Rajitha Galla1* , Vidya Rani Murthi1
, Vidya Rani Murthi1 , Yasmintaj Shaik1
, Yasmintaj Shaik1 , Saritha Karnati2
, Saritha Karnati2 , Umakanth Naik Vankadoth3
, Umakanth Naik Vankadoth3 and Umamaheswari Amineni3
and Umamaheswari Amineni3
1Institute of Pharmaceutical technology, Sri Padmavati Mahila Visvavidyalayam, Tirupati, Andhra Pradesh, India
2KVSR Siddhartha College of Pharmaceutical Sciences, Vijayawada, 520010, Andhra Pradesh, India
3Department of Bioinformatics, Sri Venkateswara Institute of Medical Sciences, Tirupati-517502, Andhra Pradesh, India
Corresponding Author E-mail: grajitha@spmvv.ac.in
DOI : https://dx.doi.org/10.13005/bpj/2908
Abstract
Since 2019, the SARS-CoV-2 infection has continued to cause significant human suffering. Numerous investigations into the viral pathogenesis have led to converging conclusions on how the virus enters and spreads within the host. The main protease (Mpro) of coronaviruses has been considered as an attractive therapeutic target because of its important role in processing polyproteins translated from viral RNA. Many studies discovered that phytoconstituents possess potent antiviral activities. Hence, in the present work, 439 co-crystal ligands of SARS-CoV-2 Mpro were collected and docked with Mpro of SARS-CoV-2 (PDB ID:7AEH) to identify best crystal ligand. Among all the crystal ligands collected, HF0 (7-O-methyl-dihydromyricetin) showed good XP G score -7.872 Kcal/Mol and it was selected as reference to compare the docking scores of phytoconstituents. Then, molecular docking study was performed for 274 antiviral phytoconstituents from various medicinal plants against Mpro of SARS-CoV-2. Molecular docking studies found that seven phytoconstituents exhibited better docking scores than best co-crystal ligand HF0. Among the seven best docked phytoconstituents, 3,4-dicaffeoylquinic acid showed good interactions with key amino acid residues in substrate binding site of Mpro with XPG Score –9.721 Kcal/Mol. Qikprop results indicated that the most phytoconstituents have demonstrated favourable pharmacological characteristics. Interaction fingerprint analysis revealed that all the seven best docked phytoconstituents of the present study bound to Glu166, key residue situated in the centre of the substrate binding site of Mpro resulting in the reduction of the catalytic activity of main protease thus blocking the replication of SARS-CoV-2.
Keywords
Antiviral; Interaction fingerprint; Molecular docking; Main protease; Phytoconstituents; SARS-CoV-2
Download this article as:| Copy the following to cite this article: Galla R, Murthi V. R, Shaik Y, Karnati S, Vankadoth U. N. Amineni U. Computational Investigation of Bioactive Phytoconstituents as Sars-Cov-2 Main Protease Inhibitors Through Molecular Docking and Interaction Fingerprint Studies. Biomed Pharmacol J 2024;17(2). |
| Copy the following to cite this URL: Galla R, Murthi V. R, Shaik Y, Karnati S, Vankadoth U. N. Amineni U. Computational Investigation of Bioactive Phytoconstituents as Sars-Cov-2 Main Protease Inhibitors Through Molecular Docking and Interaction Fingerprint Studies. Biomed Pharmacol J 2024;17(2). Available from: https://bit.ly/4c21an4 |
Introduction
The COVID-19 pandemic caused by SARS-CoV-2 (severe acute respiratory syndrome-coronavirus-2) is a worldwide health emergency. Since last two decades, the two main members of the Coronaviridae family that periodically cause pneumonia and respiratory syndromes, SARS-CoV and MERS-CoV, have drawn attention worldwide. SARS-CoV-2 belongs to Coronavirinae subfamily of Coronaviridae and is a very significant and dreadful virus 1-4. According to the World Health Organization’s global situation update, as of 10 Jan 2023, there have been 660,131,952 COVID-19 confirmed cases, including 6,690,473 deaths, reported to WHO 5. These findings demonstrate that the rise of this viral contagious disease, which now holds a significant position in global incidence of transmissible diseases, is continuing in developing nations. Despite all the improvements in conventional and contemporary medicine, many attempts to control this pandemic had negative health effects on people. As a result, conventional medicine has become more interested in providing healthcare services. Moreover, at least 25% of all modern medications are thought to be derived directly or indirectly from traditional medicines, primarily through the integration of cutting-edge technologies with age-old knowledge. A wide range of natural extracts and phytoconstituents have been investigated for their ability to act as drug-like molecules against the SARS-CoV-2 proteases and found to possess good inhibitory activities 6-10. Although few drugs like remdesivir gained urgent approval, search for more safer & efficient treatment is still required 11, 12.
Mpro, papain-like protease, RdRp (RNA dependent RNA polymerase) are few of the SARS-CoV-2 druggable targets that have been identified. The papain-like protease is capable of recognizing ubiquitin’s C-terminal region. So, papain-like protease inhibitors would also inhibit deubiquitinases of host cell, which would make drug-discovery against papain-like protease complicated. In contradistinction, the main protease particularly cleaves polypeptide sequence after glutamine. Enzymes like RdRp cannot completely operate without previous proteolytic release, thus making Mpro as a main enzyme in virus replication cycle. The SARS-CoV-2 Mpro is a cysteine protease, that shares 96% amino acid identity with SARS-CoV Mpro 13-19. Mpro forms a homodimer that consists of 306 amino acid residues in each monomer. Each monomer consists of three domains: Domain I (8– 101 residues), domain II (102–184 residues) and Domain III (201–306 residues). Domain I & II were primarily made up of antiparallel β-barrel whereas domain III was made up of α-helices. Domains II & III are connected by loop of 15 residues (residues 185– 200). The protomers attach to one another through an N-terminal finger (residues 1-7) that forms a substrate-binding site in a cleft between domains I and II and connects domains II & III. Four pockets (S1′, S1, S2 & S3) make up the substrate-binding site of Mpro, with the S10 pocket consisting of a catalytic dyad. This catalytic dyad is composed of Cys145 & His41and placed in a gap between domains I & II. Domain III promotes the formation of the dimer, by salt-bridge interaction of Glu290 form one monomer with Arg4 of the other. Dimerization is essential for enzyme’s catalytic activity, as the N-finger of each monomer interacts with Glu166 of the other to shape the S1 pocket of substrate-binding site. The N-finger is compressed between domains II & III of the parent protomer and domain II of the other in order to reach the interaction site. Mpro (nsp5) auto cleaves between non-structural protein 4 (nsp 4) & nsp6, and then cleaves polyproteins 1a & 1ab at 11 specific sites; with a unique cleavage specificity Leu-Gln↓ (Ser, Ala, Gly), to generate 12 mature nsp and probably, functional intermediates for viral RNA replication and transcription 20-25.
Molecular docking is important method in structural molecular biology and computer-aided drug discovery that predicts the predominant interactions between a ligand and protein with a known three-dimensional structure 26. Accurate ADME property predictions can prevent the needless testing of compounds that will ultimately fail before costly experimental processes, thus allowing for informed decisions about a molecule’s suitability. In contrast to fragment-based techniques, QikProp can predict attributes for both molecules with new scaffolds and analogues of well-known medications with comparable accuracy. QikProp bases its predictions on the whole 3D molecular structure 27.
Hence, in order to explore potent Mpro inhibitors, molecular docking study was performed for various phytoconstituents from different antiviral medicinal plants against SARS-CoV-2 Mpro and the phytoconstituents pharmacological descriptors and ADME properties were also predicted. In addition, interaction fingerprint was also generated for best docked phytoconstituents and SARS-CoV-2 Mpro complexes and compared with that of reference crystal ligand to identify any similarities in binding interactions.
Materials and Methods
Protein preparation
A total of 602 crystal structures of SARS-CoV-2 main protease bound with various inhibitors are currently uploaded to the Protein Data Bank (PDB) database, containing 439 crystal ligands. Among the available structures of SARS-CoV-2 Mpro, the three-dimensional structure of SARS-CoV-2 main protease in a covalent complex with a pyridine derivative of ABT-957, compound 1 (PDB: 7AEH) with resolution 1.30 A0; was considered in this study to propose novel SARS-CoV-2 Mpro inhibitors through molecular docking studies (https://www.rcsb.org/).
Target protein structure was imported to Maestro v11.1 (Schrödinger LLC, 2019) (Sun Microsystems, Schrodinger, New York, USA) workstation running on CentOS 6. Protein preparation tasks were performed with the protein preparation wizard and prepared before docking to add hydrogen atoms, adjustment of protonation states for ionizable molecules, formal charge and bond order correction, expelling atomic clashes, modification of tautomeric forms and repositioning of reorientable hydrogens and other operations which were not part of X-ray crystal structure refinement process. At neutral pH, the protein structure minimization was done using the OPLS-2005 force field, by converging the heavy atoms to RMSD of 0.3A0. Based on the already bound inhibitor in the crystal structure, the binding site on the receptor molecule was identified and a grid box of 10A0 ×10 A0 × 10 A0 was generated around the substrate binding site residues of Mpro using Glide v7.1., residues were cross validated using PDBsum 28.
Identification of best crystal ligand through molecular docking studies
All the 439 crystal ligands of covid-19 Mpro crystal structures which were available in PDB, were collected and prepared using ligprep module of schrodinger. All the prepared and optimized ligands were subjected to molecular docking studies with selected target by Glide XP docking 28.
Collection and preparation of phytoconstituents
A total of 274 phytoconstituents inhibitors, known for their antiviral activity were sourced from literature and were extracted from PubChem and DrugBank v5.0 databases 29-176. DrugBank v5.0, a specialized bioinformatics and drug cheminformatics resource, provided comprehensive data encompassing chemical, pharmacological, and pharmaceutical details for the identified compounds. Subsequently, the structures of the selected phytoconstituents were meticulously prepared, involving the creation of three-dimensional geometries, assignment of proper bond orders, and the generation of accessible tautomer and ionization states. This preparatory phase was executed using the LigPrep module. For molecular docking studies, the Schrödinger Epik module was employed in conjunction with LigPrep to ensure a robust analysis of the interactions between the phytoconstituents and their target molecules 177.
