Manuscript accepted on :
Published online on: 16-11-2015
Plagiarism Check: Yes
Donya Esmaeiligoudarzi1, Fahimeh Safarnezhad Tameshkel2, Hossein Ajdarkosh2*, Mojtaba Arsalani3, Mohammad Hesam Sohani4, Vahid Behnod5
1Department of Microbiology, Faculty of Biology Sciences, Islamic Azad University of Tonekabon Branch, Tonekabon, Iran 2Gastrointestinal and Liver Disease Research Center (GILDRC), Firoozgar Hospital, Iran University of Medical Sciences, Tehran, Iran 3Iran University of Medical Sciences, Tehran, Iran 4Department of Microbiology, Science and Research Branch, Islamic Azad University, Tehran, Iran 5BaqiyatallahUniversity of Medical Sciences, Tehran, Iran Corresponding author Email : Ajdarkosh1345@yahoo.com
DOI : https://dx.doi.org/10.13005/bpj/597
Abstract
The hygienic quality of milk has an important public health importance. Helicobacter pylori has a foodborne route and especially food with animal origin. This present study was carried out in order to isolation the H.r pylori from cow milk samples using culture and PCR method. A total of 120 bovine milk and 120 dairy product samples were collected from the supermarkets of various parts of Iran. The animals whose milk samples collected for this study were clinically healthy.All samples were cultured and those that were H. pylori-positive were subjected for PCR method fordetection of the H. pyloriureC gene.Of 240 samples studied, 33 (13.75%) were positive for the H. pylori using the culture method. Raw bovine milk were the most contaminated (16.66%) but traditional cream were the less contaminated (7.5%) samples. Significant differences was seen for the prevalence of H. pylori between raw bovine milk and traditional cream samples (P =0.027). All of the positive colonies of H. pylori were confirmed using the ureC gene based-PCR method. This study showed that cow milk and traditional dairy product samples are the sources of H. pylori infection for humans.
Keywords
Helicobacter pylori; Isolation; Cow milk; Culture; Polymerase Chain Reaction
Download this article as:| Copy the following to cite this article: Esmaeiligoudarzi D, Tameshkel F. S, Arsalani M, Sohani M. H, Behnod V, Ajdarkosh H. Prevalence of Helicobacter pyloriinIranian milk and dairy products using culture and ureC based-PCR techniques. Biomed Pharmacol J 2015;8(1) |
| Copy the following to cite this URL: Esmaeiligoudarzi D, Tameshkel F. S, Arsalani M, Sohani M. H, Behnod V, Ajdarkosh H. Prevalence of Helicobacter pyloriinIranian milk and dairy products using culture and ureC based-PCR techniques. Biomed Pharmacol J 2015;8(1). Available from: http://biomedpharmajournal.org/?p=788 |
Introduction
Milk is raised as a complete food especially for children and seniors. Its high value for proteins, minerals, fats and vitamins is undeniable and in a day, millions of people use the milk and dairy products. Therefore, hygienic quality of milk has a high importance in public health but sometimes it will be changed and several infections and illness are occurred.
The Helicobacter pylori(H. pylori) is a microaerophilic Gram negative bacterium with curved spiral shape which known as a causative agent of type B gastritis, peptic ulcer disease, gastric adenocarcinoma and Mucosa Associated Lymphoid Tissue (MALT) lymphoma (1). The bacterium has been classified as a Class I carcinogen by the World Health Organization (2).The worldwide prevalence of infection has a vast range from 40% to 80% (3, 4).The main protocol for treatment of diseases caused by H. pylori, is antibiotic therapy but the antibiotic therapy fails in about 20% of the patients (4), mainly due to antibiotic resistance (5). During the last two decades, the role of H. pylori as potential pathogens in both human and veterinary medicine has been investigated intensively, and evidence suggests possible zoonotic transmission of animal helicobacter to humans.
Therefore, accurate, sensitive and rapid detection of H. pylori in samples with animal origin plays an important role in control of diseases. There are various methods for diagnosis of brucellosis such as culture, serological and molecular methods. Culture methods are well established for H. pylori and there is a study which confirmed its application (6).The diagnosis of H. pylori by serological responses, which can be unspecific due to cross-reaction or sub-sensitive reactions in samples from areas with a low or sub clinical prevalence of helicobacteriosis (7). Several studies showed that the molecular diagnosis of H. pylori is an applied form of its diagnosis. Several Polymerase Chain Reaction (PCR) methods have been developed for rapid, sensitive and accurate diagnosis of H. pylori in clinical samples (8-10).
