Manuscript accepted on :25 May 2018
Published online on: 21-06-2018
Plagiarism Check: Yes
Shri Natrajan Arumugam1 , Akarsh Chickamagalur Rudraradhya1
, Akarsh Chickamagalur Rudraradhya1 , Sathish Sadagopan1
, Sathish Sadagopan1 , Sunilkumar Sukumaran1
, Sunilkumar Sukumaran1 , Ganesh Sambasivam1
, Ganesh Sambasivam1 and Nachimuthu Ramesh2
and Nachimuthu Ramesh2
1Anthem Biosciences Pvt Ltd., Bangalore, Karnataka, India.
2Antibiotic Resistance and Phage Therapy Laboratory, School of Biosciences and Technology, Vellore Institute of Technology, Vellore Tamil Nadu, India.
Corresponding Author E-mail: shrinatrajan.a@anthembio.com
DOI : https://dx.doi.org/10.13005/bpj/1471
Abstract
Pseudomonas aeruginosa is known to be a major cause of Hospital Acquired Infections leading to high mortality in immune-compromised patients. Due to precipitous rise in antibiotic resistance, bacteriophages are significant alternative therapeutic approach for treatment and to combat resistance development. Objective of the current study was to identify MDR Pseudomonas aeruginosa from clinical isolates and to isolate bacteriophages from sewage samples against these MDR Pseudomonas aeruginosa strains. One hundred and forty-four Pseudomonas isolates were tested for their susceptibility pattern with 13 different antibiotics by micro-broth dilution method. Frequency of multidrug resistant (MDR) and Extensive Drug resistant (XDR) of Pseudomonas aeruginosa were found to be 35.5% and 23.6%, respectively. 7.61% isolates were identified as Pan drug resistant (PDR). Rate of susceptibility pattern were Piperacillin/Tazobactam 75%, Polymyxin B 74.6%, Meropenem 73.6%, Colistin 69.2%, Cefepime 54.9%, Ciprofloxacin 54.2%, Gentamicin 54.2%, Aztreonam 53.5%, Tobramycin 47.9%, Ticarcillin/Clavulanic acid 46.9%, Ertapenem 45.8%, Ceftazidime 40.3% and Imipenem 39.2%. Ninety-four bacteriophages were isolated from sewage samples against Pseudomonas aeruginosa PAO1/ATCC9027/clinical strains and host range testing study was carried out with all MDR clinical isolates. Among 51 MDR strains 34 strains were infected by phages. Phage infectivity rate were calculated for individual phages based on their host range infectivity results. AP025 and AP006 phages exhibited good infectivity rate of 39% and 30% respectively against MDR strains. Combination of 5 phages (AP002, AP006, AP011, AP025 and AP067) lysed 62.7% of the strains. Based on the obtained results, phages could be employed for treatment of infections caused by MDR strains with substantiated in-vivo experiments.
Keywords
MDR Bacteriophages; Pseudomonas Aeruginosa; TEM Caudovirales
Download this article as:| Copy the following to cite this article: Arumugam S. N, Rudraradhya A. C, Sadagopan S, Sukumaran S, Sambasivam G, Ramesh N. Analysis of Susceptibility Patterns of Pseudomonas Aeruginosa and Isolation, Characterization of Lytic Bacteriophages Targeting Multi Drug Resistant Pseudomonas Aeruginosa. Biomed Pharmacol J 2018;11(2). |
| Copy the following to cite this URL: Arumugam S. N, Rudraradhya A. C, Sadagopan S, Sukumaran S, Sambasivam G, Ramesh N. Analysis of Susceptibility Patterns of Pseudomonas Aeruginosa and Isolation, Characterization of Lytic Bacteriophages Targeting Multi Drug Resistant Pseudomonas Aeruginosa. Biomed Pharmacol J 2018;11(2). Available from: http://biomedpharmajournal.org/?p=20832 |
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen of humans. It rarely causes infections in healthy individuals but is well documented to be a major cause of Hospital Acquired Infections (HAI) leading to severe morbidity in immunocompromised patients. Pseudomonas aeruginosa is an extremely virulent pathogen and the source of numerous types of infection(s), including pneumonia, bacteremia, urinary tract infections (UTI), and wound infection. P. aeruginosa is associated with high death/mortality in hospital-acquired pneumonia cases. In most of the cases, infections due to P. aeruginosa occur in a nosocomial setting in patients with a co-morbid ailment and compromise from contaminated catheters, tubes, and surgeries.
