Merry Hailu1 , Radhakrishnan Narayanaswamy2
, Radhakrishnan Narayanaswamy2 *, Solomon Abrehame1 and Valsa Remony Manoj1
*, Solomon Abrehame1 and Valsa Remony Manoj1
1Department of Biotechnology, Vel Tech Rangarajan Dr. Sagunthala R and D Institute of Science and Technology, 400 Feet Outer Ring Road, Avadi, Chennai-600 062, Tamil Nadu, India.
2Department of Biochemistry, St. Peter’s Institute of Higher Education and Research, Avadi, Chennai-600 054, Tamil Nadu, India.
Corresponding Author Email: nrkishnan@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1774
Abstract
Aflatoxins are secondary metabolites of certain fungi like Aspergillus flavus, Aspergillus niger, Aspergillus nomius and Aspergillus parasiticus. Food products like cereals, milk, milk products, nuts, oilseeds and spices are reported for aflatoxins contamination. Among the fourteen aflatoxin types reported so far, aflatoxins B1, B2, G1, G2, M1, and M2 are commonly studied. Aflatoxins are known to cause various diseases including aflatoxicosis in livestock and domestic animals and cancer in humans. Recently aflatoxin B1 has reported binding with Glycogen synthase kinase 3-beta (GSK-3β). This prompted to carry out the present study, where Glycogen synthase kinase 3-beta (GSK-3β) was evaluated on the docking behaviour of 13 aflatoxin analogues using Patch Dock. In addition, molecular physicochemical, drug-likeness, ADME (Absorption, Distribution, Metabolism and Excretion analyses) were also carried out. The molecular physiochemical analysis revealed that aflatoxin analogue showed nil violation and complied well with the Lipinski’s rule of five. ADME analysis indicates all thirteen aflatoxin analogue predicated to have high gastro-intestine (GI) absorption property. Docking studies, with GSK-3β, revealed that aflatoxin G2 analogue showed the largest atomic contact energy (-224.82 kcal/mol) and Aflatoxin P1 analogue had the least (-160.33 kcal/mol). In addition, aflatoxin P1 analogue has interacted with Asp200 amino acid residue of GSK-3β. Thus, the present study showed the potential of GSK-3β as aflatoxin binder.
Keywords
ADME; Aflatoxins Contamination; Aflatoxin Analogues; Docking; Glycogen Synthase Kinase 3-Beta (GSK-3β); Ligand
Download this article as:| Copy the following to cite this article: Hailu M, Narayanaswamy R, Abrehame S, Manoj V. R. In Silico Analysis of Glycogen Synthase Kinase 3-Beta (GSK-3β) as Aflatoxin Binder. Biomed Pharmacol J 2019;12(3). |
| Copy the following to cite this URL: Hailu M, Narayanaswamy R, Abrehame S, Manoj V. R. In Silico Analysis of Glycogen Synthase Kinase 3-Beta (GSK-3β) as Aflatoxin Binder. Biomed Pharmacol J 2019;12(3). Available from: http://biomedpharmajournal.org/?p=28714 |
Introduction
Mycotoxins are natural fungal secondary metabolites with low-molecular-weight (i.e., small molecules) produced by filamentous fungi and the most important genera of mycotoxin-producing fungi are Aspergillus, Fusarium, Stachybotrys, Claviceps, Penicillium and Alternaria (1). The principal classes of mycotoxins, which cause health problems in animals and human beings, include aflatoxin, trichothecenes, fumonisins, zearalenone and ochratoxin A (2). Aflatoxins are potent mycotoxin (fungal toxin) capable of causing death on humans and animals. There are several types of aflatoxins, among them, aflatoxins B1, B2, G1, G2, M1, and M2 are commonly studied (3). It is highly pathogenic and leads to carcinogenic, mutagenic, hepatotoxic and immunosuppressant effects in animals and humans (4–6). Particularly in children, it causes stunted growth, immunosuppression, liver cancer and even death; when they consume milk from dairy cattle that feed an aflatoxin contaminated feed (7,8). Moreover, aflatoxin B1 (AFB1) is the most potent carcinogenic (biological) agent which has been reported by several researchers throughout the world (9). Furthermore, in dairy cattle, after the consumption of Aflatoxin contaminated feed, AFB1 and AFB2 will be biotransformed into hydroxylated metabolites like aflatoxin M1 (AFM1) and M2 (AFM2) respectively; that later contaminates milk and milk products (10–12).