Molecular docking of phytoconstituents with Mpro of SARS-CoV-2
The binding affinities of selected phytoconstituents with SAS-CoV-2 Mpro were analyzed by performing molecular docking studies for identification of the best phytoconstituent with good binding affinity. The prepared phytoconstituents were docked into 7AEH. The binding affinity between the target and phytoconstituents was studied using the grid-based ligand docking with energetic (GLIDE) XP (extra precision) docking technique. Then the prepared phytoconstituents were docked into the grid utilizing Monte Carlo-based simulated algorithm minimization approach. Glide Scores (Gscore) and Molecular Mechanics-Generalized Born Surface Area (MM-GBSA) were used to analyze calculations for the binding free energies, affinities, orientation, and ranking of the protein ligand complex utilizing the Prime module of Schrodinger suite 2021-2 that incorporates the OPLS3 force field and VSGB dissolvable model to look through calculations. Ten poses were created for each ligand during XP docking, and the best pose was preserved after post-docking minimization 178.
Prediction of pharmacological descriptors and ADME properties
Selected phytoconstituents were subjected to QikProp module of Schrödinger suite to predict pharmacological, ADME properties. Additionally, SASA and other related values were also predicted by Schrödinger suite 179, 180.
Generation of Interaction fingerprints for best docked phytoconstituents and best crystal ligand
Docking interactions of best docked phytoconstituents were further analyzed using interaction fingerprint analysis to observe if they shared any similarities with the best co-crystal ligand’s interactions. For the best docked compounds and co-crystal ligand docked complexes, an interaction fingerprint was created that translates the three-dimensional structural binding information from a protein-ligand complex into a one-dimensional binary string. Each fingerprint reflects “structural interaction profile” of complex that can be utilized to organize, analyze, and visualize the extensive amount of information encoded in ligand receptor complexes. Value 1 indicates that the specified interaction is established, while 0 indicates that there is no such interaction 180.
Results and Discussion
Protein preparation
The three-dimensional co-crystal structure of SARS-Cov-2 Mpro (PDB I’D: 7AEH) was retrieved from PDB and prepared. The substrate-binding site residues were specified within the 4 Å region of co-crystal ligand using PDBsum. The substrate-binding pocket of Mpro complex comprises residues such as Thr 25, Thr 26, His 41, Phe 140, Leu 141, Asn 142, Gly 143, Ser 144, Cys 145 His 163, His 164, Glu 166, Gln 189 and Thr 190 within the 4 Å region around the crystal ligand 181.
Molecular docking study for the identification of best co-crystal ligand of Mpro
All the prepared crystal ligands were docked with SARS-Cov-2Mpro and results were represented in Table 1. Among all the crystal ligands, HF0 exhibited good docking score -7.872 Kcal/Mol and selected as reference crystal ligand.
Table 1: List of best docked crystal ligands of SARS-CoV-2 and docking scores with SARS-CoV-2 Mpro (PDB: 7AEH).
|
S.No. |
Crystal ligand Code |
Crystal ligand Name |
PDB I’D |
Docking score |
Interacting Amino acids
|
N |
|
|
XP G Score |
MMGBSA |
||||||
|
1. |
HF0 |
7-O-methyl-dihydromyricetin |
7DPV |
-7.872 |
-67.5446 |
Hie 164, Gln 189 |
2 |
|
2. |
MYC |
Myricetin |
7B3E |
-7.812 |
-60.0807 |
Hie 164, Glu 166, Gln 189 |
3 |
|
3. |
HER |
7-O-methyl-myricetin |
7DPU |
-7.576 |
-67.1016 |
Hie 164, Gln 189 |
2 |
|
4. |
RVW |
(2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-sulfanyl-oxane-3,4,5-triol |
7ARF |
-6.512 |
-45.2359 |
Glu 166, Thr 190 |
2 |
|
5. |
93J |
Pelitinib |
7AXM |
-6.423 |
-81.9294 |
Glu 166 |
2 |
*N: No. of interactions
Ligand preparation
The structures of selected phytoconstituents were preparedprior to molecular docking using Ligprep module of the Schrodinger. The preparation was conducted at a pH of 7.0 ± 2, employing the OPLS_3 forcefield, and involved the enhancement of protonation states and consideration of ligand stereochemical nature. Energy minimization was performed as part of the ligand preparation process.
Molecular Docking
Molecular docking study was performed for selected phytoconstituents against SARS-CoV-2 Mpro to identify potent inhibitors. All the selected phytoconstituents and crystal ligands were docked into the substrate-binding site of Mpro and binding energies were represented in Table 2. Furthermore, protein-ligand binding energies revealed that seven phytoconstituents strongly bind to substrate binding site of main protease with more binding affinity than best co-crystal ligand HF0. Among all the selected phytoconstituents, 3,4-dicaffeoylquinic acid showed better XP G score -9.721 Kcal/Mol than best co-crystal ligand HF0 (-7.872kcal/Mol). It was clearly observed thatall the best docked phytoconstituents and best crystal ligand HF0 showed at least two H-bond interactions with substrate binding site.
Table 2: Docking scores of best docked phytoconstituents and best crystal-ligand HF0 against SARS-CoV-2 Mpro
|
S. No. |
Phytoconstituents |
Docking scores with Mpro (Kcal/Mol) |
Interacting Amino Acids |
N |
|
|
XP G Score |
MMGBSA (∆G) |
||||
|
1. |
3,4-dicaffeoylquinic acid |
-9.721 |
-77.4562 |
Glu 166, Gln 189, Thr 190, Ala 191 |
5 |
|
2. |
Epigallocatechingallate |
-8.925 |
-73.8888 |
Asn 142, Hie 164, Glu 166, Gln 189, Thr 190 |
6 |
|
3. |
Isomangiferin |
-8.747 |
-55.4927 |
Ser 46, Asn 142, Hie 164, Glu 166, Gln 189 |
6 |
|
4. |
Rosemarinic acid |
-8.477 |
-60.0924 |
Glu 166, Gln 189 |
3 |
|
5. |
Rutin |
-8.405 |
-52.1528 |
Hie 164, Glu 166, Pro 168 |
4 |
|
6. |
Gnetupendin |
-7.99 |
-61.1349 |
Glu 166, Thr 190 |
3 |
|
7. |
Amarogentin |
-7.989 |
-73.8429 |
Asn 142, Glu 166, Gln 189, Thr 190 |
7 |
|
8. |
HF0 (Best crystal ligand) |
-7.872 |
-67.5446 |
Hie 164, Gln 189 |
2 |
*N: No. of interactions
Prediction of Pharmacological descriptors and ADME properties
Absorption, distribution, metabolism, excretion, and toxicity (ADMET) characteristics were estimated using qikprop module to measure the phytoconstituents’ potential as therapeutics. Various properties like mol. weight; volume; globularity descriptor; hydrophobic component of SASA; total solvent accessible surface area; weakly polar component of SASA; π (carbon and attached hydrogen) component of SASA; ionization potential; H-Bond acceptors & donors; Log Po/w; brain/blood partition coefficient; human serum albumin binding; No. of violations of Lipinski’s rule of five; No. of violations of Jorgensen’s rule of three; Predicted skin permeability etc. These results indicated that pharmacological descriptors of the most of phytoconstituents were found to be with in the acceptable range for 95% of known drugs (Table 3 & 4).
Table 3: Pharmacological descriptors of best docked phytoconstituents and best crystal ligand HF0
|
Entry Name |
MW |
volume |
glob |
SASA |
WPSA |
PISA |
FOSA |
IP(eV) |
HBD |
HBA |
|
3,4-dicaffeoylquinic acid |
516.4 |
1516.8 |
0.74617 |
855.6 |
0 |
292.77 |
133.5 |
9.08 |
7 |
11.45 |
|
Epigallocatechin gallate |
458.3 |
1255.9 |
0.80723 |
697.3 |
0 |
223.98 |
50.6 |
9.02 |
8 |
8.75 |
|
Isomangiferin |
422.3 |
1134.0 |
0.83403 |
630.5 |
0 |
147.25 |
97.6 |
8.74 |
7 |
13 |
|
Rosemarinic acid |
360.3 |
1106.8 |
0.80032 |
646.5 |
0 |
251.26 |
61.8 |
8.82 |
5 |
7 |
|
Rutin |
610.5 |
1583.3 |
0.79977 |
821.4 |
0 |
196.71 |
219.2 |
9.19 |
9 |
20.5 |
|
Gnetupendin B |
380.3 |
1169.5 |
0.81961 |
655.0 |
0 |
267.79 |
138.4 |
8.57 |
5 |
4.5 |
|
Amarogentin |
586.5 |
1558.2 |
0.83405 |
779.3 |
0 |
259.95 |
218.0 |
9.18 |
5 |
16.4 |
|
HF0 |
334.2 |
953.3 |
0.84958 |
551.3 |
0 |
161.38 |
118.6 |
9.05 |
4 |
7.2 |
*MW: Mol. Weight (130-725); Volume (500-2000); glob: Globularity (0.75-0.95); SASA: Total solvent accessible surface area (300-1000); WPSA: Weakly polar component of SASA (0-175); PISA: π (carbon and attached hydrogen) component of SASA (0-450); FOSA:Hydrophobic component of SASA (0-750) ; IP: Ionization potential; HBD: H-Bond donors (0-6); HBA: H-bond acceptors (2-20) 186
Table 4: ADME properties of best docked phytoconstituents and best crystal ligand HF0
|
Entry Name |
QP logPo/w |
QPlog BB |
QPlog Khsa |
Rule Of Five |
Rule Of Three |
QPlog Kp |
QPlogHERG |
QPPCaco |
|
3,4-dicaffeoylquinic acid |
0.973 |
-5.481 |
-0.598 |
3 |
1 |
-6.96 |
-5.228 |
0.213 |
|
Epigallocatechingallate |
-0.261 |
-4.364 |
-0.441 |
2 |
2 |
-7.561 |
-5.719 |
0.971 |
|
Isomangiferin |
-1.814 |
-3.676 |
-0.877 |
2 |
2 |
-7.245 |
-4.92 |
2.179 |
|
Rosemarinic acid |
1.183 |
-3.571 |
-0.544 |
0 |
1 |
-5.724 |
-4.154 |
1.726 |
|
Rutin |
-2.329 |
-4.567 |
-1.327 |
3 |
2 |
-6.86 |
-5.71 |
1.414 |
|
Gnetupendin B |
2.508 |
-2.568 |
0.085 |
0 |
1 |
-4.105 |
-5.728 |
43.341 |
|
Amarogentin |
0.263 |
-3.108 |
-0.687 |
3 |
2 |
-4.909 |
-5.457 |
13.753 |
|
HF0 |
0.278 |
-2.304 |
-0.402 |
0 |
1 |
-5.376 |
-4.735 |
26.471 |
* QP logPo/w: Octanol/water partition coefficient (-2.0 to 6.5); QPlog BB: Brain/blood partition coefficient (-3.0 to 1.2); QPlog Khsa: Binding to human serum albumin Prediction (-1.5 to 1.5); Rule Of Five: No. of violations of lipinski’s rule (Max 4); Rule of three: No. of violations of Jorgensen’s rule (Max 3); QPlog Kp:Predicted skin permeability (-8.0- -1) ; QPlog HERG: Predicted value of IC50 for blockage of HERG K+ channels (concern below -5); QPPCaco: Predicted apparent Caco-2 cell permeability (<25 poor, >500 great) mm/sec 186
Generation of Interaction fingerprints for best docked phytoconstituents and best crystal ligand
To describe the 3D protein-ligand interactions of the best docked phytoconstituents with SARS-CoV-2 Mpro, a 9-bit interaction fingerprint was generated. Pharmacophore feature is represented by each bit of the fingerprint, which is denoted by the numbers 0 (absence of specified interaction) or 1 (presence of specified interaction) (Table 5).