Pseudomonas aeruginosa is notorious for its resistance to antibiotics and is, therefore, a particularly dangerous and feared pathogen.1 It has a natural affinity for the development of resistance to antibiotics. This limits a defined therapeutic use of antibiotics against this bacterium and contributes to the increased rates of mortality. Its resistance to many antibiotics including Tetracycline and Benzyl penicillin is reportedly due to the permeability barrier afforded by its outer membrane lipopolysaccharide.2
Pseudomonas spp. has the unique property of forming biofilms which helps in making the cells impervious to antibiotics. Moreover, P. aeruginosa harbours antibiotic resistant plasmids, and is able to transfer these genes by elegant processes of transduction and conjugation. Only a few antibiotics are effective against Pseudomonas, including some beta-lactams, aminoglycosides, carbapenems and fluoroquinolones, and even these are not effective against all strains. The futility of treating Pseudomonas infections with antibiotics is most dramatically illustrated in cystic fibrosis patients, virtually all of whom eventually become infected with an incurable antibiotic resistant strain.3
The emergence of resistance in P. aeruginosa to currently available antibiotics drugs has become a serious issue in the modern treatment, mainly because of the associated increase in immunosuppressed patients. Non-antibiotic therapies to treat bacterial infections are now under serious consideration and one of the preferable, potential choice is using specific phage that targets bacterial pathogen. Bacteriophages are viruses which can infect bacteria and replicating within the bacterial cell.4 Bacteriophages were first identified independently by Frederick Twort in 1915 and by Félix d’Herelle in 1917.5
The specificity and virulence of these phages against the bacteria reap a great importance in various industries such as the food and other bio-processing industries. Though bacteriophages have been used for treatment since long and many reports have been published on phage therapy and considering phages as a permanent therapeutic agent is still a question. Based on their life cycle, phages classified into two classes: Virulent phages (lytic) and temperate phages. Obligate lytic phages are more interested in therapeutic use. Because the bacteria that have been infected by lytic phages are incapable to recover their viability.6
According to the data available, 94.2% of the phages targeting the Pseudomonas aeruginosa belonging to the Caudovirales order which comprises three families of double-standard DNA phages. Based on the tail structure, Caudovirales classified into Podoviridae, with a short and non-contractile tail; Myoviridae, with a long contractile tail, and Siphoviridae, with long and concontractile tail.6
Fu et al (2010) studied the effect of phages in the reduction of P. aeruginosa biofilm formation in catheters with pre-treatment and post treatment of phages. They observed 2.81 and 2.5 log reduction in phage pre-treated and post treatment catheters respectively compared with untreated control catheters after 24 h of biofilm formation. However, biofilm regrowth was observed after 24 h in phage treated catheters due to phage resistant biofilm cells. A cocktail of five phages was developed to encounter the phage resistance, showed 3 log reduction of biofilm populations after 48h, compared with control catheters.7
Benjamin K. Chan et al (2016) reported that phage OMKO1 is infecting the Pseudomonas aeruginosa by utilizing the outer membrane porin M (OprM) of the multi-drug efflux systems MexAB and MexXY as a receptor-binding site. Phage resistance selection increasing the sensitivity to antibiotics from different classes, whereby development of bacterial resistance to phage attack changes the efflux pump mechanism.8
Rotem Edgar et al (2012) demonstrated a proof of principle that reversing antibiotic efficiency of the MDR P. aeruginosa strain pathogens by lysogenization, introducing the temperate phage’s. Unique selective pressure was generated using non antibiotic toxic compounds and demonstrated that streptomycin and nalidixic acid resistance was reversed by the proposed system. Also they trust that antibiotic-treatable pathogens will be significantly increase by transferring of sensitizing cassette through phage infection in environment.9
Combination of phages of Podoviridae phage LUZ7 and streptomycin antimicrobials was attempted by Torres-Barceló et al in treatment of bacterial Pseudomonas aeruginosa infection. aeruginosa PAO1 was challenged and treated in single and in combination of phages and compared with two doses of the streptomycin. Bacterial population density was tracked over 70 h to reduce the antibiotic doses below the MIC. The combination treatment of phages and antimicrobials revealed a positive synergism resulting lower the bacterial density than additional of respective single treatments and also resistance levels were equal or even lower those of each single treatments.10
There were many in vivo studies reported on bacteriophage efficacy against P. aeruginosa. Eric Morello et al investigated the protective and curative efficacy of the phage against MDR Pseudomonas aeruginosa strain isolated from cystic fibrosis patient in immunocompetent mice. Bacteriophages and bacteria were applied intranasally and they observed 95% survival in single treatment administered 2 hr after infection (curative) 100% survival in single preventive dose, 4 days before infection.11
The objective of the current study was identifying the MDR Pseudomonas aeruginosa from clinical strains and to isolate the lytic bacteriophages from sewage samples targeting P. aeruginosa. The isolated phages were tested against MDR Pseudomonas aeruginosa strains for their host range specificity and characterized the bacteriophages having broad host range activity.
Materials and Methods
Clinical Bacterial Isolates
A total of 144 Pseudomonas aeruginosa clinical isolates were collected from super speciality and tertiary care hospitals in south India from the period 2012 to 2015. The isolates were confirmed using standard microbiological method which includes Gram staining, colony morphology and growth on selective media. Biochemical tests like catalase, oxidase, indole and citrate utilization were performed to confirm identity of P. aeruginosa. The isolates were sub-cultured and preserved for the experiment.12
The following anti-bacterial agents were procured from Sigma-Aldrich; Gentamicin, Tobramycin, Ceftazidime, Cefepime, Imipenem, Meropenam, Ertapenem, Aztreonam, Ciprofloxacin, Piperacillin/Tazobactam, Ticarcillin/Clavulanic acid and colistin. Polymyxin B was procured from TCI chemicals. Aztreonam was procured from Molekula biokemix. ATCC 27853 P. aeruginosa was included in each batch of experiment as a quality control strain.