Glycogen Synthase Kinase 3-beta (GSK-3β) was first identified as a negative regulator of glycogenesis and subsequently reported to regulate varies signalling pathway and cellular function. GSK-3β is an important enzyme in the process of neurogenesis, neuronal migration, differentiation and survival of immature brain (13,14). Numerous studies have been reported for the binding ability of GSK-3β with certain chemical compounds and plant extracts. In addition to this few potential inhibitors have been reported for this enzyme (15-16). Recently aflatoxin B1 has reported binding with GSK-3β (17). This incited to carry out the present study, where GSK-3β was evaluated on the docking behaviour of 13 aflatoxin analogues using Patch Dock as well as physicochemical, drug-likeness and ADME (Absorption, Distribution, Metabolism and Excretion) analyses were also carried out.
Material and Methods
Ligand preparation
Thirteen types of Aflatoxins were prepared as a ligand for this study, as illustrated in Table 1. The ligands were namely (I) Aflatoxin B1; (II) Aflatoxin G1; (III) Aflatoxin M1; (IV) Aflatoxin B2; (V) Aflatoxin G2; (VI) Aflatoxin M2; (VII) Tetrahydrodeoxy aflatoxin B1; (VIII) Compound 2; (IX) Compound 8; (X) Compound 11; (XI) Aflatoxin Q1; (XII) Aflatoxin P1; (XIII) Aflatoxin B1-8,9-epoxide (18). The chemical structures of these thirteen Aflatoxin analogues were first drawn in ChemBio Draw Ultra 12.0 (www.cambridgesoft.com) and subsequently the molecular mechanics (MM2) energy minimization of ligands were carried out by ChemBio 3D Ultra 12.0, according to the reported procedure (19). These minimized energy structures were further utilized for Patch Dock study.
Identification and preparation of target protein
The three dimensional (3D) structure of the target protein Glycogen synthase kinase 3-beta (GSK-3β), PDB 3F88 with resolution of 2.6 A0 was obtained from the Research Collaborator for Structural Bioinformatics (RCSB) Protein Data Bank (www.rcsb.org) (20). The first task was preparing the protein by removing another chains (B, C and D) on it other than the A chain, by using UCSF Chimera software (www.cgi.ucsf.edu/chimera), followed by removing water particles (without hydrogen bonds) which are crystallographically observed (21). Then prepared protein was uploaded to the Patch Dock server with each pre-prepared ligands for docking analysis.
Molecular descriptors calculation
Smiles of our prepared ligands are then copied to an online software to calculate the thirteen descriptors by using the online database of Molinspiration (www.molinspiration.com). The molecular descriptors studied in this study are log P, molecular weight, polar surface area, number of atoms, number of rotatable bond, number of O or N, number of OH or NH, ion channel modulator, drug-likeness including G protein coupled receptors ligand, kinase inhibitor, nuclear receptor ligand, and number of violations to Lipinski’s rule (20).
Analysis of ADME
ADME (Absorption, Distribution, Metabolism and Excretion) analysis was carried out for all the 13 aflatoxin analogues using Swiss ADME. A standard default protocol was adopted for the same (20,22).
Docking studies
To conduct docking studies all the ligand and protein were uploaded to an online server called PatchDock (http://bioinfo3d.cs.tau.ac.il/PatchDock). The docking results were sent to our pre-registered email address and all the data are obtained from it. Patch Dock uses geometry-based molecular docking algorithm method to recognize the binding scores, by binding residues atomic contact energy of the given ligands. The ACE and other information are copied and the first solution selected from the top of lists of several solutions (the docked protein-ligand complex) and downloaded in a database (pdb) file format. Finally, analysis of the protein for having a binding site for the Aflatoxin analogues were done by PyMOL software (www.pymol.org), by opening and measuring the bond distance and also identification and labelling the amino acids which binds with the particular ligand (20,21).