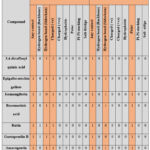 |
Table 5: Interaction fingerprint for docked complexes of the best docked phytoconstituents and best co-crystal ligand HF0 with Mpro of SARS-CoV-2 |
Interactions of best docked phytoconstituents were compared with that of best crystal ligand HF0. It was observed that all the best docked phytoconstituents binds to Mpro through at least one hydrogen (H)-bond with Glu 166 residue with better binding affinity than HF0 (Fig 1-7). In addition, 3,4-dicaffeoylquinic acid, epigallocatechingallate, isomangiferin, rosemarinic acid and gnetupendin B showed H-bond interactions with Gln 189 similar to that of best co-crystal ligand HF0. Docking results indicated that these H-bond interactions with residues Glu166 and Gln189 are very important as any interaction with Glu166 can lead to inactive monomer formation that interferes with Mpro catalytic activity; and also, Glu166 anchor holds the ligand firmly to the central region of binding site, that facilitates the multiple interactions with remaining residues 182-184. Further, the H-bond interaction with Gln189 helps in inhibitor recognition through increasing S2 subsite plasticity 185. In addition to interaction with Glu 166 and Gln 189, 3,4-dicaffeoylquinic acid exhibited H-bond interactions with backbone residues of Thr 190 and Ala 191 (Fig 1). Epigallocatechingallate displayed H-bond interactions with backbone residues of Hie 164, Thr 190 and with side chain residues of Asn 142 (Fig 2). Isomangiferin showed H-bond interactions with the backbone residues of Hie 164; with side chain residues of Ser 46, Asn 142 (Fig 3). Rutin displayed H-bond interactions with backbone residues of Hie 164, Pro 168 (Fig 5). Gnetupendin B exhibited H-bond interaction with backbone residue of Thr 190 (Fig 6). Amarogentin showed H-bond interactions with side chain residues of Asn 142 and with backbone residue of Thr 190 (Fig 7). Best crystal ligand HF0 exhibited H-bond interactions with backbone residue of Hie 164; side chain residue of Gln 189 of Mpro (Figure 8). The results of the present study indicated that these best docked phytoconstituents can efficiently bind with key amino acid residues such as Glu 166, Gln 189 in the substrate binding site of SARS-CoV-2 Mpro with more binding affinity than the reference HF0, which can result in the formation of inactive monomer thus inhibiting the catalytic activity of main protease in virus replication. The best docked phytoconstituents also exhibited interactions with more than two amino acids in SARS-CoV-2 Mpro substrate binding site. However, in addition to aforementioned hits, experimental validation of computational studies by in vitro and in vivo methods is required to discover the therapeutic efficacy of 3,4-dicaffeoylquinic acid as novel SARS-CoV-2 Mpro inhibitor.
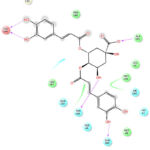 |
Figure 1: Docking Interactions of 3,4-dicaffeoylquinic acid with SARS-CoV-2 Mpro
|
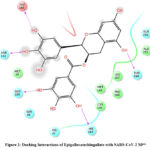 |
Figure 2: Docking Interactions of Epigallocatechingallate with SARS-CoV-2 Mpro
|
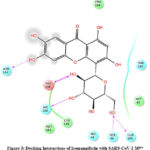 |
Figure 3: Docking Interactions of Isomangiferin with SARS-CoV-2 Mpro
|
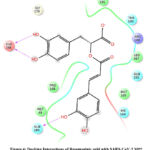 |
Figure 4: Docking Interactions of Rosemarinic acid with SARS-CoV-2 Mpro
|
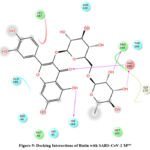 |
Figure 5: Docking Interactions of Rutin with SARS-CoV-2 Mpro
|
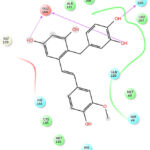 |
Figure 6: Docking Interactions of Gnetupendin B with SARS-CoV-2 Mpro
|
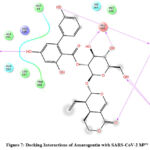 |
Figure 7: Docking Interactions of Amarogentin with SARS-CoV-2 Mpro
|
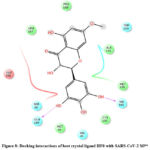 |
Figure 8: Docking interactions of best crystal ligand HF0 with SARS-CoV-2 Mpro
|
Conclusion
The SARS-CoV-2 Mpro is considered to be a potential drug target, because it differs from human proteases and plays important role in viral replication. Hence, the present study explored the inhibitory potentials of 274 antiviral phytoconstituents from medicinal plants against SARS-CoV-2 Mpro. Then, the best docked crystal ligand HF0 (-7.872kcal/Mol) was selected as reference among 439 crystal ligands of Mpro. Among the phytoconstituents, 3,4-dicaffeoylquinic acid was found to show good binding affinity with XPG score -9.721 kcal/Mol than standard HF0. From ADMET properties prediction, it was found that most of the phytoconstituents showed acceptable pharmacological properties. Interaction fingerprint analysis revealed that Glu 166 was present in seven best docked phytoconstituents, either as close contact or as participant in H-bond which diminishes the catalytic activity of SARS-CoV-2 Mpro resulting in inhibition of viral replication. Thus the present study provided an insight about possible mechanism of 3,4-dicaffeoylquinic acid in inhibition of catalytic function of Mpro which would help in the further development of SARS-CoV-2 Mpro inhibitors. From above results, it has been observed that the 3,4-dicaffeoylquinic acid has potential to act as novel SARS-CoV-2 Mpro inhibitors. Further in vitro studies will be required to understand the efficacy of 3,4-dicaffeoylquinic acid in inhibition of COVID-19 main protease.
Acknowledgements
Authors are thankful to Department of Bioinformatics, Sri Venkateswara Institute of Medical Sciences, Tirupati for their support to perform the molecular docking studies.
Conflict of Interest
Authors declare no competing interests to disclose.
Funding Sources
There is no funding of Sources
Ethical approval
Not applicable
References
- Weiss S.R and Leibowitz J.L. Corona pathogenesis. Adv. Virus Res.,2011; 81: 85-164.
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res., 2006; 66:193-292.
- Wit E, Doremalenvan D, Falzarano N.D and Munster V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol., 2016; 14:523-534.
- WHO. Novel coronavirus – China. Jan 12, 2020. http://www.who.Int/csr/don/12-january-2020-novel-coronavirus-china/en/ accessed on June 2022.
- WHO Corona virus Dashboard, 2023. https://covid19.who.int/ accessed on January 2023
- Manish K.T, Pushpendra S, Sujata S, Tej PS, Ethayathulla A.S and Punit K. Identification of bioactive molecule from Withania somnifera (Ashwagandha) as SARS-CoV-2 main protease inhibitor. J. Biomol. Struct. Dyn., 2021; 39(15):668–681.
- Oso B.J, Oluwaseun A.A and Olaoye I.F. Pharmacoinformatics and hypothetical studies on allicin, curcumin, and gingerol as potential candidates against COVID-19-associated proteases. J. Biomol. Struct. Dyn., 2020; 40(1):389-400. https://doi.org/10.1080/07391102.2020.1813630.
- Rupesh V.C, Shailendra S.G, Rajesh B.P, Saurabh K.S, Satyendra K.P, Anshul S, Sushant K.S, Nilambari S.G and Rupali S. Prasad. SARS-CoV-2 host entry and replication inhibitors from Indian ginseng: an in-silico approach. J. Biomol. Struct. Dyn., 2021; 39(12):4510–4521.
- Chowdhury P. In silico investigation of phytoconstituents from Indian medicinal herb ‘Tinospora cordifolia (giloy)’ against SARS-CoV-2 (COVID-19) by molecular dynamics approach. J. Biomol. Struct. Dyn., 2020; 39(17): 6792-6809. https://doi.org/10.1080/07391102.2020.1803968.
- Lakhera S, Devlal K, Ghosh A and Rana M. In Silico Investigation of Phytoconstituents of Medicinal Herb ‘Piper Longum’ Against SARS-CoV-2 by Molecular Docking and Molecular Dynamics Analysis. Results Chem., 2021; 3:100199. https://doi.org/10.1016/j.rechem.2021.100199.
