Manuscript accepted on :February 18, 2018
Published online on: --
Plagiarism Check: Yes
Solomon Oladapo Rotimi, Iyanuoluwa Temitayo Olugbemi and Oluwakemi Anuoluwapo Rotimi
Biochemistry of Biochemistry and Molecular Biology Research Laboratory, Covenant University, Ota, Ogun State, Nigeria.
Corresponding Author E-mail: Ola.rotimi@covenantuniversity.edu.ng
DOI : https://dx.doi.org/10.13005/bpj/1411
Abstract
Gatifloxacin is a fourth-generation fluoroquinolone that induces oxidative stress in the liver. Hence, this study investigated the alterations in the expression of genes involved in epigenetics and apoptosis associated with gatifloxacin-induced oxidative stress in rat liver. To achieve this, adult rats were exposed to oral doses (10 mg/kg, 20 mg/kg, 40 mg/kg and 80 mg/kg) of gatifloxacin for five days. Thereafter, biomarkers of oxidative stress were assessed spectrophotometrically while the levels of expression of Bcl2l1, caspases 3, 8 and 9 as well as Dnmt1, Hdac5, Prdm2, Eid3, Suv39h1 and Ehmt2 were assessed using reverse transcription polymerase chain reaction technique. The results showed that the hepatic oxidative stress was associated with increase in the expression of proapoptotic genes. Also, gatifloxacin resulted in significant (p < 0.05) upregulation of genes involved in DNA and histone methylation. These alterations observed at the lowest dose of 10 mg/kg showed that gatifloxacin exposure could induce hepatic apoptosis and epigenetic changes.
Keywords
Apoptosis Word; Epigenetics; Gatifloxacin; Oxidative Stress;
Download this article as:| Copy the following to cite this article: Rotimi S. O, Olugbemi I. T, Rotimi O. A. Alterations of Genes Involved in Apoptosis and Epigenetic Modulation Associated with Gatifloxacin-Induced Oxidative Stress in Rat Liver. Biomed Pharmacol J 2018;11(1). |
| Copy the following to cite this URL: Rotimi S. O, Olugbemi I. T, Rotimi O. A. Alterations of Genes Involved in Apoptosis and Epigenetic Modulation Associated with Gatifloxacin-Induced Oxidative Stress in Rat Liver. Biomed Pharmacol J 2018;11(1). Available from: http://biomedpharmajournal.org/?p=19712 |
Introduction
Gatifloxacin (1-Cyclopropyl-1,4-dihydro-6-fluoro-8-methoxy-7-(3-methyl-1-piperazinyl)-4-oxo-3-quinolinecarboxylic acid, DB01044) is a member of the fourth-generation fluoroquinolone antibiotic family that is used in treating infection caused by a broad range of microorganisms. It functions by inhibiting the bacterial enzymes DNA gyrase and topoisomerase IV in Gram-positive and Gram-negative organisms, including anaerobes such as, Mycoplasma, Chlamydia, and Legionella and mycobacteria.1 Fluoroquinolones, including gatifloxacin, have been reported to produce several side effects including hepatotoxicity, joint defects and phototoxicity with complications like liver damage, purpura and dysglycemia.2-4 In particular, gatifloxacin has been reported to induce fulminant hepatic failure.3 Olayinka et al reported that exposure of rats to graded doses of gatifloxacin resulted in liver damage characterized by hepatic portal congestion and cellular infiltration by mononuclear cells as well as elevation in the activities of plasma biomarkers of liver damage like alkaline phosphatase, alanine transaminase, aspartate aminotransferase and gamma-glutamyl transferase. These side effects like phototoxicity, cartilage damage and liver damage have been linked to the generation of reactive oxygen species (ROS) leading to oxidative stress.6-8 Fluoroquinolones penetrate neutrophils and enhance their antimicrobial activity by generating ROS.1 Although studies have shown the potential of gatifloxacin to induce oxidative stress, there is dearth of information on whether the induced oxidative stress alters the expression of genes involved in oxidative DNA damage/repair.
Evidences are now emerging that oxidative stress is accompanied with changes in epigenetic signature of the DNA in the liver and that xenobiotics can modulate these changes.9,10 Epigenetic modifications are modifications affecting the expression of DNA without affecting the DNA sequence. These modifications include DNA methylation and histone modifications.11,12 Although it is becoming well-established that various agents can cause epigenetic changes, there is still a dearth of information on the ability of pharmaceuticals to induce epigenetic changes. A recent study has suggested gatifloxacin as an agent that can alter pluripotency by interfering with histone modification signature.13
Therefore, to further elucidate the mechanism of gatifloxacin-induced toxicity in the liver, this study investigated the effect of gatifloxacin on oxidative stress and expression of genes associated with apoptosis, DNA methylation and histone modification in rat liver.