Results and Discussion
Aflatoxins are potent mycotoxins which are now threatening both the feed and livestock sector throughout the world. In recent years’, numbers of aflatoxin binders have been reported in the literature (23). For instance, few commercial available aflatoxin binders are aflabalan, astra-ben-20, anzymit, flow guard, formycin, mycoflix plus, mycosorb, red crown and SA-20 (24–26). Thus, in the present study Glycogen synthase kinase 3-beta (GSK-3β) was evaluated on the docking behaviour of 13 aflatoxin analogues using PatchDock. Table 1 represents the thirteen ligands structure and SMILES selected for the present study.
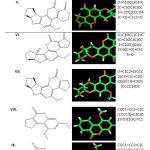 |
Table 1: Thirteen aflatoxin analogues selected for the present study |
Note: *- (I) Aflatoxin B1; (II) Aflatoxin G1; (III) Aflatoxin M1; (IV) Aflatoxin B2; (V) Aflatoxin G2; (VI) Aflatoxin M2; (VII) Tetrahydrodeoxy aflatoxin B1; (VIII) Compound 2; (IX) Compound 8; (X) Compound 11; (XI) Aflatoxin Q1; (XII) Aflatoxin P1; (XIII) Aflatoxin B1-8,9-epoxide. **- Simplified molecular input line entry system (SMILES)
To study the physiochemical and drug-likeness properties of these thirteen ligands (aflatoxin analogues) Lipinski’s rule of five/Thumb of five was applied. According to previous studies violation of the Lipinski’s rule of five is when logA is >5, MW >500, number of N, O (hydrogen bond receptor) is >10, number of OH and NH (hydrogen bond donor) is >5 and number of the rotatable bond (rotb) is >15 (21). With regard physiochemical properties, all the 13 aflatoxin analogues showed nil violation and complied well with the Lipinski’s rule of five as shown in Table 2.
Table 2: Molecular physicochemical descriptors analysis of thirteen aflatoxin analogues using Molinspiration online software tool.
| Ligand Names | Log Aa | TPSAb | Natomsc | MWd | nONe | nOHNHf | Nviolationsg | Nrotbh | Volumei |
|
|
1.48 | 74.98 | 23 | 321.28 | 6 | 0 | 0 | 1 | 253.24 |
| 1.52 | 84.22 | 24 | 328.28 | 7 | 0 | 0 | 1 | 262.22 | |
| -0.90 | 95.21 | 24 | 328.28 | 7 | 1 | 0 | 1 | 260.93 | |
| 1.57 | 74.98 | 23 | 314.29 | 6 | 0 | 0 | 1 | 259.42 | |
| 1.61 | 84.22 | 24 | 330.29 | 7 | 0 | 0 | 1 | 268.41 | |
| 1.00 | 95.21 | 24 | 330.29 | 7 | 1 | 0 | 1 | 267.12 | |
| 2.23 | 57.91 | 22 | 300.31 | 5 | 0 | 0 | 1 | 257.24 | |
| 2.38 | 48.68 | 18 | 246.26 | 4 | 0 | 0 | 2 | 219.24 | |
| 1.72 | 65.75 | 19 | 260.25 | 5 | 0 | 0 | 2 | 221.42 | |
| 1.72 | 65.75 | 19 | 260.25 | 5 | 0 | 0 | 2 | 221.42 | |
| 0.49 | 95.21 | 24 | 328.28 | 7 | 1 | 0 | 1 | 261.28 | |
| 1.20 | 85.98 | 22 | 298.25 | 6 | 1 | 0 | 0 | 235.71 | |
| 1.11 | 87.51 | 24 | 328.28 | 7 | 0 | 0 | 1 | 257.62 |
Note: a- Octanol-Water partition coefficient, b- Polar surface area, c -Number of non-hydrogen atoms, d- Molecular weight, e- Number of hydrogen bond acceptors [ O and N atoms], f -Number of hydrogen bond donors [ OH and NH groups], g- Number of Rule of 5 violations, h- Number of rotatable bonds, i- Molecular volume.
In the case of drug-likeness if the score > 0 was considered active, -5.0 to -0.0 as moderate active and < -5.0 as inactive (21). All the 13 ligands showed active to moderate active score towards all the six descriptions. Interestingly, none of them showed inactive score as shown in Table 3.