This present study was carried out in order to detection the H. pylori in bovine milk samples using culture and PCR techniques in Isfahan, Iran.
Materials and Methods
Samples
A total of 120 bovine milk samples and 120 traditional dairy products including cheese (n=80) and cream (n=40) were randomly collected from various parts of Iran. Samples were collected from 24 randomly selected dairy herds at spring of 2012. The animals selected for this study were clinically healthy and the milk samples showed normal physical characteristics. Samples were collected under sterile hygienic conditions and were immediately transported at 4°C to laboratory in a cooler with ice packs. All milk samples were kept at –20°C until processing.
Isolation of H. pylori
Twenty five milliliter of each sample were added to 225 mL of Wilkins Chalgren anaerobe broth (Oxoid, UK) supplemented with 5% of horse serum (Sigma, St.
Louis, MO, USA) and colistinmethanesulfonate (30 mg/L), cycloheximide (100 mg/L), nalidixic acid (30 mg/L), trimethoprim (30 mg/L), and vancomycin (10 mg/L) (Sigma, St.
Louis, MO, USA) and incubated for 7 days at 37°C with shaking under microaerophilic conditions. Then, 0.1 mL of the enrichment selective broth was plated onto Wilkins Chalgren anaerobe agar (Oxoid, UK) supplemented with 5% of defibrinated horse blood and 30 mg/L colistinmethanesulfonate, 100 mg/L cycloheximide, 30 mg/L nalidixic acid, 30 mg/L trimethoprim, and 10 mg/L vancomycin(Sigma, St.
Louis, MO, USA) (6) and incubated for 7 days at 37°C under microaerophilic conditions. Suspected colonies were identified as H. pylori on the basis of the morphology of the colonies, Gram staining, and oxidase, catalase, and urease production (11). The isolates were identified as H. pylori by using conventional bacteriological methods, were also positive using the PCR assay. For comparison, a reference strain of H. pylori (ATCC 43504) was employed.
Detection of H. pylori using PCR method
DNA from 1 mL of each milk samples was extracted by a DNA isolation kit for cells and tissues (Roche Applied Science, Germany, 11814770001) according to the manufacturer’s instructions and its density was assessed by optic densitometry. Extracted genomic DNA was amplified for the ureC gene and detected with the specific primers HP-F: 5′-GAATAAGCTTTTAGGGGTGTTAGGGG-3’, HP-R: 5’GCTTACTTTCTAACACTAACGCGC-3′. The gene product was 294 bp. PCR reactions were performed in a final volume of 50 µL containing 5 µL 10 × buffer + MgCl2, 2 mMdNTP, 2 unit Taq DNA polymerase, 100 ng genomic DNA as a template, and 25 picomole of each primer. PCR was performed using a thermal cycler (Eppendorf Co., Germany) under the following conditions: an initial denaturation for 10 minutes at 94°C; 35 cycles for 1 minute at 94°C, 1 minute at 55°C, 1 minute at 72°C, and a final extension at 72°C for 10 minutes. The PCR products were electrophoresed through a 1.5% agarose gels (Fermentas, Germany) containing ethidium bromide. A DNA ladder (Fermentas Co., Germany) used to detect the molecular weight of observed bands under a UV lamp. All tests were performed in triplicate. Samples inoculated with H. pylori were used as positive controls.
Statistical analysis
Data were transferred to Microsoft Excel spreadsheet (Microsoft Corp., Redmond, WA, USA) for analysis. Using SPSS 16.0 statistical software (SPSS Inc., Chicago, IL, USA), Chi-square test and Fisher’s exact two-tailed test analysis was performed and differences were considered significant at values of p< 0.05.