Material and Methods
Chemicals and Reagents
Gatifloxacin was obtained from Sigma-Aldrich, St. Louis, MO. EASYspin Plus® was obtained from Aidlab Biotechnologies Co., Ltd, Beijing, China while RNAhold® and EasyScript® one-step RT-PCR kit was obtained from TransBionovo Co., Ltd. Beijing, China. Other chemicals and reagents were of analytical standard and purchased from Sigma-Aldrich.
Experimental Animals and Procedure
Twenty-five25 inbred male Wistar rats (130±30 g) were used for this research. The animals were maintained on standard 12-h light and dark cycles and granted access to water and feed, ad libitum. The animals were allowed to acclimatize for three weeks before commencement of the experiment. The experiment was approved by the Covenant University Ethical Committee (CU/BIOSCRECU/BIO/2016/004) and carried out according to the guidelines of the committee. Thereafter, the animals were randomly allotted into five5 experimental groups after the initial 2 weeks of acclimatization. Group 1 served as control, while the remaining groups received varying doses of gatifloxacin thus: group 2 (10 mg/kg bw), group 3 (20 mg/kg bw), group 4 (40 mg/kg bw) and group 5 (80 mg/kg bw) orally for 5 days. Twenty-four (24) hours after the last dosage, the rats were anaesthesized under light ether and sacrificed. The liver was excised immediately and its portion for oxidative stress assays were processed appropriately14, while other portions were cryopreserved in RNAhold® for RNA analysis.
Biochemical Analysis
The level of lipid peroxidation was quantified by assessing the concentration of thiobarbituric acid reactive substances (TBARS) as described by Buege and Aust15. Glutathione-S-transferase’s activity was assayed using as described by Habig,16 by measuring the rate of conjugation of glutathione and 1-Chloro-2,4-dinitrobenzene at 340 nm. Superoxide dismutase’s activity was determined as described by Marklund and Marklund,17 by measuring the rate of autooxidation of pyrogallol at 420 nm. The level of reduced glutathione (GSH) concentration was quantified according to the method of Ellman.18 Nitric oxide (NO) concentration was assayed as described by Yucel et al.,19 using the Griess reaction method. The Lowry method was used for the determination of protein concentration as described by Gallagher and Desjardins.20
The tissue level of hydrogen sulfide (H2S) was assayed using the methylene blue formation method as described Shen et al.21. Briefly, 75 μL of liver homogenate was mixed with 250 µL Zn acetate (1%) and 450 µL distilled water for 10 min at room temperature. TCA (10%; 250 µL) was then added, centrifuged at 14,000 g for 10 min. The supernatant was reacted with N,N-dimethyl-p-phenylenediamine sulfate (20 mM/L; 133 μL) and FeCl3 (30 mM/L; 133 μL) and the absorbance was read at 670 nm after 20 min.
Gene Expression Analysis
The expression level of certain apoptotic, DNA methylating and chromatin modifying genes
(Table 1) were quantified using relative reverse transcriptase polymerase chain reaction (RT-PCR) techniques as described by Chaudhry,22with appropriate modifications. In brief, RNA was extracted from the liver using Aidlab® EASYspin Plus® kit following the manufacturer’s guideline. About 500 ng of RNA was used for the RT-PCR using the Transgen® EasyScript® one-step RT-PCR reagent. Briefly, the cDNA synthesis was carried out at 45oC for 30 minutes. This was followed by 35 cycles of PCR amplification, using gene specific primers (GSP) (Table 1), in a C1000 TouchTM Thermal Cycler (BioRad, CA, USA). The cycles consisted of 94oC for 30s, 5min at the annealing temperature of GSP and 1min at 72oC.