- Zuckerman D.M. Emergency Use Authorizations (EUAs) Versus FDA Approval: Implications for COVID-19 and Public Health. Am. J. Public. Health., 2021;111(6):1065–1069.
- Mohamed E, Ayman A.E, Ahmed M, Noura M, Abo S, Eman Y.S, Bahaa E and Ahmed A. Ligand-based design, synthesis, computational insights, and in vitro studies of novel N-(5-Nitrothiazol-2-yl)-carboxamido derivatives as potent inhibitors of SARS-CoV-2 main protease. J. Enzyme. Inhib. Med. Chem., 2022; 37:1, 2112-2132. DOI: 10.1080/14756366.2022.2105322.
- Mouffouk C, Mouffouk S, Mouffouk S, Hambaba L and Habab H. Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CLpro and PLpro),spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2). Eur. J. Pharmacol.,2020; 891: 173759. https://doi.org/10.1016/j.ejphar.2020.173759.
- Kandeel M and Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci.,2020; 257:117627. https://doi.org/10.1016/j.lfs.2020.117627.
- Zmudzinski M, Rut M, Olech G, Granda J, Giurg M, Grabowska M.B, Zhang L, Sun X, Zongyang L, Nayak D, Brodacka M.K, Olsen S.K, Hilgenfeld R and Drag M. Ebselen derivatives are very potent dual inhibitors of SARS-CoV-2 proteases -PLpro and Mpro in vitro studies. BioRxiv., 2020. [online] https://doi.org/10.1101/2020.08.30.273979.
- Cavasotto C.N and Filippo J.I.D. In silico Drug Repurposing for COVID-19: Targeting SARS-CoV-2 Proteins through Docking and Consensus Ranking. Mol. Inform., 2021; 41(1): 2000115. https://doi.org/10.1002/minf.v40.110.1002/minf.202000115
- Yadav R, Chaudhary J.K, Jain N, Chaudhary P, Khanra S, Dhamija P, Sharma A, Kumar A and Handu S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Molecules., 2021; 10(4):821.
- Zaki A.A, Al-Karmalawy A.A, El-Amier Y.A and Ashour A.J.N.J.C. Molecular docking reveals the potential of Cleome amblyocarpa isolated compounds to inhibit COVID-19 virus main protease. New J. Chem., 2020; 44:16752–8.
- Joshi R.S, Jagdale S.S and Bansode S.B. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn., 2021; 39:3099–114.
- Snijder E.J, Decroly E and Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res., 2016; 96:59–126, doi: 10.1016/bs.aivir.2016.08.008.
- Anand K, Ziebuhr J, Wadhwani P, Mesters J.R and Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science., 2003; 300(5626): 1763-1767.https://doi.org/10.1126/science.1085658.
- Khodadadi E, Maroufi P, Khodadadi E, Esposito I, Ganbarov K, Espsoito S, Yousefi M, Zeinalzadeh E and Kafil H.S. Study of combining virtual screening and antiviral treatments of the Sars-CoV-2 (Covid-19). Microb. Pathog., 2020; 146: 104241. https://doi.org/10.1016/j.micpath.2020
- Sven U and Christoph N. The SARS-CoV-2 main protease as drug target.Bioorg. Med. Chem. Lett., 2020; 30(17): 127377. https://doi.org/10.1016/j.bmcl.2020.127377.
- Ahmad B, Batool M, Ain Qu, Kim M.S and Choi S. Exploring the Binding Mechanism of PF-07321332 SARS-CoV-2 Protease Inhibitor through Molecular Dynamics and Binding Free Energy Simulations.Int. J. Mol. Sci., 2021; 22:9124. https://doi.org/10.3390/ ijms22179124.
- Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K and Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science., 2020; 368(6489):409-412. doi:10.1126/science.abb3405.
- Kukol A. Molecular Modeling of Proteins Volume 443 || Molecular Docking, 2008:365–382. doi:10.1007/978-1-59745-177-2_19.
- https://www.schrodinger.com/products/qikprop
- Sasikala M and Rajitha G.Cytotoxic oxindole derivatives: in vitro EGFR inhibition, pharmacophore modeling, 3D-QSAR and molecular dynamics studies. J, Recept. Signal Transduct., 2019; 39(5-6):460-469.
- Kuban J.A, Sahu K.K, Gorska M, Tuszynski J.A and Wozniak M. Chicoric acid binds to two sites and decreases the activity of the YopH bacterial virulence factor. Oncotarget., 2016; 7(3):2229-2238.
- Yu S, Guo Q, Jia T, Zhang X, Guo D, Jia Y, Li J and Sun J. Mechanism of Action of Nicotiflorin from Tricyrtis maculata in the Treatment of Acute Myocardial Infarction: From Network Pharmacology to Experimental Pharmacology. Drug Des. Devel. Ther., 2021; 15:2179-2191.
- Swarup V, Ghosh J, Ghosh S, Saxena A and Basu A. Antiviral and anti-inflammatory effects of rosmarinic acid in an experimental murine model of Japanese encephalitis. Antimicrob. Agents Chemother., 2007; 51(9):3367-3370.
- Nyandoro S.S. In vivoantiviral activity of the constituents of Artabotrys monteiroae and Artabotrysmodestusagainst Infectious Bursal Disease and Newcastle Disease Viruses. Int. J. Biol. Chem. Sci., 2018; 11(6): 3075-3085.
- Rosmalena R, Elya B, Dewi B.E, Fithriyah F, Desti H, Angelina M, Hanafi M, Lotulung P.D, Prasasty V.D and Seto D. The Antiviral Effect of Indonesian Medicinal Plant Extracts Against Dengue Virus In Vitro and In Silico. Pathogens., 2019; 8(2): 85. https://doi.org/10.3390/pathogens8020085.
- Syed L.B, Shah F, Akhtar M, Benjamin G.P, Abdul H.E and Mariusz J. Antiviral activities of flavonoids.Biomed. Pharmacother., 2021; 140:111596. https://doi.org/10.1016/j.biopha.2021.111596.
- Kaihatsu K, Yamabe M and Ebara Y. Antiviral Mechanism of Action of Epigallocatechin-3-O-gallate and Its Fatty Acid Esters. Molecules., 2018; 23(10):2475. https://doi.org/10.3390/molecules23102475
- Oliveira D.A, Prince D, Lo C.Y, Lee L.H and Chu T.C. Antiviral activity of theaflavin digallate against herpes simplex virus type 1. Antiviral Res., 2015; 118:56-67.
- Churiyah, Olivia B.P, Elrade R and Tarwadi. Antiviral and Immunostimulant Activities of Andrographis paniculata. HAYATI. J. Biosci., 2015; 22(2): 67-72.
- Zhang G, Zhang B, Zhang X and Bing F. Homonojirimycin, an alkaloid from dayflower inhibits the growth of influenza A virus in vitro. Acta. Virol., 2013; 57(1):85-6.
- Kumar V and Van S. A Review of Swertia chirayita (Gentianaceae) as a Traditional Medicinal Plant.Front. Pharmacol., 2016; 6:308. https://doi.org/10.3389/fphar.2015.00308.
- Medda S, Mukhopadhyay S and Basu M.K. Evaluation of the in-vivo activity and toxicity of amarogentin, an antileishmanial agent, in both liposomal and niosomal forms. J. Antimicrob. Chemother., 1999; 44:791–794.
- Zanello P.R, Koishi A.C, Rezende Júnior C, Oliveira L.A, Pereira A.A, Almeida M.V, Duarte S.C.N and Bordignon J. Quinic acid derivatives inhibit dengue virus replication in vitro. VirolJ.,2015; 12:223. https://doi.org/10.1186/s12985-015-0443-9.
- Zheng M.S and Lu Z.Y. Antiviral effect of mangiferin and isomangiferin on herpes simplex virus. Chin. Med. J., 1990; 103(2):160-165.
- Liu A.L, Liu B, Qin H.L, Lee S.M, Wang Y.T and Du G.H. Anti-influenza virus activities of flavonoids from the medicinal plant Elsholtzia rugulosa. Planta. Med., 2008; 74(8):847-851
- Song X, Tan L, Wang M, Ren C, Guo C, Yang B, Ren Y, Cao Z, Li Y and Pei J (2021). Myricetin: A review of the most recent research. Biomed. Pharmacother., 2021; 134:111017.https://doi.org/10.1016/j.biopha.2020.111017
- Murali R, Srinivasan S and Ashokkumar N. Antihyperglycemic effect of fraxetin on hepatic key enzymes of carbohydrate metabolism in streptozotocin-induced diabetic rats. Biochimie., 2013; 95(10):1848–1854.
- Kratz J.M, Andrighetti-Fröhner C.R, Kolling D.J, Leal P.C, Cirne-Santo C.C, Yunes R.A, Nunes R.J, Trybala E,; Bergström T, Frugulhetti I.C.P.P, Barardi C.R.M and Simões C.M.O. Anti-HSV-1 and anti-HIV-1 activity of gallic acid and pentyl gallate. Mem. Inst. Oswaldo. Cruz., 2008; 103(5):437–442.
- Xue W, Tang X, Zhang C, Chen M, Xue Y and Liu T. Synthesis and Antiviral Activity of Novel Myricetin Derivatives Containing a Ferulic Acid Amide Scaffolds. New J. Chem., 2020; 44:2374-2379. https://doi.org/10.1039/C9NJ05867B.
- Saleem H.N, Batool F, Mansoor H.J, Shahzad-ul-Hussan S and Saeed M. Inhibition of dengue virus protease by Eugeniin, Isobiflorin, and Biflorin Isolated from the Flower Buds of Syzygium aromaticum (Cloves). ACS omega., 2019; 4(1):1525-1533.
- Lani R, Hassandarvish P, Shu M..H, Phoon WH, Chu J.J, Higgs S, Vanlandingham D, Abu Bakar S and Zandi K. Antiviral activity of selected flavonoids against Chikungunya virus. Antiviral Res., 2016; 133:50-61.
- Petrillo D.A, Orrù G, Fais A and Fantini M.C. Quercetin and its derivates as antiviral potentials: A comprehensive review. Phytother Res 2022; 36(1):266-278.