Antibiotics Susceptibility test
MIC studies were performed according to the CLSI guideline.13 The following anti-bacterial agents were tested for susceptibility; Gentamicin, Tobramycin, Ceftazidime, Cefepime, Imipenem, Meropenam, Ertapenem, Aztreonam, Ciprofloxacin, Piperacillin/Tazobactam (TZP), Ticarcillin/Clavulanic acid (TCC), Colistin and Polymyxin B. The isolates were tested in duplicate and results were read visually and spectrophotometrically after overnight incubation at 35°C. All reference antibiotics were tested from 0.125 µg/mL to 128 µg/mL. The resistance and susceptibility limit of the clinical isolates were decided as per breakpoints available in CLSI guidelines.
Bacteriophage Isolation
Thirty-two sewage samples were collected from Bangalore, India, with a surrounding distance of 120 km, between February 2015 to April 2016. Approximately 200 ml of sewage samples were collected in a sterile containers and transported to the laboratory and stored at 4°C, until processing.
Bacteriophages were isolated from the collected sewage samples by enrichment technique.14 Under sterile conditions, 80 ml of sewage sample was mixed with 20 ml of 5X Luria broth (LB broth) and 1 ml of log-phase broth culture (optical density measured at 600 nm) of Pseudomonas aeruginosa PAO1/Pseudomonas aeruginosa ATCC9027/clinical isolate.
Bacterial and sewage suspension were incubated overnight at 37°C, with shaking (120 rev/minutes) and centrifuged at 10,000 g for 10 minutes at 4°C to remove cellular debris. Supernatant was filtered through a 0.22 μm syringe filter (Millipore, India) to obtain a bacterial free phage lysate (BFP) and further analysed for the presence of phages by plating method.
Modified double layer plaque technique was used for bacteriophage isolation from phage lysate.15 Three millilitre of 0.6% of Luria bertani agar (LBA: LB broth – 1 mM CaCl2, 1 MgSO4, 0.2% glucose, Agar – 0.6 g, D.H2O – 100 ml) was mixed with 100 μL of 3-4 hrs log-phase Pseudomonas aeruginosa PAO1 strain / Pseudomonas aeruginosa ATCC9027 and 100 μl of the BFP. Suspension was plated on freshly prepared 1.5% LB agar plate and incubated in an upright position at 37°C overnight. Formation of plaques or zones of clearance indicated the presence of phages.16 Based on the plaque morphology (Size, turbidity, plaque edge appearance) plaques were isolated and inoculated into respective host to prepare the Monophages suspension to avoid cross contamination. Monophage suspension of bacteriophages were prepared through 3 successive rounds of single plaque picking and culturing them again with respective host. All phage lysates were stored at 4°C.
Purification and Concentration of Phage’s
The phage lysate in LB was centrifuged at 10,000 x g for 15 minutes and further filtered through 0.22 μm filter to remove bacterial debris. Bacteriophages were purified from the phage lysate by using simplified polyethylene glycol (PEG) precipitation method.17
To 10 ml of phage lysate, 0.58 g of NaCl was added to make a final concentration of 1 M. The culture was swirled until the salt dissolved and was incubated in ice for 1 hr. The supernatant was centrifuged at 8000 x g for 10 minutes at 4°C to eliminate cell debris. Then 0.5 g of solid PEG 8000 was added to a final concentration of 10% w/v and dissolved. The PEG/phage solution was stored at 4°C for at least 1 hr to allow the phage particles to precipitate. The precipitated bacteriophages were recovered by centrifugation at 8000xg for 10 minutes at 4°C. The supernatant was discarded and the pellet was dried at room temperature for 5-10 minutes. The pellet was suspended in SM buffer (50 mM Tris – HCL (pH 7.5), 100 mM NaCl, 10 mM MgSO4, and 2% gelatin) and incubated at room temperature for 1 hr. The PEG and cell debris were removed from the suspension by adding equal volume of chloroform and followed by vortexing for 30 seconds. The organic and aqueous phases were separated by centrifugation at 4300xg for 15 minutes at 4°C and the aqueous phase containing the bacteriophages was recovered.14
Host Range testing
The isolated phage’s (n=94) were then tested against a collection of MDR Pseudomonas aeruginosa (n=51) to check the host range extensiveness by spot test method.