Table 1: List of genes studied and the sequences of Gene Specific Primers
| Gene Code | Gene name | Primer Sequence (5′->3′) | Template |
| Prdm2 | PR/SET domain 2 methyltransferase | Forward: CGGATTGGTGTCTGGGCTAC | NM_001077648.1 |
| Reverse: AAGCCAAAGGCCTCTCATCC | |||
| Hdac5 | Histone deacetylase 5 | Forward: TTGCTTGGGCCCTATGACAG | NM_053450.1 |
| Reverse: GGTGAGGTGCGAGTTGGTAA | |||
| Eid3 | EP300 interacting inhibitor of differentiation 3 | Forward: CGCCCAGTTTCTGGTTTTGG | NM_001044304.1 |
| Reverse: TTGGCTCGAGAATTGGCAGT | |||
| Suv39h1 | Suppressor of variegation 3-9 homolog 1 | Forward: GGCGACTCTAGGTTGCAGTG | NM_001106956.1 |
| Reverse: GGCCTTCTGCACCAGGTAAT | |||
| Ehmt2 | Euchromatic histone lysine methyltransferase 2 | Forward: GTCCCTTGTCTCCCCTCCC | NM_212463.1 |
| Reverse: AGAGCCACTCCTGTCTGACT | |||
| Dnmt1 | DNA methyltransferase 1 | Forward: AGAACGGAACACTCTCTCTCACTCA | NM_053354.3 |
| Reverse: AAGCTTCAATCATGGTCTCACTGTC | |||
| Bcl2l1 | Bcl-2-like 1 | Forward: TTTTGCTGAGTTACCGGCGA | NM_001033672.1 |
| Reverse: GCCACAAGGGTAGCCAGAAT | |||
| Casp3 | Caspase 3 | Forward: GAGCTTGGAACGCGAAGAAA | NM_012922.2 |
| Reverse: TAACCGGGTGCGGTAGAGTA | |||
| Casp8 | Caspase 8 | Forward: AGAGAAGCAGCCTATGCCAC | NM_022277.1 |
| Reverse: CCCCGAGGTTTGCTCTTCAT | |||
| Casp9 | Caspase 9 | Forward: GCGCGACATGATCGAGGATA | NM_031632.1 |
| Reverse: TCTCCATCAAAGCCGTGACC | |||
| β-ACTIN | Actin, Beta | Forward: GTCAGGTCATCACTATCGGCAAT | NM_031144.3 |
| Reverse: AGAGGTCTTTACGGATGTCAACGT |
The level of transcription of the genes relative to β-actin was quantified using Image J® software.23,24
Statistical Analysis
Data were expressed as mean ± SEM and analysis of variance was carried out to test for the level of homogeneity at p ˂ 0.05 among the groups. Heterogeneous groups were subjected to Duncan’s multiple range post hoc test.
Results
Gatifloxacin Induced Oxidative Stress in Rat Liver
The levels of GSH, H2S, TBARS and NO as well as the activities of GST and SOD were assessed in the liver of the rats (Figure 1, a-f). Gatifloxacin resulted in a dose-dependent significant (p<0.05) reduction in the levels of hepatic GSH and H2S with a concomitant significant (p<0.05) dose-dependent increase in the levels of TBARS and NO. Although the activity of SOD also followed a dose-dependent significant (p<0.05) decrease, only 40 mg/kg and 80 mg/kg resulted in significant (p<0.05) decrease in GST activity.
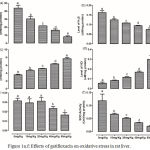 |
Figure 1a-f: Effects of gatifloxacin on oxidative stress in rat liver.
|
(a) levels of liver reduced glutathione, (b) levels of liver hydrogen sulfide (c) level of liver thiobaribituric acid reactive substances, (d) the level of liver nitric oxide, (e) the activity of liver gluthathione-s-transferase and (f) the activity of superoxide dismutase.
Bars represent mean ± SEM (n=6). Bars with different statistical markers are significantly different at p<0.05.
Gatifloxacin Modulated the Expression of Genes Involved in Epigenetic Regulations in Rat Liver
The level of expression of Dnmt1 was significantly (p<0.05) increased only in the liver of rats treated with 80 mg/kg (Figure 2a). However, gatifloxacin administration resulted in significant (p<0.05) decrease in the expression of Hdac5 at 10 mg/kg; though, none of the higher dosages significantly altered its expression (Figure 2b). While a dose-dependent significant (p<0.05) increase was observed in level of expression of Ehmt2 and Suv39h1, only 80 mg/kg significantly (p<0.05) increased the level of expression of Eid3 and Prdm2 (Figure 2, c-f).
 |
Figure 2a.f: Effects of gatifloxacin on genes involved in epigenetic regulations.
|
(a) The levels of expression of Dnmt1 in the liver, (b) the levels of expression of Hdac5 in the liver, (c) the levels of expression of Ehmt2 in the liver, (d) the levels of expression of Eid3 in the liver, (e) the levels of expression of Prdm2 in the liver and (f) the levels of expression of Suv39h1 in the liver.