Table 3: Drug-likeness property analysis of thirteen aflatoxin analogues using Molinspiration online software tool.
| Ligand Names | GPCR* ligand | Ion channel modulator | Kinase inhibitor | Nuclear receptor ligand | Protease inhibitor | Enzyme inhibitor |
| -0.26 | -0.60 | -0.43 | 0.09 | -0.36 | 0.09 | |
| -0.24 | -0.51 | -0.38 | 0.01 | -0.38 | -0.01 | |
| -0.16 | -0.52 | -0.42 | 0.21 | -0.29 | 0.18 | |
| -0.22 | -0.60 | -0.53 | 0.10 | -0.19 | 0.23 | |
| -0.19 | -0.49 | -0.47 | 0.03 | -0.02 | 0.14 | |
| -0.13 | -0.48 | -0.47 | 0.26 | -0.16 | 0.29 | |
| -0.37 | -0.72 | -0.59 | -0.05 | -0.36 | 0.11 | |
| -0.78 | -0.79 | -0.82 | -0.26 | -0.85 | -0.19 | |
| -0.61 | -0.71 | -0.69 | -0.05 | -0.65 | -0.04 | |
| -0.55 | -0.63 | -0.70 | -0.03 | -0.59 | 0.00 | |
| -0.07 | -0.52 | -0.34 | 0.21 | -0.22 | 0.17 | |
| -0.23 | -0.53 | -0.41 | 0.23 | -0.33 | 0.22 | |
| -0.23 | -0.56 | -0.36 | 0.17 | -0.23 | 0.28 |
Note: *GPCR- G Protein coupled receptors
Similarly, Absorption, Distribution, Metabolism and Excretion (ADME) is simple and essential analysis tool. Recent days, it is commonly accepted in the early stage of drug development program, because of its unique characteristic nature. Table 4 shows the ADME profile of the thirteen selected aflatoxin analogues; all the ligands are predicted to have high gastrointestinal (GI) absorption effect.
Table 4: ADME (Absorption, Distribution, Metabolism and Excretion) analysis of thirteen aflatoxin analogues using Swiss ADME online tool.
| Ligand Names | GI absorption | BBB permeability | P-gp substrate | *CYP1A2 inhibitor | *CYP2C19 inhibitor | *CYP2C9 inhibitor | *CYP2D6 inhibitor | *CYP3A4 inhibitor | # Log Kp |
| High | Yes | No | No | Yes | No | Yes | Yes | -7.05 | |
| High | No | No | No | Yes | No | No | No | -7.05 | |
| High | No | No | No | No | No | Yes | No | -7.92 | |
| High | Yes | No | No | Yes | No | Yes | Yes | -7.27 | |
| High | No | No | No | Yes | No | Yes | No | -7.41 | |
| High | No | Yes | No | No | No | Yes | Yes | -8.14 | |
| High | Yes | Yes | Yes | Yes | No | Yes | Yes | -6.56 | |
| High | Yes | No | Yes | Yes | No | No | No | -6.20 | |
| High | Yes | No | Yes | Yes | No | No | No | -6.95 | |
| High | Yes | No | Yes | No | No | No | No | -6.92 | |
| High | No | No | No | No | No | No | No | -7.94 | |
| High | No | No | No | No | No | Yes | No | -7.20 | |
| High | No | No | No | No | No | Yes | Yes | -7.71 |
Note: *CYP-Cytochrome P450, #Log Kp -Skin Permeation (cm/s).
The docking study and binding site analysis with GSK-3β, showed that ligand of Aflatoxin G2 exhibited the highest atom contact energy (ACE) of -224.82 kcal/mol, on other hand Aflatoxin P1 showed the minimum ACE (-160.33 kcal/mol). The binding energy calculation showed the following order: Aflatoxin G2 ˂Aflatoxin M1˂Aflatoxin B2˂ AFB1-8,9-epoxy˂Tetrahydreoxy Aflatoxin B1 ˂ Aflatoxin G1˂ Compound 2= AFQ1˂ Compound 8˂Aflatoxin B1=Aflatoxin M2 ˂ Compound 11 ˂ AFP1. Further, five ligands (Aflatoxin G1, G2, M2, Tetrahydreoxy Aflatoxin B1 and AFQ1) have shown to interact with Gln185 amino acid residue of GSK-3β. Interesting another five ligands (Aflatoxin B1, M1 and Compound 2, 8, 11) does not show interaction with amino acid residue of GSK-3β (Table 5). Present result infers that the interaction is better for the bio transformed analogues of Aflatoxin (Aflatoxin Q1, Aflatoxin P1, Aflatoxin M2, Tetrahydrodeoxy aflatoxin B1 and AFB1-8, 9-epoxide), rather than the naturally occurring like AFB1, AFGB2 and AFG2. Thus, the present finding was in good agreement with earlier reports (17,27).