MDR bacterial lawns were prepared on LB agar plates and 10 μl of purified phages were spotted onto the bacterial lawns. Phage suspensions were adjusted to 104 -105 PFU per spot. After 18-22 hrs of incubation, the effect of the phage suspensions on the lawn was investigated. A phage infection was accounted if there was ≥ 20 plaques or full lysis (clear or opaque) in the spot.18
TEM Analysis
The morphological analyses of selected phages were done by Transmission Electron Microscopic (TEM) analyses. TEM analyses showed the distinct features of each phages. Thin 200 mesh carbon-coated copper grids of 3 mm diameter were used for the analyses. Negative staining technique using Phosphotungstic acid was preferred and the phage titer was set at around 1010-1011 PFU/mL. 3-5 µl of the sample was added on to the grid and allowed for 30sec and excess sample was removed. Then few drops of 2% Phosphotungstic acid was added to the grid and allowed for few secs and the excess stain was removed. The grids were dried for 30-45mins and observed in Tecnai, G2 20 Twin High-Resolution Transmission Electron Microscope at 80kV.18
Adsorption Assay
The adsorption assay was done to find adsorption time and percentage of adsorption of the selected phages. Assay estimates the time taken for the phages to get adsorbed into the bacterial cell. Phages range of 108 – 1010 PFU/mL were added to the exponentially grown PA01 at a MOI of 0.1 – 10 and incubated at 37o C for 15mins. After incubation the aliquots of 100 µl were drawn at an interval of 2-3 min up to 15-20 mins and non-adsorbed phage particles was calculated by double overlay technique.20
One-Step Growth and Burst Size
The one-step growth determines the latent period and burst of the phages. This gives the latent period of the particular phage and number of phages that burst out of a single cell. The phages (108 – 1010 PFU/mL) were added to exponentially grown PA01 at an MOI of 0.1-10 and incubated at 37o C for 15 minutes. The mixture was centrifuged at 10000 rpm for 10 mins at room temperature(RT). Pellet was re-suspended in supplemented LB broth and incubated at RT. Samples were taken at 5 mins interval for 60 mins. Test samples were diluted and titrated by double layer technique.20
Results
Antibiotic Susceptibility test Results
Antibiotic susceptibility pattern of 13 antibiotics from 7 different antimicrobial classes for the 144 P. aeruginosa were listed in the table 1. Among 144 clinical isolates of P. aeruginosa, 35.4% (51) of the isolates were identified as multi drug resistant (MDR), 23.6%(34) isolates were extensively drug resistant (XDR) and about 4.9% (7) of the isolates displayed possible pan drug resistance (PDR) Percentage susceptibility of P. aeruginosa against antibiotics were given in the Figure 1.
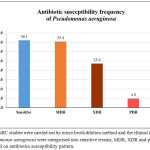 |
Figure 1: MIC studies were carried out by micro broth dilution method and the clinical isolates of Pseudomonas aeruginosa were categorised into sensitive strains, MDR, XDR and possible PDR based on antibiotics susceptibility pattern.
|
Results are expressed as percentage of isolates
Piperacillin/Tazobactam was observed to be most effective antimicrobial agent with 75% susceptibility, followed by Polymyxin B (74.6%), Meropenem (73.6%), Colistin (69.2%,) Cefepime (54.9%), Ciprofloxacin (54.2%), Gentamicin (54.2%) and Aztreonam (53.5%). Drugs including Tobramycin (47.9%), Ticarcillin/Clavulanic acid (TCC) (46.9%), Ertapenem (45.8%), Ceftazidime (40.3%) and Imipenem (39.2%) which are also the drug of choice against P. aeruginosa have not shown higher susceptibility. The MIC90 and MIC50 value for all the tested antibiotics were presented in the Table 1.
Table 1: Antibiotic susceptibility test results for 144 clinical strains. Percentage Resistance (R), Susceptibility(S) and Intermediate (I) were determined as per CLSI guideline
| S. No | Antibiotic name | Antibiotic class | Breakpoints | Number tested | % of Isolates
(No of isolates) |
µg/mL | |||
| %R | %I | %S | MIC50 | MIC90 | |||||
| 1 | Gentamicin | Aminoglycosides | S≤4 R≤16 | 144 | 45.1 | 0.7 | 54.2 | 4 | 128 |
| 2 | Tobramycin | Aminoglycosides | S≤4 R≤16 | 144 | 45.1 | 6.9 | 47.9 | 8 | 128 |
| 3 | Ceftazidime | Cephems | S≤8 R≤32 | 144 | 55.6 | 4.2 | 40.3 | 32 | 128 |
| 4 | Cefepime | Cephems | S≤8 R≤32 | 144 | 38.9 | 6.2 | 54.9 | 8 | 128 |
| 5 | Imipenem | Penems | S≤2 R≤8 | 143 | 42.0 | 18.9 | 39.2 | 4 | 64 |
| 6 | Meropenem | Penems | S≤2 R≤8 | 144 | 20.1 | 6.2 | 73.6 | 1 | 32 |
| 7 | Ertapenem | Penems | S≤4 R≤16 | 144 | 36.1 | 18.1 | 45.8 | 8 | 64 |
| 8 | Aztreonam | Monobactams | S≤8 R≤32 | 144 | 35.4 | 11.1 | 53.5 | 8 | 128 |
| 9 | Ciprofloxacin | Quinolones | S≤1 R≤4 | 144 | 36.1 | 9.7 | 54.2 | 1 | 128 |
| 10 | Piperacillin/Tazobactam | Beta-lactam + Inhibitors | S≤64 R≤128 | 144 | 25.0 | 0 | 75.0 | 16 | 128 |
| 11 | Ticarcillin/Clavulanic acid | Beta-lactam + Inhibitors | S≤64 R≤128 | 143 | 53.1 | 0 | 46.9 | 128 | <256 |
| 12 | Colistin | Lipopeptides | S≤2 R≤8 | 143 | 24.5 | 6.3 | 69.2 | 1 | 128 |
| 13 | Polymyxin B | Lipopeptides | S≤2 R≤8 | 142 | 23.9 | 1.4 | 74.6 | 0.5 | 128 |
Bacteriophage Isolation and Host Range Study Results
In total, 94 bacteriophages were isolated from Bangalore, Karnataka, India from 32 different sewage samples, collected at different time. Pseudomonas aeruginosa PAO1, Pseudomonas aeruginosa ATCC9027 and clinical strains were used as host strains for isolation of bacteriophages. The purified bacterial free phage lysates were mixed with respective host and poured on to agar plates to analyse their infectivity. All 32 sewage samples had phages with distinct morphology in plaque size, turbidity and plaque edge. Based on the plaque morphology the bacteriophages were purified by double layer plaque method.