Bars represent mean ± SEM (n=6). Bars with different statistical markers are significantly different at p<0.05.
Gatifloxacin Modulated the Expression of Genes Involved in Apoptosis in Rat Liver
The expression of Bcl2l1, Casp3, Casp8 and Casp9 are depicted in figure 3 (a-d). There was a significant (p < 0.05) increase in the expression of Bcl2l1 in the liver of rats treated with 20 mg/kg gatifloxacin with a further increase in group treated with 80 mg/kg. Although a significant (p < 0.05) dose-dependent increase was observed in the levels of expression of Casp8 and Casp9, the increase in the dosage of gatifloxacin beyond 10 mg/kg had no significant (p > 0.05) effect on the expression of Casp3.
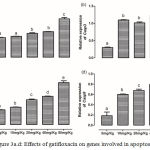 |
Figure 3a.d: Effects of gatifloxacin on genes involved in apoptosis.
|
(a) The levels of expression of Bcl2l1 in the liver, (b) the levels of expression of caspase 3 in the liver, (c) the levels of expression of caspase 8 in the liver and (d) the levels of expression of caspase 9 in the liver.
Bars represent mean ± SEM (n=6). Bars with different statistical markers are significantly different at p<0.05.
Discussion
The ability of gatifloxacin to induce hepatic oxidative stress in rats was investigated by analyzing the levels of TBARS, H2S, NO and GSH as well as the activities of GST and SOD. Our findings showed that gatifloxacin induced oxidative stress in a dose-depend manner. Kumbhar et al.,6 reported a similar dose-dependent induction of oxidative stress in rabbits treated with gatifloxacin. In this study, as well as that of Talla and Veerareddy,1 oxidative stress was characterized by decreased GSH and H2S levels, and activities of GST and SOD with an associated increase in the level of nitric oxide and TBARS. As part of their bactericidal mechanism, fluoroquinolones trigger the transcriptional activation of iron transport genes and enhance the Fenton reaction resulting in the production of ROS.25 Also, a recent report by Pan et al26 showed that fluoroquinolones could decrease SOD activity by forming a complex through hydrogen bonds and van der Waals forces resulting in inhibition and subsequent oxidative stress. Nitric oxide (NO) and H2S are biological messengers that contribute to many physiological processes and play important roles in response to xenobiotics.27 Although NO is a potent antioxidant that rapidly neutralizes superoxide anion, it is subsequently converted to prooxidant and its biphasic action of protection at low concentrations and oxidative killing of cells at high concentration has been reported.28 On the other hand, H2S regulates GSH biosynthesis from GSSG.29 The depletion of hepatic H2S metabolism has been implicated in the pathogenesis of many liver diseases29 and our findings suggests that it could also be involved in the pathogenesis of gatifloxacin-induced liver damage.
The interaction between fluoroquinolones and iron also alters the epigenetic signature of the cell through inhibition of dioxygenases that require iron as a co-factor30. Such epigenetic alterations may include DNA methylation and histone modifications. Our findings showed that gatifloxacin altered the expressions of Dnmt1, Hdac5, Prdm2, Eid3, Suv39h1 and Ehmt2. The Dnmt1 is responsible for methylating cytosine residues of DNA and aberrant methylation patterns, resulting from increased Dnmt1 expression, are associated with etiology of certain diseases, especially liver disorders.31,32 On the other hand, histone modification could occur via methylation or deacetylation. Histone methylation is achieved by an array of methyltransferases which include Prdm2, Eid3, Suv39h1 and Ehmt2 33,34 that methylate the histone lysine residues. Therefore, these methyltransferases are key components in cellular processes, and alteration in their expression is associated with pathogenesis.34 Histone deacetylase is another protein involved in this mechanism and it deacetylates the lysine residues on the N-terminal of core histones.35,36 Previous studies have reported certain quinolones to inhibit this enzyme 35 and such inhibition or decrease in expression of Hdac5 has been reported to induce growth arrest, differentiation, and/or apoptotic cell death.36,37
Interestingly, the induction of apoptosis by certain fluoroquinolones has been reported.38,39 In this present study, gatifloxacin administration resulted in a dose-dependent upregulation of Bcl2l1and caspases 3,8 and 9. Previous studies have reported increase in expression of these proteins by a novel bis-fluoroquinolone compound,40 levofloxacin41 and ciprofloxacin.42
Conclusion
Our findings therefore demonstrated that gatifloxacin-induced oxidative stress is associated with alterations in expression of epigenetic and proapoptotic genes. These alterations in gene expression could be part of the underlining mechanisms resulting in hepatotoxicity of gatifloxacin.