Results
A total of 240 raw milk and traditional dairy product samples were analyzed for the presence of H. pylori using culture and PCR technique. All samples were cultured immediately. Table 1 shows the total distribution of H. pylori in various types of raw milk and dairy product samples. Results showed that 33 out of 120 milk and dairy products (13.75%) were positive for the H. pylori using the culture method. Raw bovine milk were the most contaminated (16.66%) but traditional cream were the less contaminated (7.5%) samples. Significant differences was seen for the prevalence of H. pylori between raw bovine milk and traditional cream samples (P =0.027). All of the positive colonies of H. pylori were confirmed using the ureC gene based-PCR method. Figure 1 shows the results of the gel electrophoresis for PCR products of the ureC gene of the Helicobacter pylori isolates from raw milk and dairy products. The results of the culture technique were also confirmed by the PCR method. No significant differences in detection of H. pylori in milk and dairy samples were observed between culture and PCR techniques.
Discussion
The results of the present study showed that the Iranian raw bovine milk and traditional dairy product samples were contaminated with H. pylori. Total prevalence rate of H. pylori in the samples of our investigation was 13.75% which was considerable high. One possible explanation for the high prevalence of H. pylori in our study is maybe that the animals which their milk samples were collected for this study were infected with H. pylori. Another reason is the fact that the workers of and stuffs of the traditional companies of dairy production were maybe infected with H. pylori and transmit this pathogen to these products. Infection of the water sources used for washing of boxes, dishes and containers of milk and dairy preparation maybe another sources of high prevalence of H. pylori.
As far as we know, the H. pylori has been detected in various types of foods including milk (9), meat (12) and ready to eat foods (13).Previous study from Iran showed that three of 447 milk samples (0.67%), including two sheep (2.2%) and one buffalo (1.6%) milk samples, were found to be contaminated with H. pylori using the culture method while the H. pyloriureC gene was detected in 56 (12.5%) of milk samples, including 19 cow (14.1%), 11 sheep (12.2%), nine goat (8.7%), two camel (3.6%), and 15 buffalo (23.4%) milk samples using the PCR method (9) was entirely lower that Japan (72.2%) (14)and Iran (previous study) (14.1%) (9). Also, previous study from Iran showed that 16% of raw milk samples were contaminated with H. pylori (15).In central Iran, the prevalence of H. pylori is very high (78%) (15). One of the major sources of infection in humans could be cow, sheep, and goat’s milk contaminated with H. pylori (16). The consumption of milk and its products vary considerably in different regions in the world. Bovine milk and dairy products have a long tradition in human nutrition. Iranian people especially children and senior drink cow’s milk commonly.Therefore, consumption of raw milk has been considered as the main source for human helicobacter infection. Also, the H. pylori has been isolated from pasteurized milk samples, previously (55% prevalence rate) (14).The Greek study showed that the prevalence of H. pylori in cow’s milk was 20% which was higher than our percentage (17).The study in USA showed that 60% of sheep milk samples were contaminated with H. pylori (18). Funnily, the results of our study are similar with the results of Azevedo et al. (2007) which could not prove the existence of H. pylori in milk through culturing. These findings are very important to explain the way of transmission of H. pylori to humans through milk and food (19).
Table 1: Distribution of Helicobacter pylori in various types of raw milk and dairy product samples.
| Types of samples | No. samples collected | Positive results for H. pylori in culture (%) | PCR confirmation (%) |
| Bovine milk | 120 | 20 (16.66) | 20 (16.66) |
| Traditional cheese | 80 | 10 (12.5) | 10 (12.5) |
| Traditional cream | 40 | 3 (7.5) | 3 (7.5) |
| Total | 240 | 33 (13.75) | 33 (13.75) |
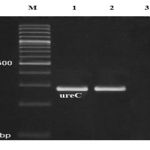 |
Figure 1: Results of the gel electrophoresis for PCR products of the ureC gene of the Helicobacter pylori isolates from raw milk and dairy products. M is 100bp DNA marker, 1 is positive sample, 2 is positive control and 3 is negative control. |
Conclusion
All of the above studies from Iran showed that the H. pylori is endemic in this country and it is essential to use from well-boiled milk and traditional dairies for human consumption. It is tempting to speculate that cow were the ancestral host of the H. pylori and that it entered the human population after domestication of cow. It will be of interest to examine cows and dairy products in different regions to test the hypothesis that cows are natural hosts of H. pylori.