Sample collection details of the bacteriophage isolation was given in the Table 2.
Table 2: Sewage sample collection details for the bacteriophage isolation
| Nature of Sample | Location | No of Samples collected | Host strains | No. of phage’s Isilated |
| Sewage and Sewage treatment plant | Bangalore, Karnataka | 32 | Pseudomonas aeruginosa PAO1, ATCC9027 and clinical isolate | 94 |
Host range studies were executed for each purified phages with 51 MDR strains to identify the broad host infectivity. Among 51 MDR strains, 53% (n=27) of Pseudomonas aeruginosa strains were infected by at least one of the phage tested.
Five Pseudomonas aeruginosa MDR strains were more susceptible to phage infection, the strains showed 40 – 80% susceptibility for phage infection.
Phage infectivity rate was calculated for individual phages based on their broad host range infectivity with all 51 MDR strains. Percentage infectivity of individual phages, AP001 to AP097 by spot test method against 51 MDRs P. aeruginosa are given in Figure 2 and Figure 3. Among 94 tested phages, 2 phages exhibited good infectivity rate of 39.2% (AP025) and 30% (AP006) against MDR strains. AP002 and AP011 phages showed moderate infectivity rate of 27.5%. AP067 phage exhibits 23.5% infectivity rate against MDR P. aeruginosa.
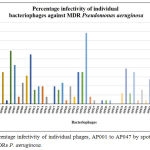 |
Figure 2: Percentage infectivity of individual phages, AP001 to AP047 by spot test method against 51 MDRs P. aeruginosa.
|
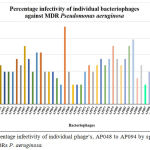 |
Figure 3: Percentage infectivity of individual phage’s, AP048 to AP094 by spotting method against 51 MDRs P. aeruginosa.
|
Transmission Electron Microscope (TEM) Analysis
Based on the host range identification study, bacteriophages that showed broad host infectivity were selected for characterization analysis including Transmission Electron Microscopic studies. TEM analysis picture, Adsorption rate, latent period and burst size of the bacteriophages are presented in Table 3.
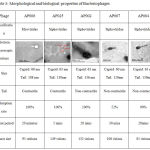 |
Table 3: Morphological and biological properties of Bacteriophages
|
Bacteriophage AP025 had a head of 68 nm diameter, a clear distinct neck region of 9 nm, body and tail of 108 nm. The neck region and a long contractile tail represents that the phage falls under the family Myoviridae of Order Claudoviriales. Phage AP006 had a head of 63nm a short neck and a long contractile tail of 80-130nm. The morphological characteristics of the phage resembled that of a member of Myoviridae.
AP002 had a head size of 63 nm, no distinct neck region and a long non-contractile tail of length 136 nm resembling a members of family Siphoviridae. AP067 and AP084 had a Icosahedral head of 60 nm and a long non-contractile tail of 110 nm. The characteristics feature resembled that of a member of Siphoviridae.
Phage AP084 has a head size of 63 nm and a long non-contractile tail of 130 nm which denotes that this phage is a member of the family Siphoviridae.
Discussion
Resistance to antibiotics among pathogenic bacteria remains a challenge for clinicians. There are four known mechanisms through which organisms develop resistance 1. Restricted uptake 2. Antibiotic efflux 3. Drug inactivation and 4. Change in target sites.21 Routine antimicrobial resistance surveillance is imperative to update the activity levels of various drugs that are commonly used. The current study results clearly indicate that the development of resistance is predominantly due to indiscriminate use of antibiotics. Penicillin was considered as one of best discoveries of the 20th century and with the over enthusiastic use of this antibiotic most of the common pathogens developed resistance. However, the results from the current study underlines the impact of using any antibiotic extensively.
In this current study, a total of 144 P. aeruginosa isolates were screened to identify their resistance pattern and the results showed that the frequency of MDR P. aeruginosa, XDR and PDR were 35.4, 23.6% and 4.9% respectively. A similar study carried out by Gill et al. (2011) reported 22.7% of MDR, 11% of XDR and 4.3% of PDR percentage while screening 180 clinical isolates in Army medical college, Rawalpindi from January to September 201022 and Mona wassef et al (2015) also reported about 12.3% P. aeruginosa were MDR from 2593 clinical isolates in Egypt.23
Studies by Farhatullah et al (2009) have also alluded to the point of increasing percentage of multi-drug resistance in P. aeruginosa infections.24
In this current study, 80 of P. aeruginosa showed highest resistance to ceftazidime accounting for 55.6% of the isolates. A high rate of ceftazidime resistance was also reported in India by Prashant et al (2011) where 53% of P. aeruginosa isolates were resistant which was similar to the observations made by Diwivedi et al (2005), who reported that ceftazidime resistance was about 63%25,26. Higher rate of ceftazidime resistance was also reported in an Egyptian study in which 43.8 % of P. aeruginosa isolates showed resistance to ceftazidime.23 These highlight the point that the incidence of multidrug resistance is increasing with time.