- Utsunomiya H, Ichinose M, Ikeda K, Uozaki M, Morishita J, Kuwahara T, Koyama A.H and Yamasaki H. Inhibition by caffeic acid of the influenza A virus multiplication in vitro. Int. J. Mol. Med., 2014; 34(4):1020-1024.
- Sahil K and Souravh B. A Review on Protocatechuic Acid and Its Pharmacological Potential. ISRN Pharmacol.,2014. https://doi.org/10.1155/2014/952943.
- Tutunchi H, Naeini F, Ostadrahimi A and Hosseinzadeh-Attar M.J. Naringenin, a flavanone with antiviral and anti-inflammatory effects: A promising treatment strategy against COVID-19. Phytother Res., 2020; 34(12):3137-3147.
- Ferreira P.G, Ferraz A.C, Figueiredo J.E, Lima C.F, Rodrigues V.G, Taranto A.G, Ferreira J.M.S, Brandão G.C, Vieira-Filho S.A, Duarte L.P, de Brito Magalhães C.L and de Magalhães J.C. Detection of the antiviral activity of epicatechin isolated from Salacia crassifolia (Celastraceae) against Mayaro virus based on protein C homology modelling and virtual screening. Arch. Virol., 2018; 163(6):1567-1576.
- Barnard D.L, Huffman J.H, Morris J.L, Wood S.G, Hughes B.G and Sidwell R.W. Evaluation of the antiviral activity of anthraquinones, anthrones and anthraquinone derivatives against human cytomegalovirus. Antiviral Res., 1992; 17(1):63-77.
- Xu J, Xu Z, Zheng W. A Review of the Antiviral Role of Green Tea Catechins. Molecules., 2017. 22(8): 1337. https://doi.org/10.3390/molecules22081337.
- Ninfali P, Antonelli A, Magnani M and Scarpa E.S. Antiviral Properties of Flavonoids and Delivery Strategies. Nutrients., 2020; 12(9):2534. https://doi.org/10.3390/nu12092534.
- Ding Y, Cao Z, Cao L, Ding G, Wang Z and Xiao W. Antiviral activity of chlorogenic acid against influenza A (H1N1/H3N2) virus and its inhibition of neuraminidase. Sci. Rep.,2017; 7:45723. https://doi.org/10.1038/srep45723.
- Santhi V.P, Masilamani P, Sriramavaratharajan V, Murugan R, Gurav S.S, Sarasu V.P, Parthiban S and Ayyanar M. Therapeutic potential of phytoconstituents of edible fruits in combating emerging viral infections. J. food biochem.,2021; 45(8):13851. https://doi.org/10.1111/jfbc.13851.
- Jennings M.R and Parks R.J. Curcumin as an Antiviral Agent. Viruses., 2020; 12(11):1242. https://doi.org/10.3390/v12111242.
- Abyari M, Nasr N, Soorni J and Sadhu D. Enhanced Accumulation of Scopoletin in Cell Suspension Culture of Spilanthes acmella Murr. Using Precursor Feeding. Braz. Arch. Biol. Technol., 2016; 59. doi:10.1590/1678-4324-2016150533.
- Warowicka A, Nawrot R and Goździcka-Józefiak A. Antiviral activity of berberine. Arch. Virol., 2020; 165(9):1935-1945
- Yogesh A.D, Sameer C, Rajesh S, Sapana S.C and Bhushan A.D. Plant Based Molecules for the Management of Covid-19. J. Infect. Dis. Ther., 2020; S2:008.
- Badary O.A, Hamza M.S and Tikamdas R. Thymoquinone: A Promising Natural Compound with Potential Benefits for COVID-19 Prevention and Cure. Drug. Des. Devel. Ther., 2021; 15:1819-1833.
- Shilpi S, Pratima G, Abha M and Suaib L. Acacetin, a flavone with diverse therapeutic potential in cancer, inflammation, infections and other metabolic disorders. Food. Chem. Toxicol., 2020; 145:111708. https://doi.org/10.1016/j.fct.2020.111708.
- Bajpai V.K, Kim N.H, Kim K and Kang S.C. Antiviral potential of a diterpenoid compound sugiol from Metasequoia glyptostroboides. Pak. J. Pharm. Sci., 2016; 29(3):1077-1080.
- Umashankar V, Deshpande SH, Harsha HV, Ishwar S and Chattopadhyay D. Phytochemical Moieties from Indian Traditional Medicine for Targeting Dual Hotspots on SARS-CoV-2 Spike Protein: An Integrative in-silico Approach. Front. Med., 2021; 8. https://doi.org/10.3389/fmed.2021.672629.
- Cao T.W, Geng C.A, Ma Y.B, He K, Zhou N.J, Zhou J, Zhang X.M and Chen J.J. Chemical constituents of Swertia delavayi and their anti-hepatitis B virus activity. Fitoterapia.,2015; 40(5):897-902.
- Keivan Z, Boon T.T, Sing-Sin S, Pooi-Fong W, Mohd R.M and Sazaly A. In vitro antiviral activity of Fisetin, Rutin and Naringenin against Dengue virus type-2. J. Med. Plants Res., 2011; 5(23):5534-5539.
- Kalló G, Kunkli B, Győri Z, Szilvássy Z, Csősz É and Tőzsér J. Compounds with Antiviral, Anti-Inflammatory and Anticancer Activity Identified in Wine from Hungary’s Tokaj Region via High Resolution Mass Spectrometry and Bioinformatics Analyses. Int. J. Mol. Sci., 2020; 21(24):9547. https://doi.org/10.3390/ijms21249547.
- Kaul R, Paul P, Kumar S, Büsselberg D, Dwivedi V.D and Chaari A. Promising Antiviral Activities of Natural Flavonoids against SARS-CoV-2 Targets: Systematic Review. Int. J. Mol. Sci., 2021; 22(20):11069. https://doi.org/10.3390/ijms222011069.
- Huan C, Xu Y, Zhang W, Guo T, Pan H and Gao S. Research Progress on the Antiviral Activity of Glycyrrhizin and its Derivatives in Liquorice. Front. Pharmacol., 2021; 12. https://doi.org/10.3389/fphar.2021.680674.
- LeCher J.C, Diep N, Krug P.W and Hilliard J.K. Genistein Has Antiviral Activity against Herpes B Virus and Acts Synergistically with Antiviral Treatments to Reduce Effective Dose. Viruses., 2019. 11(6):499. https://doi.org/10.3390/v11060499.
- Yazawa K, Kurokawa M, Obuchi M, Li Y, Yamada R, Sadanari H, Matsubara K, Watanabe K, Koketsu M, Tuchida Y and Murayama T. Anti-influenza virus activity of tricin, 4′,5,7-trihydroxy-3′,5′-dimethoxyflavone. Antivir. Chem. Chemother.,2011; 22(1):1-11.
- Yu C, Zhang P, Lou L and Wang Y. Perspectives Regarding the Role of Biochanin A in Humans. Front. Pharmacol., 2019; 10. https://doi.org/10.3389/fphar.2019.00793.
- López L.L.I, Nery F.S.D, Silva B.S.Y and Sáenz G.A. Naphthoquinones: Biological Properties And Synthesis Of Lawsone And Derivatives – A Structured Review. Vitae. Revista. De. La. Facultad. De. Química. Farmacéutica., 2014; 21(3):248-258.
- Zandi K, Teoh B.T, Sam S.S, Wong P.F, Mustafa M.R and AbuBakar S. Antiviral activity of four types of bioflavonoid against dengue virus type-2. Virol. J., 2011; 8:560. https://doi.org/10.1186/1743-422X-8-560.
- Nagoor M.M.F, Goyal S.N, Suchal K, Sharma C, Patil C.R and Ojha S.K. Pharmacological Properties, Molecular Mechanisms, and Pharmaceutical Development of Asiatic Acid: A Pentacyclic Triterpenoid of Therapeutic Promise. Front. Pharmacol.,2018; 9. https://doi.org/10.3389/fphar.2018.00892.
- Moghaddam E, Teoh B.T, Sam S.S, Lani R, Hassandarvish P, Chik Z, Yueh A, Abubakar S and Zandi K. Baicalin, a metabolite of baicalein with antiviral activity against dengue virus. Sci. Rep., 2014; 4:5452. https://doi.org/10.1038/srep05452.
- Hassan S.T, Berchová-Bímová K and Petráš J. Plumbagin, a Plant-Derived Compound, Exhibits Antifungal Combinatory Effect with Amphotericin B against Candida albicans Clinical Isolates and Anti-hepatitis C Virus Activity. Phytother. Res., 201630(9):1487-92.
- Fan W, Qian S, Qian P and Li X. Antiviral activity of luteolin against Japanese encephalitis virus.Virus Res., 2016; 220:112-116.
- Scott R.B, Inna G, Sergei G, Antony W, Marion K.B, Michael A.P, Robert J.C, James B.M, John A.B and Stuart F.J.L. Selective inhibition of HIV-1 reverse transcriptase-associated ribonuclease H activity by hydroxylated tropolones. Nucleic Acids Res., 2005; 33(4):1249–1256.
- Gilling D.H, Kitajima M, Torrey J.R and Bright K.R. Antiviral efficacy and mechanisms of action of oregano essential oil and its primary component carvacrol against murine norovirus. J. Appl. Microbiol.,2014; 116(5):1149-1163.
- Hayati R.F, Better C.D, Denis D, Komarudin A.G, Bowolaksono A, Yohan B and Sasmono R.T. [6]-Gingerol Inhibits Chikungunya Virus Infection by Suppressing Viral Replication. Biomed. Res. Int., 2021. https://doi.org/10.1155/2021/6623400.
- Choi J.G, Lee H, Hwang Y.H, Lee J.S, Cho W.K and Ma J.Y. Eupatorium fortunei and Its Components Increase Antiviral Immune Responses against RNA Viruses. Front. Pharmacol., 2017; 8. https://doi.org/10.3389/fphar.2017.00511.