References
- Hegarty, J.P., Dowd, M.T., Baker, K.H. (Occurrence of Helicobacter pylori in surface water in the United States. J ApplMicrobiol., 1999; 87: 697–701.
- Aruin, L.I. Helicobacter pylori infection is carcinogenic for humans. ArkhPato., 1997; l59: 74–78.
- EmadYahaghi, FahamKhamesipour, FatemehMashayekhi, et al., “Helicobacter pylori in Vegetables and Salads: Genotyping and Antimicrobial Resistance Properties,” BioMed Research International, vol. 2014, Article ID 757941, 11 pages, 2014. doi:10.1155/2014/757941
- Kusters, J.G., van Vliet, A.H., Kuipers, E.J. Pathogenesis of Helicobacter pylori infection. ClinMicrobiol Rev., 2006; 19: 449–490.
- Megraud, F. H. pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut., 2004; 53: 1374–1384.
- Poms, R.E., Tatini, S.R. Survival of Helicobacter pylori in ready-toeat foods at 4 degrees celsiusInt. J Food Microbiol., 2001; 63: 281–286.
- Herbrink, P., van Doorn, L.J. Serological methods for diagnosis of Helicobacter pylori infection and monitoring of eradication therapy. Eur J ClinMicrobiol Infect Dis., 2000; 19(3): 164-73.
- Kabir, S. Detection of Helicobacter pylori in faeces by culture, PCR and enzyme immunoassay. J Med Microbiol., 2001; 50(12): 1021-9.
- Rahimi, E., Kheirabadi, E.K. Detection of Helicobacter pylori in bovine, buffalo, camel, ovine, and caprine milk in Iran. Foodborne Pathog Dis., 2012; 9(5): 453-6.
- Weiss, J., Tsang, T.K., Meng, X., Zhang, H., Kilner, E., Wang, E., Watkin, W. Detection of Helicobacter pylori gastritis by PCR: correlation with inflammation scores and immunohistochemical and CLO test findings. Am J ClinPathol., 2008; 129(1): 89-96.
- Dunn, B.E., Cohen, H., Blaser, M.J. Helicobacter pylori. ClinMicrobiol Rev., 1997; 10: 720–741.
- Meng, X., Zhang, H., Law, J., Tsang, R., Tsang, T. Detection of Helicobacter pylori from food sources by a novel multiples PCR assay. J Food Safe., 2008; 28: 609–619.
- Yvonne, T.H.P., Duynhoven, V., De Jonge, R. Transmission of Helicobacter pylori: a role for food? Bull World Health Organ., 2001; 79: 455–460.
- Fujimura, S., Kawamura, T., Kato, S., Tateno, H., Watanabe, A. Detection of Helicobacter pylori in cow’s milk. LettApplMicrobiol., 2002; 35(6): 504-7.
- GhasemianSafaei, H., Rahimi, E., Zandi. A., Rashidipour, A. Helicobacter pylori as a zoonotic infection: the detection of H. pylori antigens in the milk and faeces of cows. J Res Med Sci., 2011; 16(2): 184–187.
- Vale, F.F., Vitor, J.M. Transmission pathway of Helicobacter pylori: does food play a role in rural and urban areas? Int J Food Microbiol.,2010; 138(1-2): 1–12.
- Angelidis, A.S., Tirodimos, I., Bobos, M., Kalamaki, M.S., Papageorgiou, D.K., Arvanitidoum M. Detection of Helicobacter pylori in raw bovine milk by fluorescence in situ hybridization (FISH). Int J Food Microbiol., 2011; 151(2):252-6.
- Dore, M.P., Sepulveda, A.R., El-Zimaity, H., Yamaoka, Y., Osato, M.S., Mototsugu, K., Nieddu, A.M., Realdi, G., Graham, D.V. Isolation of Helicobacter pylori From Sheep—Implications for Transmission to Humans. The American Journal of Gasterointestinol., 2001; 96: 1396-1401.
- Azevedo, N.F., Guimaraes, N., Figueiredo, C., Keevil, C.W., Vieira, M.J. A new model for the transmission of Helicobacter pylori: role of environmental reservoirs as gene pools to increase strain diversity. Crit Rev Microbiol., 2007; 33(3):157–69.