References
- Talla V., Veerareddy P. Oxidative stress induced by fluoroquinolones on treatment for complicated urinary tract infections in Indian patients. J Young Pharm. 2011;3:304-9.
CrossRef - Park-Wyllie L. Y., Juurlink D. N., Kopp A., et al. Outpatient gatifloxacin therapy and dysglycemia in older adults. N Engl J Med. 2006;354:1352-61.
CrossRef - Coleman C. I., Spencer J. V., Chung J. O., Reddy P. Possible gatifloxacin-induced fulminant hepatic failure. Ann Pharmacother. 2002;36:1162-7.
CrossRef - Masood I., Bhargava R., Ahmed Z., Sharma D., Rehman S., Amin S. Gatifloxacin-induced purpura—an unusual adverse drug reaction. J Indian Acad Clin Med. 2005;6:239-40.
- Olayinka E., Ore A., Adeyemo O. Alterations in biochemical indices and antioxidant status in rats following treatment with gatifloxacin. British Journal of Pharmaceutical Research. 2015;6:293-305.
CrossRef - Kumbhar G., Khan A., Rampal S. Evaluation of gatifloxacin for its potential to induce antioxidant imbalance and retinopathy in rabbits. Human & experimental toxicology. 2014:0960327114530743.
- Afolabi I. S., Osikoya I. O., Fajimi O. D., et al. Solenostemon monostachyus, Ipomoea involucrata and Carica papaya seed oil versus Glutathione, or Vernonia amygdalina methanolic extracts of novel plants for the management of sickle cell anemia disease. BMC complementary and alternative medicine. 2012;12:262.
CrossRef - Adebayo H. A., Song F. H., Liu X. T., et al. Citrullus Ianatus Extract Reverses Oxidative and Haematological Dysfuntion in Carbon Tetrachloride Induced Liver Damaged Rats. International Journal of Pharmacology. 2014;10:218-24.
CrossRef - Nishida N., Kudo M. Oxidative stress and epigenetic instability in human hepatocarcinogenesis. Digestive Diseases. 2013;31:447-53.
CrossRef - Shukla S. D., Lim R. W. Epigenetic effects of ethanol on the liver and gastrointestinal system. Alcohol Res. 2013;35:47-55.
- Csoka A. B., Szyf M. Epigenetic side-effects of common pharmaceuticals: a potential new field in medicine and pharmacology. Med Hypotheses. 2009;73:770-80.
CrossRef - Osamor V. C., Chinedu S. N., Azuh D. E., Iweala E. J., Ogunlana O. O. The interplay of post-translational modification and gene therapy. Drug Des Devel Ther. 2016;10:861-71.
CrossRef - Bhanu N. V., Sidoli S., Garcia B. A. Histone modification profiling reveals differential signatures associated with human embryonic stem cell self‐renewal and differentiation. Proteomics. 2016;16:448-58.
CrossRef - Graham J. Homogenization of mammalian tissues. The Scientific World Journal. 2002;2:1626-9.
CrossRef - Buege J. A., Aust S. D. [30] Microsomal lipid peroxidation. Methods Enzymol. 1978;52:302-10.
CrossRef - Habig W. H., Pabst M. J., Jakoby W. B. Glutathione S-transferases the first enzymatic step in mercapturic acid formation. Journal of biological Chemistry. 1974;249:7130-9.
- Marklund S., Marklund G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur J Biochem. 1974;47:469-74.
CrossRef - Ellman G. L. Tissue sulfhydryl groups. Archives of biochemistry and biophysics. 1959;82:70-7.
CrossRef - Yucel H., Ozaydin M., Dogan A., et al. Plasma concentrations of asymmetric dimethylarginine, nitric oxide and homocysteine in patients with slow coronary flow. Scandinavian journal of clinical and laboratory investigation. 2012;72:495-500.
CrossRef - Gallagher S. R., Desjardins P. Quantitation of nucleic acids and proteins. Current Protocols Essential Laboratory Techniques. 2011;2(36)1-2.
CrossRef - Shen X., Pattillo C. B., Pardue S., Bir S. C., Wang R., Kevil C. G. Measurement of plasma hydrogen sulfide in vivo and in vitro. Free Radical Biology and Medicine. 2011;50:1021-31.
CrossRef - Chaudhry M. A. An exercise to estimate differential gene expression in human cells. Biochemistry and Molecular Biology Education. 2006;34:116-20.