The present study showed a percentage susceptibility of 75% against piperacillin/tazobactam. Supporting this observation, Singh et al., (2012) reported 10.3 % resistance against piperacillin/tazobactam when he screened 107 clinical isolates of P. aeruginosa isolated from northern part of India.27 Pseudomonas aeruginosa antimicrobial susceptibility studies by Pathmanathan et al. (2009) reported a percentage susceptibility of 92.8% against piperacillin/tazobactam.28
In this current study, we observed that P. aeruginosa was highly susceptible to carbapenems group of antibiotic meropenem (73.6%). Pseudomonas antimicrobial susceptibility by Wright et al (2016) reported 77.4% in Asia/South Pacific, when screened 1,392 samples29 and similar study by Viren et al (2008) reported susceptibility of 69.6%.30
We also observed 69.2% sensitivity to colistin. This was surprising as colistin is used as the third level of treatment and is not a very commonly prescribed or an over the counter medication. Polymyxin B exhibited highest sensitivity rate of 74.6% against P. aeruginosa which is nearly same as TZP. Among the susceptible strains, 17% and 11% displayed resistance to colistin and polymyxin B, respectively.
The MIC90 values for meropenem and imipenem were much lower than the maximum tested concentrations indicating that majority of the strains are still susceptible to these two antibiotics at higher concentrations. Nevertheless, continuous use of antimicrobial agents might lead to resistance development and similar antibiotics sensitivity profiling study if carried out after 2-3 years might increase the MIC90 values for these antibiotics as well.
Accumulated evidence suggests that patients infected with MDR organisms are likely to experience a delay in the effective antimicrobial therapy, and reports suggest that this risk could be minimized with addition of a second agent. Kumar et al (2010) evaluated the therapeutic benefit of combining beta lactams with aminoglycosides, fluoroquinolones, and macrolides in 4,662 cases of culture positive bacterial septic shock patients. Even though there was a decrease in mortality with combination therapy, anti-pseudomonal penicillin, cephalosporin’s and carbapenems failed to exhibit significant benefit with the addition of a second agent. It was inferred that the combination could have proven effective if it was selected with the nature of infection in mind. Resorting to empirical therapy in an effort to provide quick and rapid relief inadvertently could be a significant contributing factor in the development of drug resistance among pathogen. The selection of empiric combination therapy for presumed infections with multi drug resistant strains should be made after considering the surveillance data available in the respective hospital along with the patient’s antibiotics treatment history.31 Clinicians should work in close liaison with the microbiology laboratory to understand the resistance pattern within the hospital before prescribing anti-microbial therapy.
The current study indicates that the resistance rates for the clinical isolates of Pseudomonas aeruginosa in southern parts of India appeared to have increased marginally for all antimicrobials. Piperacillin/Tazobactam and Meropenem appeared to show maximum susceptibility when tested against these isolates. Failure to see more than 90% susceptibility is a clear indication of the over enthusiastic use of broad spectrum antibiotics. Serious measures or alternate approaches should be initiate to avoid development and spread of multidrug resistant pathogens.
The continuous increase of MDR P. aeruginosa in addition to low rate of new antimicrobial development, immediately demands a novel and new approaches to combat infections by pathogens. The massive diversity of bacteriophages, easy isolation of new phages from environment like sewage and soil, low production cost, and their more efficiency against the target pathogens make them a very attractive alternate choice to antibiotics to treat the drug resistance pathogens.
There are very few reports have been published so far in India, mostly on isolation and characterization of the phages targeting the MDR Pseudomonas aeruginosa. Abundant bacteriophages were present in sewage sample when compared to other samples like soil and sea waters. Obligate lytic phages were more promising for the phage therapy. Because they result in rapid killing of their target host cell, rapid increase in numbers and persist by releasing the phages that migrate to other site of infection. Transferring of antibiotic resistant gene by transduction is relatively rare in lytic phages.32
In this present study, 94 lytic phages were isolated from 32 different sewage samples, collected on different time period and further characterized by utilizing MDRs clinical strains of P. aeruginosa as a bacterial lawn for finding the host-phage interaction. Isolated phages were spotted on the MDR Pseudomonas aeruginosa strains to find out the broad host infectivity. Similarly, Zhabiz Golkar et al (2013) isolated bacteriophage from clinical specimen, urine from the UTI patients, targeting clinical MDR Pseudomonas aeruginosa and identified as Myoviridea based on the electron microscopic examination. The efficacy of the isolated phage was evaluated in murine superficial infection with MDR P. aeruginosa strain showed complete healing of the wounds by oral and i.p route of administrations.33
In present study, 53% (27 nos) of Pseudomonas aeruginosa strains were infected by at least one of the isolated phages and it indicates that MDR Pseudomonas aeruginosa strains more susceptible to phages though they are highly resistance to antibiotics. In present study, AP025 and AP006 phages lysed 20 (39.2%) and 15 (30%) strains of MDR P. aeruginosa out of 51 strains, respectively. 27.5% of infectivity rate was observed with other two phages namely AP002 and AP011 against MDRs. AP067 phage exhibits 23.5% infectivity rate against MDR P. aeruginosa. (Figure 3).