- Meyer J.J, Afolayan A.J, Taylor M.B and Erasmus D. Antiviral activity of galangin isolated from the aerial parts of Helichrysum aureonitens. J. Ethnopharmacol., 1997; 56(2):165-169.
- Benencia F and Courrèges M.C. In vitro and in vivo activity of eugenol on human herpesvirus. Phytother. Res., 2000; 14(7):495-500.
- Jeong G.S, Lee D.S, Kim D.C, Jahng Y, Son J.K, Lee S.H and Kim Y.C. Neuroprotective and anti-inflammatory effects of mollugin via up-regulation of heme oxygenase-1 in mouse hippocampal and microglial cells. Eur. J. Pharmacol., 2011; 654(3):226–234.
- Chung C.Y, Liu C.H, Burnouf T, Wang G.H, Chang S.P, Jassey A, Tai C.J, Tai C.J, Huang C.J, Richardson C.D, Yen M.H, Lin C.C and Lin L.T. Activity-based and fraction-guided analysis of Phyllanthus urinaria identifies loliolide as a potent inhibitor of hepatitis C virus entry. Antiviral Res., 2016; 130:58-68.
- Kwon J.E, Lee Y.G, Kang J.H, Bai Y.F, Jeong Y.J, Baek N.I, Seo Y.J and Kang S.C. Anti-viral activity of compounds from Agrimonia pilosa and Galla rhois extract mixture. Bioorg Chem.,2019; 93:103320. https://doi.org/10.1016/j.bioorg.2019.103320.
- Badam L, Bedekar S.S, Sonawane K.B and Joshi S.P. In vitro antiviral activity of bael (Aegle marmelos Corr) upon human coxsackieviruses B1-B6. J. Commun. Dis., 2002; 34(2):88-99.
- Unal M.A, Bitirim C.V, Summak G.Y, Bereketoglu S, Cevher Zeytin I, Besbinar O, Gurcan C, Aydos D, Goksoy E, Kocakaya E, Eran Z, Murat M, Demir N, Aksoy Ozer Z.B, Somers J, Demir E, Nazir H, Ozkan SA, Ozkul A, Azap A, Yilmazer A and Akcali K.C. Ribavirin shows antiviral activity against SARS-CoV-2 and downregulates the activity of TMPRSS2 and the expression of ACE2 in vitro. Can. J. Physiol. Pharmacol. 2021; 99(5):449-460.
- Ranit R, Banani R.C, Progya M, Debanjan M, Sohom B and Tapajyoti R. Study on Antiviral Activities of some Immunity Boosting Herbs- Extraction, Encapsulation and Development of Functional Food. Int. J. Innov. Sci. Res. Technol., 2021; 6(8):168-176.
- Mishra S, Pandey A and Manvati S. Coumarin: An emerging antiviral agent. Heliyon., 2020; 6(1):e03217. https://doi.org/10.1016/j.heliyon.2020.e03217.
- Krystyn Z.B, Nigel B, Shirley F.R and Lawrence R.S. Plant products as topical microbicide candidates: assessment of in vitro and in vivo activity against herpes simplex virus type 2. Antiviral Res., 1999; 42(3):219–226.
- Liu A.L and Du G.H. Antiviral Properties of Phytochemicals. Dietary Phytochem. Microbes., 2012; 18:93-126. https://doi.org/10.1007/978-94-007-3926-0_3.
- Godoy D.L.R, Barros M.T and Silva L.R. Medicinal Attributes of Lignans Extracted from Piper Cubeba: Current Developments. Chemistry Open 2018; 7(2):180–191.
- Kowalczyk A, Przychodna M, Sopata S, Bodalska A and Fecka I. Thymol and Thyme Essential Oil-New Insights into Selected Therapeutic Applications. Molecules., 2020. 25(18):4125. https://doi.org/10.3390/molecules25184125.
- Dai J.P, Li W.Z, Zhao X.F, Wang G.F, Yang J.C, Zhang L, Chen X.X, Xu Y.X and Li K.S. A drug screening method based on the autophagy pathway and studies of the mechanism of evodiamine against influenza A virus. PLoS One., 2012. 7(8):e42706. https://doi.org/10.1371/journal.pone.0042706
- Ben-Shabat S, Yarmolinsky L, Porat D and Dahan A. Antiviral effect of phytochemicals from medicinal plants: Applications and drug delivery strategies.Drug Deliv. Transl. Res., 2020; 10(2):354-367.
- Saranya N, Jayakanthan M, Ravikumar C, Kandavelmani A, Bharathi N, Kumaravadivel N, Ramasamy G, Muthurajan R, Subbarayalu M, Neelakandan K. Exploring Phytochemicals of Traditional Medicinal Plants Exhibiting Inhibitory Activity Against Main Protease, Spike Glycoprotein, RNA-dependent RNA Polymerase and Non-Structural Proteins of SARS-CoV-2 Through Virtual Screening. Front Pharmacol.,2021; 12. doi:10.3389/fphar.2021.667704.
- Tutik S.W, Dzul A, Adita A.P, Myrna A, Lydia T, Tri W, Utsubo C.A, Widyawaruyanti A, Achmad F and Hak H. Anti-Viral Activity of Phyllanthus niruri against Hepatitis C Virus. Malays. Appl. Biol.,2019; 48(3):105-111.
- Patel K, Gadewar M, Tahilyani V and Patel D.K. A review on pharmacological and analytical aspects of diosmetin: a concise report.Chin. J. Integr. Med., 2013; 19(10):792-800.
- Paulpandi M, Kannan S, Thangam R, Kaveri K, Gunasekaran P and Rejeeth C. In vitro anti-viral effect of β-santalol against influenza viral replication.Phytomedicine.,2011; 19(3-4):231-235.
- Adriana C.C.R, Gabriel M.V, Breno de M.S, Cíntia L.B.M, Markus K and Geraldo C.B. Anti-arboviral activity and chemical characterization of hispidulin and ethanolic extracts from Millingtonia hortensis L.f. and Oroxylum indicum (L.) Kurz (Bignoniaceae). Nat. Prod. Res.,2022; 37(4). https://doi.org/10.1080/14786419.2022.2065485.
- Alam F, Mohammadin K, Shafique Z, Amjad S.T and Asad M.H.H.B. Citrus flavonoids as potential therapeutic agents: A review. Phytother. Res., 2022; 36(4):1417-1441.
- Sokolova N, Yarovaya O, Baranova D, Galochkina A, Kireeva M, Shtro A, Borisevich S, Gatilov Y, Zarubaev V and Salakhutdinov N. Quaternary ammonium salts based on (-)-borneol as effective inhibitors of influenza virus. Arch Virol., 2021; 166(7):1965-1976. https://doi.org/10.1007/s00705-021-05102-1.
- Peele K.A, Chandrasai P, Srihansa T, Krupanidhi S, Sai A, Vijaya, Babu D.J, Indira M, Reddy A.R and Venkateswarulu T.C. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: A computational study. Inform. Med. Unlocked., 2020; 19:100345. https://doi.org/10.1016/j.imu.2020.100345.
- Kasaian J and Mohammadi A. Biological activities of farnesiferol C: a review. J. Asian. Nat. Prod. Res., 2017; 20(1):27-35.
- Moradi J, Abbasipour F, Zaringhalam J, Maleki B, Ziaee N, Khodadoustan A and Janahmadi M. Anethole, a Medicinal Plant Compound, Decreases the Production of Pro-Inflammatory TNF-α and IL-1β in a Rat Model of LPS-Induced Periodontitis. Iran. J. Pharm. Res. Fall.,2014; 13(4):1319-1325.
- Fabra M.J, Castro-Mayorga J.L, Randazzo W, Lagarón J.M, López-Rubio A, Aznar R and Sánchez G. Efficacy of Cinnamaldehyde Against Enteric Viruses and Its Activity After Incorporation Into Biodegradable Multilayer Systems of Interest in Food Packaging.Food Environ. Virol., 2016; 8(2):125-132.
- Gavanji S, Sayedipour S.S, Larki B and Bakhtari A. Antiviral activity of some plant oils against herpes simplex virus type 1 in Vero cell culture. J. Acute Med.,2015; 5(3):62–68.
- Zhou B, Yang Z, Feng Q, Liang X, Li J, Zanin M, Jiang Z and Zhong N. Aurantiamide acetate from baphicacanthus cusia root exhibits anti-inflammatory and anti-viral effects via inhibition of the NF-κB signaling pathway in Influenza A virus-infected cells. J. Ethnopharmacol., 2017; 199(6):60–67.
- Choi H.J. Chemical Constituents of Essential Oils Possessing Anti-Influenza A/WS/33 Virus Activity.Osong Public Health Res. Perspect., 2018; 9(6):348-353
- Ghildiyal R, Prakash V, Chaudhary V.K, Gupta V and Gabrani R. Phytochemicals as Antiviral Agents: Recent Updates. Plant-derived Bioactives., 2020; 12:279–295.
- Matveeva T, Khafizova G, Sokornova S. In Search of Herbal Anti-SARS-Cov2 Compounds. Front. Plant Sci., 2020; 11. https://doi.org/10.3389/fpls.2020.589998.
- Hoffmann M, Hofmann-Winkler H, Smith J.C, Krüger N, Sørensen L.K, Søgaard O.S, Hasselstrøm J.B, Winkler M, Hempel T, Raich L, Olsson S, Yamazoe T, Yamatsuta K, Mizuno H, Ludwig S, Noé F, Sheltzer J.M, Kjolby M and Pöhlmann S. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. BioRxiv [online] 2020; https://doi.org/10.1101/2020.08.05.237651.
- Kumar V, Dhanjal J.K, Bhargava P, Kaul A, Wang J, Zhang H, Kaul SC, Wadhwa R and Sundar D. Withanone and Withaferin-A are predicted to interact with transmembrane protease serine 2 (TMPRSS2) and block entry of SARS-CoV-2 into cells. J. Biomol. Struct. Dyn., 2022; 40(1):1-13.
- Rawah H.E, Zeinab N.A.S, Mohamed A.B and Salwa A.A. Antiviral activity of castor oil plant (Ricinus communis) leaf extracts. J. Ethnopharmacol.,2021; 271:113878. https://doi.org/10.1016/j.jep.2021.113878.