CrossRef - Abràmoff M. D., Magalhães P. J., Ram S. J. Image processing with Image. J. Biophotonics international. 2004;11:36-42.
- Rotimi S. O., Bankole G. E., Adelani I. B., Rotimi O. A. Hesperidin prevents lipopolysaccharide-induced endotoxicity in rats. Immunopharmacol Immunotoxicol. 2016:1-8.
CrossRef - Ferrándiz M., Martín-Galiano A., Arnanz C., Zimmerman T., dela Campa A. Reactive oxygen species contribute to the bactericidal effects of the fluoroquinolone moxifloxacin in Streptococcus pneumoniae. Antimicrobial agents and chemotherapy. 2016;60:409-17.
CrossRef - Pan X., Qin P., Liu R., Li J., Zhang F. Molecular mechanism on two fluoroquinolones inducing oxidative stress: evidence from copper/zinc superoxide dismutase. RSC Advances. 2016;6:91141-9.
CrossRef - Magierowski M., Magierowska K., Kwiecien S., Brzozowski T. Gaseous mediators nitric oxide and hydrogen sulfide in the mechanism of gastrointestinal integrity protection and ulcer healing. Molecules. 2015;20:9099-123.
CrossRef - Joshi M. S., Ponthier J. L., Lancaster J. R. Cellular antioxidant and pro-oxidant actions of nitric oxide. Free Radical Biology and Medicine. 1999;27:1357-66.
CrossRef - Mani S., Cao W., Wu L., Wang R. Hydrogen sulfide and the liver. Nitric Oxide. 2014;41:62-71.
CrossRef - Badal S., Her Y. F., Maher L. J. Nonantibiotic effects of fluoroquinolones in mammalian cells. Journal of Biological Chemistry. 2015;290:22287-97.
CrossRef - Kondo Y., Kanai Y, Sakamoto M., Mizokami M., Ueda R., Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis a comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970-9.
CrossRef - Robertson K.. D. DNA methylation and human disease. Nature Reviews Genetics. 2005;6:597-610.
CrossRef - Trievel R. C. Structure and function of histone methyltransferases. Critical Reviews™ in Eukaryotic Gene Expression. 2004;14.
- Mozzetta C., Boyarchuk E., Pontis J., Ait-Si-Ali S. Sound of silence: the properties and functions of repressive Lys methyltransferases. Nature Reviews Molecular Cell Biology. 2015;16:499-513.
CrossRef - Meinke P. T., Colletti S. L., Doss G., et al. Synthesis of apicidin-derived quinolone derivatives: parasite-selective histone deacetylase inhibitors and antiproliferative agents. Journal of medicinal chemistry. 2000;43:4919-22.
CrossRef - Haberland M., Montgomery R. L., Olson E. N. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Reviews Genetics. 2009;10:32-42.
CrossRef - Sharma P. C., Chaudhary M., Sharma A., Piplani M., Rajak H., Prakash O. Insight view on possible role of fluoroquinolones in cancer therapy. Current topics in medicinal chemistry. 2013;13:2076-96.
CrossRef - Song M., Wu H., Wu S., et al. Antibiotic drug levofloxacin inhibits proliferation and induces apoptosis of lung cancer cells through inducing mitochondrial dysfunction and oxidative damage. Biomedicine & Pharmacotherapy. 2016;84:1137-43.
CrossRef - Liang H. X., Fan Y. Y., Zhang Y., Huangfu C. S., Hu G. Q., Liu B. Benzaldehyde levofloxacin schiff baseinduced apoptosis of human hepatocarcinoma cells. Int J Clin Exp Med. 2016;9:1314-21.
- Ma Y. C., Wang Z. X., Jin S. J., et al. Dual Inhibition of Topoisomerase II and Tyrosine Kinases by the Novel Bis-Fluoroquinolone Chalcone-Like Derivative HMNE3 in Human Pancreatic Cancer Cells. PLoS One. 2016;11:e0162821.
- Bidell M. R., Lodise T. P. Fluoroquinolone‐Associated Tendinopathy: Does Levofloxacin Pose the Greatest Risk? Pharma cotherapy. The Journal of Human Pharmacology and Drug Therapy. 2016.
- Herold C., Ocker M., Ganslmayer M., Gerauer H., Hahn E., Schuppan D. Ciprofloxacin induces apoptosis and inhibits proliferation of human colorectal carcinoma cells. British journal of cancer. 2002;86:443-8.
CrossRef