For phage therapy, a broad host range phage that kills multiple species of same genus would be equivalent to a broad spectrum antibiotic. The phages were selected for further characterization based on the results obtained from host range study. (Table 3)
Morphological characteristics of the phages were analysed through TEM, revealed that AP006 and AP025 belongs to Myoviridae. AP002, AP067, AP084 were belongs to family of Siphoviridae. The present study results were correlated with study conducted by Golkar et al. and he isolated phage that targets MDR P. aeruginosa was belongs to Myoviriade.32 Also Diana et al (2015) reported that more than 90% of bacteriophages which infect P. aeruginosa were belongs to Caudovirales order which comprises three families, Myoviridae, Siphoviridae and Podoviridae.6
Experiments using phage cocktails, not to imply that single phage treatment successes have not been seen, by which phage therapy outcomes may be enhanced. Bacteriophages employed for the treatment as cocktails by mixing more than 4 to 5 broad host range phages which uses a different receptors of bacteria with in the same species. Phage cocktails could increase the activity and also reduce the resistance development against them.
In this present study, combination of AP002, AP006, AP011, AP025 and AP067 phage’s displayed an increased infectivity rate of 62.7% against MDR clinical pathogens. (Figure 4). Our results are correlated with study conducted by Alex R. Hall and they evaluated phage’s in vitro and in-vivo in single phages, pairs and combination of phages in simultaneous and sequential application. Their results suggest that compare to single, the combination of phages in treatment are most effective if its applied them all simultaneously. In combination of phages in treatment is more effective, since different phages targets different receptors present in bacteria and able to lyse.
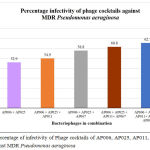 |
Figure: 4: Percentage of infectivity of Phage cocktails of AP006, AP025, AP011, AP067 and AP002 against MDR Pseudomonas aeruginosa.
|
Conclusion
The global increase of antibiotic resistance among the pathogenic bacteria makes it an imperious to exploit alternative approaches to combat this threat. Bacteriophages have numerous advantages that consider them potentially striking therapeutic agents. They are highly specific, replicates in site of infection, self-limiting, no serious side effects have been reported, Phage-resistant bacteria remain susceptible to other phages and also selecting a new phage is relatively rapid. Based on the results presented in this article strongly shows that AP006 and AP025 bacteriophages and bacteriophage cocktails efficiently constitute a potent tool to fight P. aeruginosa infections, specifically those caused by MDR bacteria. Furthermore, phages efficacy need to be evaluated by in-vivo studies to confirm the specificity and sensitivity.
Acknowledgement
We thank Mr. Ajay Bhardwaj CEO and Mr. Ravindra Chandrappa COO Anthem Biosciences, Bangalore, India for conducting and reporting findings of this study.
References
- Rayner C. F. J, Cole P. J, Wilson R. Management of chronic bronchial sepsis due to bronchiectasis. Clin Pulm Med. 1994;6:348-355.
CrossRef - Li X. Z, Livermore D. M and Nikaido H. Role of efflux pump(s) in intrinsic of Pseudomonas aeruginosa: Resistance to tetracycline, chloramphenicol and norfloxacin. Antimicrob Agents Chemother. 1994;8:1732-1741.
CrossRef - Robert Wilson, Ruth B Dowling. Pseudomonas aeruginosa and other related species. 1998;53:213–219.
- Twort FW. An investigation on the nature of ultra-microscopic viruses. Lancet. 1915;186:1241–1243.
CrossRef - Xavier Wittebole, Sophie De Roock, Steven M Opal. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence. 2014;5(1):226–235.
CrossRef - Diana P. Pires, Diana Vilas Boas, Sanna Sillankorva, Joana Azeredo. Phage therapy: a step forward in the treatment of Pseudomonas aeruginosa J Virol. 2015;89:7449-7456.
CrossRef - Fu W, Forster T, Mayer O, Curtin JJ, Lehman SM, Donlan RM. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob Agents Chemother. 2010;54:397-404.
CrossRef - Benjamin K. Chan, Mark Sistrom, John E. Wertz, Kaitlyn E. Kortright, Deepak Narayan & Paul E. Turner1. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Scientific Reports. 2016;6:2671.
CrossRef - Rotem Edgar, Nir Friedman, Shahar Molshanski-Mor, Udi Qimron. Reversing Bacterial Resistance to Antibiotics by Phage-Mediated Delivery of Dominant Sensitive. Genes Applied and Environmental Microbiology. 2012;78:744-751.
CrossRef - Torres-Barceló C, Arias-Sánchez FI, Vasse M, Ramsayer J, Kaltz O, Hochberg ME. A window of opportunity to control the bacterial pathogen Pseudomonas aeruginosa combining antibiotics and phages. PLoS One. 2014;9:10662
CrossRef - Eric Morello, Emilie Saussereau, Damien Maura, Michel Huerre, Lhousseine Touqui, Laurent Debarbieux. Pulmonary Bacteriophage Therapy on Pseudomonas aeruginosa Cystic Fibrosis Strains: First Steps Towards Treatment and Prevention. PLoS ONE. 2011:6(2).
CrossRef - Bergey D H, John G Holt. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore: Williams & Wilkins; 1994.
- Performance Standards for Antimicrobial Susceptibility Testing; approved standard-twenty sixth edition. M100S:2016.
- Duraisamy Nivas, Nachimuthu Ramesh, Vaithilingam Krishnakumar, Pandiyan Rajesh, Ebenezer King Solomon, Velu Rajesh Kannan. Distribution, isolation and characterization of lytic bacteriophages against multi-drug resistant and extended-spectrum of β-lactamase producing pathogens from hospital effluents. Asian J Pharm Clin Res. 2015;8:384-389.
