeman D.J, Falkiner F.R, Keane C. New method for detecting slime production by coagulase negative staphylococci.J. Clin. Pathol., 1989; 42:872–874.
- Singh N, Armstrong D.G, Lipsky B. Preventing Foot Ulcers in patients with Diabetes. JAMA., 2005; 293:217–228.
- Rani V, Nithyalakshmi J.A comparative study of Diabetic and Non-diabetic wound infections with special reference to MRSA and ESBL. J. Curr. Microbio.l App. Sci.,2014;3:546–554.
- Banu A, Hassan M.M, Rajkumar J, Srinivasa S. Spectrum of bacteria associated with diabetic foot ulcer and biofilm formation: a prospective study. Australa Med. J., 2015; 8: 280-285.
- Raja NS. Microbiology of diabetic foot infections in a teaching hospital in Malaysia: a retrospective study of 194 cases. Microbiol. Immunol. Infect., 2007; 14:45–49.
- Halpati, A., Desai, K. J., Jadeja, R., and Parmar, M.A study of aerobic and anaerobic bacteria in diabetic foot ulcer and in vitro sensitivity of antimicrobial agent. J.Med. Sci. Public Health., 2014;3:818–821.
- Ghafoor A, Hay D.I, Rehm A.H. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol., 2011; 77: 5238-5246.
- Esmat, M. M, and Saif Al Islam, Diabetic foot infection: Bacteriological causes and antimicrobial therapy. J. Am. Sci.2012;8:389–393.
- Leid, J. G, Shirtliff, M. E, Costerton, J. W, and Stoodley, P. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus Infect. Immun., 2002 ;70:6339–6345.
- James, G. A, Swogger, E, Wolcott, R, Pulcini, E, Secor, P, Sestrich, J., et al Biofilms in Chronic wounds. Wound Repair Regen., 2008;16:37–44.
- Swarna, S. R, Madhavan, R, Gomathi, S, and Thamaraiselvi, S. A study of Biofilm on Diabetic Foot Ulcer. J. Res. Pharm. Biomed. Sci., 2012;3:1809–1814.
- Zubair, M, Malik, A, Ahmad, J, Rizvi, M., Farooqui, K. J, and Rizvi, M. W. A study of biofilm production by gram-negative organisms isolated from diabetic foot ulcer patients. Med., 2011;3:147–157.
- Damir, A. Why Diabetic Foot Ulcers do not heal? Journal International Medical Sciences Academy., 2011;24:205.