Table 5: The interaction energy analysis of thirteen aflatoxin analogues with Glycogen synthase kinase (GSK) using Patch Dock.
| Ligand Names | ACE* (kcal/mol) | Interaction | Bonding distance (Å) |
| -168.64 | No | Nil | |
| -197.50 | Gln185 | 1.91 | |
| -216.37 | No | Nil | |
| -210.99 | Val 135 | 3.52 | |
| -224.82 | Gln 185 | 2.22 | |
| -168.64 | Gln 185 | 3.54 | |
| -197.56 | Gln 185 | 2.32 | |
| -195.41 | No | Nil | |
| -183.87 | No | Nil | |
| -163.27 | No | Nil | |
| -195.41 | Glu 137 | 2.61 | |
| Gln185 | 3.16 | ||
| -160.33 | Asp 200 | 2.60 | |
| -202.42 | Val 135 | 2.88 |
Note: *- Atomic contact energy.
Conclusion
In the present study, all the 13 tested ligands have shown to dock with the target protein Glycogen synthase kinase 3-beta (GSK-3β). Thus, the present study showed the potential of GSK-3β as aflatoxin binder, especially for the bio transformed analogues of Aflatoxin (Aflatoxin Q1, Aflatoxin P1, Aflatoxin M2, Tetrahydrodeoxy aflatoxin B1 and AFB1-8, 9-epoxide).
Acknowledgment
The authors acknowledges Vel Tech Rangarajan Dr. Sagunthala R & D Institute of Science and Technology for all the support provided during the study and we would like thank Mr. Vijayakumar veeraragavan, Research Scholar, Organic Chemistry Laboratory, Vel Tech Rangarajan Dr.Sagunthala R & D Institute of Science and Technology for his help in drawing the chemical structures
Conflict of interest statement
None declared.
References
- Pinotti, L., Ottoboni, M., Giromini, C., Dell’Orto, V., Cheli, F., Mycotoxin contamination in the EU feed supply chain: A focus on Cereal Byproducts. Toxins 8: 45 (2016).
- Vardon, P.J., McLaughlin, C., Nardinelli, C., 2003. Mycotoxins: Risks in plant, animal and human systems. Task Force Report, Council for Agricultural Science and Technology,Ames, Lowa, USA.
- Gratz, S., 2007. Aflatoxin binding by probiotics: Experimental studies on intestinal aflatoxin transport, metabolism and toxicity. Doctoral dissertation, Kuopio University Publications, Kuopio, Finland.
- Kilic, S., Cam, I.B., Tongur, T., Kilic, M., Health risk assessment of exposure to heavy metals and aflatoxins via dietary Intake of dried red pepper from market places in Antalya, Southern Turkey. Journal of Food Science 83: 2675-2681 (2018).
- Balina, A., Kebede, A., Tamiru, Y., Review on Aflatoxin and its Impacts on Livestock. Journal of Dairy and Veterinary Sciences 6: 555685 (2018).
- Eaton, D.L, Gallagher, E.P., Mechanisms of aflatoxin carcinogenesis. Annual Review of Pharmacology and Toxicology 34: 135–172 (1994).
- Gurban, A.M., Epure, P., Oancea, F., Doni, M., Achievements and prospects in electrochemical-based biosensing platforms for aflatoxin M1 detection in milk and dairy products. Sensors 17: 1–21 (2017).
- Lindahl, J., Kagera, I., Grace, D., Aflatoxin M1 levels in different marketed milk products in Nairobi, Kenya. Mycotoxin Research 34: 289-295 (2018).
- Gross-Steinmeyer, K., Eaton, D.L., Dietary modulation of the biotransformation and genotoxicity of aflatoxin B1. Toxicology 299: 69–79 (2012).
- Gizachew, D., Szonyi, B., Tegegne, A., Hanson, J., Grace, D., Aflatoxin contamination of milk and dairy feeds in the Greater Addis Ababa milk shed, Ethiopia. Food Control 59:773–779 (2016).