- Madia V.N, Marta D.A, Daniela D.V, Antonella M, Alessandro D.L, Davide I, Valeria T, Francesco S, Giovanna D.C, Stefania G, Luigi S, Anna T.P, Roberto D.S, Lucia N and Roberta C. Investigation of Commiphora myrrha (Nees) Engl. Oil and Its Main Components for Antiviral Activity. Pharmaceuticals., 2021; 14(3):243. https://doi.org/10.3390/ph14030243.
- Panagiotopoulos A, Tseliou M, Karakasiliotis I, Kotzampasi D.M, Daskalakis V, Kesesidis N, Notas G, Lionis C, Kampa M, Pirintsos S, Sourvinos G and Castanas E. P-cymene impairs SARS-CoV-2 and Influenza A (H1N1) viral replication: In silico predicted interaction with SARS-CoV-2 nucleocapsid protein and H1N1 nucleoprotein.Pharmacol. Res. Perspect.,2021; 9(4):e00798. https://doi.org/10.1002/prp2.798.
- Silva J.K.R.D, Figueiredo P.L.B, Byler K.G and Setzer W.N. Essential Oils as Antiviral Agents. Potential of Essential Oils to Treat SARS-CoV-2 Infection: An In-Silico Investigation. Int. J. Mol. Sci.,2020; 21(10):3426. https://doi.org/10.3390/ijms21103426.
- Amraiz D, Zaidi N.S.S and Fatima M. Antiviral evaluation of an Hsp90 inhibitor, gedunin, against dengue virus. Trop. J. Pharm. Res., 2017; 16(5). https://doi.org/10.4314/tjpr.v16i5.5.
- Jingping R, Wei Z, Changsheng J, Chang L, Chengjun Z, Hua C, Wentao L and Qigai H. Inhibition of Porcine Epidemic Diarrhea Virus by Cinchonine via Inducing Cellular Autophagy. Front. Cell. Infect. Microbiol.,2022; 12:856711. https://doi.org/10.3389/fcimb.2022.856711.
- Nupur M. Medicinal plants, aromatic herbs and spices as potent immunity defenders: Antiviral (COVID-19) perspectives. Ann. Phytomed., 2020; 9(2):30-49.
- Marcotullio M.C, Pelosi A and Curini M. Hinokinin, an emerging bioactive lignan. Molecules.,2014; 19(9):14862-14878.
- Lu W, Shi L, Gao J, Zhu H, Hua Y, Cai J, Wu X, Wan C, Zhao W and Zhang B. Piperlongumine Inhibits Zika Virus Replication In vitro and Promotes Up-Regulation of HO-1 Expression, Suggesting An Implication of Oxidative Stress. Virol Sin 2021; 36(3):510-520.
- Abdal D.A, Choi H.Y, Kim Y.B and Cho S.G. Antiviral effect of methylated flavonol isorhamnetin against influenza. PLoS One.,2015; 13(3): e0121610. https://doi.org/10.1371/journal.pone.0121610.
- Sania A, Mamunur R, Pollob A, Safaet A and Hossain M.A. Prospective Asian plants with corroborated antiviral potentials: Position standing in recent years. Beni-Suef. Univ. J. Basic. Appl. Sci.,2022; 11(47):1-26.
- Elsbaey M, Ibrahim M.A.A, Bar F.A and Elgazar A.A. Chemical constituents from coconut waste and their in silico evaluation as potential antiviral agents against SARS-CoV-2. S Afr. J. Bot., 2021; 141:278-289.
- Parasuraman P, S.R and P.P. Searching Antiviral Drugs For Ebola Virus from Phyto-Constituents of Azadirachta Indica: Application of Molecular Modeling Studies. Asian. J. Pharm. Clin. Res., 2017; 10(7):254-257.
- Shin H.B, Choi M.S, Ryu B, Lee N.R, Kim H.I, Choi H.E, Chang J, Lee K.T, Jang D.S and Inn K.S. Antiviral activity of carnosic acid against respiratory syncytial virus. Virol. J.,2013; 10:303. https://doi.org/10.1186/1743-422X-10-303
- Asif M, Saleem M, Saadullah M, Yaseen H.S and Al Zarzour R. COVID-19 and therapy with essential oils having antiviral, anti-inflammatory, and immunomodulatory properties. Inflammopharmacol., 2020; 28:1153–1161.
- Loe M.W.C, Hao E, Chen M, Li C, Lee R.C.H, Zhu I.X.Y, Teo Z.Y, Chin W.X, Hou X, Deng J.G and Chu J.J.H. Betulinic acid exhibits antiviral effects against dengue virus infection. Antiviral Res.,2020; 184:104954. https://doi.org/10.1016/j.antiviral.2020.104954.
- Najar B, Mecacci G, Nardi V, Cervelli C, Nardoni S, Mancianti F, Ebani V.V, Giannecchini S and Pistelli L. Volatiles and Antifungal-Antibacterial-Antiviral Activity of South African Salvia spp. Essential Oils Cultivated in Uniform Conditions. Molecules.,2021; 26(9). https://doi.org/10.3390/molecules26092826.
- Usama R.A, Amgad A, Basma S.A, Soad A.L.B, Lourin G.M, Iman SA.K, Gerhard B and Salwa F.F. Natural coumarins as potential anti-SARS-CoV-2 agents supported by docking analysis. RSC Adv., 2021; 11:16970-16979.
- Shakiba Y. Antiviral activity of Alpha-Pinene and Beta-Pinene derived from plants against Bovine Viral Diarrhea virus in vitro. Phytomedicine: Int. J. Phytother. Phytopharmacol.,2016. https://www.researchgate.net/publication/322725314.
- Pal T, Paul S and Hossen J. Investigation of Potential Activity of Some Organic Compounds and Antiviral Drugs Against COVID-19 Based on Molecular Docking. J. Sci. Res., 2022; 14:309-320.
- Hu Y, Liu M, Qin H, Lin H, An X, Shi Z, Song L, Yang X, Fan H and Tong Y. Artemether, Artesunate, Arteannuin B, Echinatin, Licochalcone B and Andrographolide Effectively Inhibit SARS-CoV-2 and Related Viruses In Vitro. Front. Cell. Infect. Microbiol., 2021; 11. https://doi.org/10.3389/fcimb.2021.680127.
- Zaher K, Ahmed W and Zerizer S. Observations on the Biological Effects of Black Cumin Seed (Nigella sativa) and Green Tea (Camellia sinensis). Glob. Vet., 2008; 2(4):198-204.
- Ryabchenko B, Tulupova E, Schmidt E, Wlcek K, Buchbauer G and Jirovetz L. Investigation of Anticancer and Antiviral Properties of Selected Aroma Samples. Nat. Prod. Commun., 2008; 3(7):1085-108.
- Ye J, Wang Z, Jia J, Li F, Wang Y, Jiang Y, Wang Y, Ren Z and Pu H. Lupeol impairs herpes simplex virus type 1 replication by inhibiting the promoter activity of the viral immediate early gene α0.Acta. Virol.,2021; 65(3):254-263.
- Yarovaya O.I, Kovaleva K.S, Zaykovskaya A.A, Yashina L.N, Scherbakova N.S, Scherbakov D.N, Borisevich S.S, Zubkov F.I, Antonova A.S, Peshkov R.Y, Eltsov I.V, Pyankov O..V, Maksyutov RA and Salakhutdinov N.F. New class of hantaan virus inhibitors based on conjugation of the isoindole fragment to (+)-camphor or (-)-fenchone hydrazonesv. Bioorg. Med. Chem. Lett., 2021; 40:127926. https://doi.org/10.1016/j.bmcl.2021.127926.
- Indah R.A, Fatchiyah F, Nashi W, Mohamad A and Muhammad S.D. Shogaol, Bisdemethoxycurcumin, and Curcuminoid: Potential Zingiber Compounds Against COVID-19. Biointerface. Res. Appl. Chem., 2021; 11(5):12869-12876.
- Liu S, Wei W, Li Y, Lin X, Shi K, Cao X and Zhou M. In vitro and in vivo anti-hepatitis B virus activities of the lignan nirtetralin B isolated from Phyllanthus niruri L. J. Ethnopharmacol., 2014; 157:62–68.
- Pavlova N.I, Savinova O.V, Nikolaeva S.N, Boreko E.I and Flekhter O.B. Antiviral activity of betulin, betulinic and betulonic acids against some enveloped and non-enveloped viruses. Fitoterapia., 2003; 74(5):489-492.
- Svrlanska A, Ruhland A, Marschall M, Reuter N and Stamminger T. Wedelolactone inhibits human cytomegalovirus replication by targeting distinct steps of the viral replication cycle. Antiviral Res., 2020; 174:104677. https://doi.org/10.1016/j.antiviral.2019.104677.
- Jha N.K, Sharma C, Hashiesh H.M, Arunachalam S, Meeran M.N, Javed H, Patil C.R, Goyal S.N and Ojha S. β-Caryophyllene, A Natural Dietary CB2 Receptor Selective Cannabinoid can be a Candidate to Target the Trinity of Infection, Immunity, and Inflammation in COVID-19. Front. Pharmacol., 2021; 12. https://doi.org/10.3389/fphar.2021.590201.
- Kaushik S, Jangra G, Kundu V, Yadav J.P and Kaushik S. Anti-viral activity of Zingiber officinale (Ginger) ingredients against the Chikungunya virus. Virus disease., 2020; 31(3):270-276.
- Ramabharathi V, Saigopal D.V.R and Rajitha G. Antiviral activity of leaf-bud gum-resin of Tarenna asiatica. Bangladesh J. Pharmacol.,2014; 9:398- 405.
- Bhattacharya R, Dev K and Sourirajan A. Antiviral activity of bioactive phytocompounds against coronavirus: An update. J. Virol. Methods., 2021; 290:114070. https://doi.org/10.1016/j.jviromet.2021.114070.
- Rouf R, Uddin S.J, Sarker D.K, Islam M..T, Ali ES, Shilpi J.A, Nahar L, Tiralongo E and Sarker S.D. Antiviral potential of garlic (Allium sativum) and its organosulfur compounds: A systematic update of pre-clinical and clinical data. TrendsFood Sci. Technol., 2020; 104:219-234.