- Sanna Sillankorva, Peter Neubauer, Joana Azeredo. Isolation and characterization of a T7-like lytic phage for Pseudomonas fluorescens. BMC Biotechnology. 2008;8:80.
CrossRef - Rene N. Beaudoin, Danielle R. DeCesaro, Debrah L. Durkee, and Susan E. Barbaro. Isolation of a bacteriophage from sewage sludge and characterization of its bacterial host cell. Rivier Academic Journal. 2007;3:1
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. New York: Cold Spring Harbour Laboratory Press. 1989:600.
- Vinni Mona Hansen, Hanne Rosenquist, Dorte Lau Baggesen, Stanley Brown, Bjarke Bak Christensen. Characterization of Campylobacter phages including analysis of host range by selected Campylobacter Penner serotypes. BMC Microbiology. 2007;7:9.
CrossRef - Erna Li, Xiao Wei, Yanyan Ma, Zhe Yin, Huan Li, Weishi Lin, Xuesong Wang, Chao Li, Zhiqiang Shen, Ruixiang Zhao, Huiying Yang, Aimin Jiang Wenhui Yang, Jing Yuan & Xiangna Zhao. Isolation and characterization of a bacteriophage phiEap-2 infecting multidrug resistant Enterobacter aerogenes, Springer nature.
- Seema Kumari, Kusum Harjai, Sanjay Chhibber. Characterization of Pseudomonas aeruginosa PAO1 Specific Bacteriophages Isolated from Sewage Samples. J. Biomed. Sci. 2009;1(2):91-102.
CrossRef - Lambert P A. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med. 2002;95(41):22-26.
- Maria Mushtaq Gill, Javid Usman, Fatima Kallem, Afreenish Hassan, Ali Khalid, Rabia Anjum, Quanita Fahim. Frequency and Antibiogram of Multi-Drug Resistant Pseudomonas aeruginosa. J Coll Physicians Surg Pak. 2011;9:531-534.
- Mona wassef, Hadir El Mahallawy, Mai Mahmoud Zafer, Doaa Ghaith and Rasha abdel hamid. Lab Based Surveillance of Multidrug Resistant Pseudomonas aeruginosa in Cairo University Hospitals. J Microbiol Exp. 2015:39-44.
- Farhatullah, Malik SA, Ahmed J. Antimicrobial susceptibility and ESBL prevalence in Pseudomonas aeruginosa isolated from burn patients in the North West of Pakistan. Burns. 2009;7:1020-1025.
- Prashant Durwas Peshattiwar and Basavaraj Virupaksappa Peerapur. ESBL and MBL mediated resistance in Pseudomonas aeruginosa: an emerging threat to clinical therapeutics. J Clin Diagn Res. 2011;5:1552-1554.
- Dwivedi M, Mishra A, Singh RK, Azim A, Baronia RK, Prasad KN. Nosocomial cross-transmission of Pseudomonas aeruginosa between patients in a tertiary intensive care unit. Indian jounal of pathology and microbiology. 2005; 52:509-513.
- Singh AH, Basu R. Antimicrobial susceptibility pattern of clinical isolates of Pseudomonas aeruginosa in an Indian tertiary care hospital. IJCRR. 2012;22:99-10
- Siva Gowri Pathmanathan, Nor Azura Samat, Ramelah Mohamed. Antimicrobial susceptibility of clinical isolates of Pseudomonas aeruginosa from a Malaysian Hospital. Malays J Med Sci. 2009;2:27-32.
- Wright W. Nichols, Boudewijn L. M. de Jonge, Krystyna M. Kazmierczak, James A. Karlowsky, and Daniel F. Sahm. In Vitro Susceptibility of Global Surveillance Isolates of Pseudomonas aeruginosa to Ceftazidime-Avibactam: INFORM. 2012-2014. Agents Chemother. 2016;60(8):4743-4749.
- Viren A. Javiya, Somsuvra B. Ghatak, Kamlesh R. Patel, Jagruti A. Patel. Antibiotic susceptibility patterns of Pseudomonas aeruginosa at a tertiary care hospital in Gujarat, India. Indian J Pharmacol. 2008;40:230-234.
CrossRef - Kumar A, Zarychanski R, Light B, Parrillo J, Maki D, Simon D, Laporta D, Lapinsky S, Ellis P, Mirzanejad Y, Martinka G, Keenan S, Wood G, Arabi Y, Feinstein D, Kumar A, Dodek P, Kravetsky L, Doucette S. Early combination antibiotic therapy yields improved survival compared with monotherapy in septic shock: a propensity-matched analysis. Crit Care Med. 2010;38(9):1773-1785.
CrossRef - Hugo Oliveira, Sanna Sillankorva, Maia Merabishvili, LeonD.Kluskens, Joana Azeredo. Unexploited opportunities for phagetherapy. Frontiers in Pharmacology. 2015;6 (180).
- Zhabiz Golkar, Omar Bagasra and Nusrat Jamil. Experimental Phage Therapy on Multiple Drug Resistant Pseudomonas aeruginosa Infection in Mice. J Antivir Antiretrovir. 2013;(10):005.