- Shigute, T., Washe, A.P., Reduction of aflatoxin M1 Levels during Ethiopian traditional fermented milk (Ergo) production. Journal of Food Quality 2018: 4570223 (2018).
- Mohammadi, H., 2011. A Review of aflatoxin M1, milk and milk products. Aflatoxins – Biochemistry and Molecular Biology, Ramon Gerardo Guevara-Gonzalez, IntechOpen Limited, London, United Kingdom.
- Luo, J., The role of GSK3beta in the development of the central nervous system. Front Biology 7: 212–220 (2012).
- Malagon, S.G.G., Muñoz, A.M.L., Doro, D., Bolger, T.G., Poon, E., Tucker, E.R., Al-Lami, H.A., Krause, M., Phiel, C.J., Chesier, L., Liu, K.J., Glycogen synthase kinase 3 controls migration of the neural crest lineage in mouse and Xenopus. Nature Communications 9:1–15 (2018).
- Bustanji, Y., Taha, M.O., Almasri, I.M., Al-Ghussein, M.A., Mohammad, M.K., Alkhatib, H.S., Inhibition of glycogen synthase kinase by curcumin : Investigation by simulated molecular docking and subsequent in vitro / in vivo evaluation. Journal of Enzyme Inhibition and Medicinal Chemistry 24: 771–778 (2009).
- Varadharaj, V., Kandakati, N., Glycogen synthase kinase-3 beta protein inhibition by selected phytocompounds in silico. Asian Journal of Pharmaceutical and Clinical Research 10: 87–90 (2017).
- Wang, Y., Liu, J., Zhang, L., He, X., Zhang, J.Z.H., Computational search for aflatoxin binding proteins. Chemical Physics Letters 685: 1–8 (2017).
- Wogan, G.N., Edwards, G.S., Newberne, P.M., Structure-activity relationships in toxicity and carcinogenicity of aflatoxins and analogs. Cancer Research 31: 1936–1942 (1971).
- Vijayakumar, V., Radhakrishnan, N., Rameshkumar, C., Molecular docking analysis of
- imidazole derivatives and Polybenzimidazole analogs as inhibitors of Superoxide Dismutase (SOD) and Xanthine Oxidase (XO). IEEE International Conference on Smart Technologies and Management for Computing, Communication, Controls, Energy and Materials proceedings, 513 (2017).
- Narayanaswamy, R., Molecular docking analysis of Alginate oligosaccharides ( Alg2-Alg6 ) as bacterial collagenase inhibitor. Journal of Applied Cosmetology 35: 15–23 (2017).
- Veeraragavan, V., Radhakrishnan, N., Chidambaram, R., Biodegradability nature of Polybenzimidazole analogs by modulating two histidine degradation enzymes (Urocanase and Formiminoglutamase): in silico Approach. Asian Journal of Chemistry 30: 2205–2209 (2018).
- Wang, J., Urban, L., The impact of early ADME profiling on drug discovery and development strategy. Drug Discovery World 5: 73–96 (2004).
- Van Kessel, T.F.M., Hiang-chek, N., Aflatoxin binders – how to get the best value for money. International Poultry Production 12: 33–35 (2018).
- Nazarizadeh, H., Pourreza, J., Evaluation of three mycotoxin binders to prevent the adverse effects of aflatoxin B 1 in growing broilers. Journal of Applied Animal Research 47: 135–139 (2019).
- Tapia-salazar, M., García-pérez, O.D., Nieto-lópez, M.G., Villarreal-cavazos, D.A., Cruz- suárez, L.E., Ricque-marie, D., Evaluating the efficacy of commercially available aflatoxin binders for decreasing the effects of aflatoxicosis on Pacific white shrimp Litopenaeus vannamei. Hidrobiológica 27: 411–418 (2017).Diaz, D.E, Hagler, W.M. Jr., Blackwelder, J.T., Eve, J.A., Brinton, A., Anderson, K.L., Jones, F.T., Whitlow, L.W., Aflatoxin Binders II : Reduction of aflatoxin M1 in milk by sequestering agents of cows consuming aflatoxin in feed. Mycopathologia 157: 233–241(2004).
- Picconi, M.A., Pico, G.A., Gatti, C.A., Fluorometric studies on the binding of aflatoxin B1
- to bovine serum albumin. Health and Environmental Research Online 44: 141–148 (1984).