- Mösbauer K, Fritsch V.N, Adrian L, Bernhardt J, Gruhlke M.C.H, Slusarenko A.J, Niemeyer D and Antelmann H. The Effect of Allicin on the Proteome of SARS-CoV-2 Infected Calu-3 Cells. Front. Microbiol.,2021; 12. https://doi.org/10.3389/fmicb.2021.746795.
- Takatsuki A, Nakatani N, Morimoto M, Tamura G, Matsui M, Arima K, Yamaguchi I and Misato T. Antiviral and antitumor antibiotics. XX. Effects of rotenone, deguelin, and related compounds on animal and plant viruses.Appl. Microbiol., 1969; 18(4):660-667.
- Ćavar Z.S, Schadich E, Džubák P, Hajdúch M and Tarkowski P. Antiviral Activity of Selected Lamiaceae Essential Oils and Their Monoterpenes Against SARS-Cov-2. Front. Pharmacol.,2022; 13. shttps://doi.org/10.3389/fphar.2022.893634.
- Marhaeny H.D, Widyawaruyanti A, Widiandani T, Fuad Hafid A and Wahyuni T.S. Phyllanthin and hypophyllanthin, the isolated compounds of Phyllanthus niruri inhibit protein receptor of corona virus (COVID-19) through in silico approach. J. Basic Clin. Physiol. Pharmacol., 2021; 32(4):809-815
- Parvez M.K, Alam P, Arbab A.H, Al-Dosari M.S, Alhowiriny T.A and Alqasoumi S.I. Analysis of antioxidative and antiviral biomarkers β-amyrin, β-sitosterol, lupeol, ursolic acid in Guiera senegalensis leaves extract by validated HPTLC methods. Saudi Pharm. J.,2018; 26(5):685-693.
- Sajjadi SE, Shokoohinia Y, Hemmati S, Gholamzadeh S and Behbahani M. Antivirial activity of elemicin from Peucedanum pastinacifolium. Res. Pharm. Sci., 2012; 7:784.
- Akram M, Tahir I.M, Shah S.M.A, Mahmood Z, Altaf A, Ahmad K, Munir N, Daniyal M, Nasir S, Mehboob H. Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: A systematic review. Phytother. Res., 2018; 32(5). https://doi.org/10.1002/ptr.6024.
- Yadalam P.K, Varatharajan K, Rajapandian K, Chopra P, Arumuganainar D, Nagarathnam T, Sohn H and Madhavan T. Antiviral Essential Oil Components Against SARS-CoV-2 in Pre-procedural Mouth Rinses for Dental Settings During COVID-19: A Computational Study. Front. Chem.,2021; 9. https://doi.org/10.3389/fchem.2021.642026.
- Loizzo M.R, Saab A.M, Tundis R, Statti G.A, Menichini F, Lampronti I, Gambari R, Cinatl J and Doerr H.W. Phytochemical analysis and in vitro antiviral activities of the essential oils of seven Lebanon species.Chem. Biodivers., 2008; 5(3):461-470.
- Gómez L..A, Stashenko E and Ocazionez RE. Comparative study on in vitro activities of citral, limonene and essential oils from Lippia citriodora and L. alba on yellow fever virus. Nat. Prod. Commun., 2013; 8(2):249-252.
- Yan H, Wang H, Ma L, Ma X, Yin J, Wu S, Huang H and Li Y. Cirsimaritin inhibits influenza A virus replication by downregulating the NF-κB signal transduction pathway. Virol. J., 2018; 15:88. https://doi.org/10.1186/s12985-018-0995-6.
- Baildya N, Khan A.A, Ghosh N.N, Dutta Tand Chattopadhyay A.P (2021) Screening of potential drug from Azadirachta Indica (Neem) extracts for SARS-CoV-2: An insight from molecular docking and MD-simulation studies. J. Mol. Struct., 2021; 1227:129390. https://doi.org/10.1016%2Fj.molstruc.2020.129390.
- Nag A and Chowdhury R.R. Piperine, an alkaloid of black pepper seeds can effectively inhibit the antiviral enzymes of Dengue and Ebola viruses, an in silico molecular docking study. Virus disease., 2020; 31(3):308-315.
- Silveira D, Prieto-Garcia J.M, Boylan F, Estrada O, Fonseca-Bazzo Y.M, Jamal C.M, Magalhães P.O, Pereira E.O, Tomczyk M and Heinrich M. COVID-19: Is There Evidence for the Use of Herbal Medicines as Adjuvant Symptomatic Therapy? Front. Pharmacol.,2020; 11. https://doi.org/10.3389/fphar.2020.581840.
- Parang K, Wiebe L.I, Knaus E.E, Huang J.S, Tyrrell D.L and Csizmadia F. In vitro antiviral activities of myristic acid analogs against human immunodeficiency and hepatitis B viruses. Antiviral Res., 1997; 34(3):75-90.
- Taylor D.J.R, Hamid S.M, Andres A.M, Saadaeijahromi H, Piplani H, Germano J.F, Song Y, Sawaged S, Feuer R, Pandol S.J and Sin J. Antiviral Effects of Menthol on Coxsackievirus B. Viruses.,2020; 12(4):373. https://doi.org/10.3390/v12040373.
- Mondal R, Negi A and Mishra M. Acalypha Indica -A Boon to Mankind. World J. Pharm. Res., 2021; 10(3):761-798. http://dx.doi.org/10.20959/wjpr20213-19890.
- Sinha A, Farooqui S, Sharma A, Mishra A and Verma V. Reactivity of allyl methyl sulphide, the in-vitro metabolite of garlic, with some amino acids and with phospholipid involved in viral infections. J. Biomol. Struct. Dyn.,2020; 40:1-7.
- Tohmé M.J, Giménez M.C, Peralta A, Colombo M.I and Delgui L.R. Ursolic acid: A novel antiviral compound inhibiting rotavirus infection in vitro. Int. J. Antimicrob. Agents., 2019; 54(5):601-609.
- Li R, Morris-Natschke S.L and Lee K.H. Clerodane diterpenes: sources, structures, and biological activities.Nat. Prod. Rep.,2016; 33(10):1166-226.
- Lin LT, Hsu WC, Lin CC. Antiviral Natural Products and Herbal Medicines. J. Tradit. Complement. Med., 2014; 4:24-35.
- Somdutt M, Ranjit K and Harwansh. In-silico bioprospecting of taraxerol as a main protease inhibitor of SARS-CoV-2 to develop therapy against COVID-19. Struct. Chem., 2022; https://doi.org/10.21203/rs.3.rs-1308726/v1.
- Márquez N, Sancho R, Bedoya L.M, Alcamí J, López-Pérez J.L, Feliciano A.S, Fiebich B.L and Muñoz E. Mesuol, a natural occurring 4-phenylcoumarin, inhibits HIV-1 replication by targeting the NF-kappaB pathway. Antiviral Res., 2005; 66:137-145.
- Pulok K.M, Venkatesan K, Mainak M and Peter J.H. Acoruscalamus.: Scientific Validation of Ayurvedic Tradition from Natural Resources. Pharm. Biol., 2007; 45(8):651-666.
- Rajitha G, Vidya R.M, Naik V.U and Umamaheswari A. Design of Novel Selective Estrogen Receptor Inhibitors using Molecular Docking and Protein-Ligand Interaction Fingerprint Studies. J. Pharm. Res. Int., 2021; 33(46A):470-483. https://doi.org/10.9734/jpri/2021/v33i46A32890.
- Soujanya M, Rajitha G, Umamaheswari A and Sudheer Kumar K. Synthesis, Biological Evaluation and Docking Studies of N-(2-benzamido feruloyl) Aryl Hydrazone Analogues. Lett. Drug Des. Discov.,2018; 15(8):875-886.
- QikProp, version 4.4, Schrödinger, LLC, New York, NY, 2015.
- Rajitha G, Rani M.V, Naik V.U and Umamaheswari A. Design of Heterocyclic Compounds as Epidermal Growth Factor Receptor Inhibitors using Molecular Docking and Interaction Fingerprint Studies.Biosci. Biotech. Res. Comm.,2022; 15. http://dx.doi.org/10.21786/bbrc/15.1.19.
- Martin A.R, David C.O, Lennart B, Amelia H.C, Petra L, Claire S.D, Sean W.R, Patrick M.C, Philipp S, Mark S, Chris J.R, Iva N.H, Daren F, Alice D, Frank-von D, Tika R.M, Laura V, Thomas V, Jan T, Pieter L, Tu-Trinh N, Mitchell H, Anthony T, David J.H, Christopher J.S, David I.S, Andrew L.H and Martin AW. Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.Sci. Rep., 2021. 11:13208. https://doi.org/10.1038/s41598-021-92416-4.
- Forrestall K.L, Burley D.E, Cash M.K, Pottie I.R and Darvesh S. 2-Pyridone natural products as inhibitors of SARS-CoV-2 main protease. Chem-Biol. Interact., 2021; 335:109348. https://doi.org/10.1016/j.cbi.2020.109348.
- Zhang L, Daizong L, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K and Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science., 2020; 368(6489):409-412. https://doi.org/10.1126/science.abb3405.
- Ahmad B, Batool M, Ain Qu, Kim MS and Choi S. Exploring the Binding Mechanism of PF-07321332 SARS-CoV-2 Protease Inhibitor through Molecular Dynamics and Binding Free Energy Simulations. Int. J. Mol. Sci.,2021; 22(17):9124. https://doi.org/10.3390%2Fijms22179124.
- Mohamed A.S, Amgad A, Mohamed A.A and Hany S.I. Importance of glutamine 189 flexibility in SARS-CoV-2 main protease: Lesson learned from in silico virtual screening of ChEMBL database and molecular dynamics. Eur. J. Pharm. Sci., 2021; 160:105744. https://doi.org/10.1016/j.ejps.2021.105744.
- http://gohom.win/ManualHom/Schrodinger/Schrodinger_2015-2_docs/qikprop/qikprop_user_manual.pdf








