Vijitra Luang-In1* , Manatchanok Yotchaisarn1, Worachot Saengha1, Piyachat Udomwong2, Sirirat Deeseenthum1 and Kedsukon Maneewan1
, Manatchanok Yotchaisarn1, Worachot Saengha1, Piyachat Udomwong2, Sirirat Deeseenthum1 and Kedsukon Maneewan1
1Natural Antioxidant Innovation Research Unit, Department of Biotechnology, Faculty of Technology, Mahasarakham University, Khamriang, Kantarawichai, Maha Sarakham, 44150, Thailand.
2International College of Digital Innovation, Chiang Mai University, Chiang Mai 50200, Thailand.
Corresponding Author E-mail: vijitra.l@msu.ac.th
DOI : https://dx.doi.org/10.13005/bpj/1735
Abstract
This study aimed to isolate and identify bacteria that can produce amylase enzyme from the unexplored Nasinuan Forest, Kantarawichai District, Mahasarakham Province, Thailand. Thirteen bacterial isolates with amylase-producing capacity on 1% starch agar were identified using 16S rRNA sequencing. Twelve bacteria were gram-positive, rod shaped and identified as Bacillus spp. and one bacterium with gram-negative and rod shaped character was Enterobacter cloacae. Their closest relatives were found in India, China, Korea, Indonesia, Argentina, Italy, Israel, USA, Argentina and South Africa. These bacteria were tested for specific amylase activity after 1-3 days enzyme induction with 1% starch at 37°C. The results showed the highest specific activity at day 2 incubation in the order: Bacillus cereus 3.5AL2 > 3.4AL1 > 1.4AL3 and thus 2-day enzyme induction was chosen for further analysis. Bacillus sp. 3.5AL2 was found to exhibit the highest specific amylase enzyme activity of 1.97 ± 0.41 U/mg protein at the optimal conditions of 60°C and pH 7.0 after 30 min incubation with 1% starch in 0.05 M PBS buffer. This amylase–producing bacterial strain offers great potential for applications in food and agricultural industries in Thailand.
Keywords
Amylase; Bacteria; Nasinuan forest; soil
Download this article as:| Copy the following to cite this article: Luang-In V, Yotchaisarn M, Saengha W, Udomwong P, Deeseenthum S, Maneewan K. Isolation and Identification of Amylase-producing Bacteria from Soil in Nasinuan Community Forest, Maha Sarakham, Thailand. Biomed Pharmacol J 2019;12(3). |
| Copy the following to cite this URL: Luang-In V, Yotchaisarn M, Saengha W, Udomwong P, Deeseenthum S, Maneewan K. Isolation and Identification of Amylase-producing Bacteria from Soil in Nasinuan Community Forest, Maha Sarakham, Thailand. Biomed Pharmacol J 2019;12(3). Available from: http://biomedpharmajournal.org/?p=28497 |
Introduction
The starch hydrolytic amylases (α-amylase, β-amylase and glucoamylase) are ones of the most widely used enzymes in present-day biotechnology. Glucoamylases (1,4-α-D-glucan glucanohydrolase; EC 3.2.1.3) or amyloglucosidases, are exo-acting amylases that produce glucose from the non-reducing end of starch and corresponding oligosaccharides. Glucoamylases are implemented commercially for the transformation of malto-oligosaccharides into glucose1. Although amylases are produced from different sources (microorganisms, plants and animals), amylases from microbes are most suitable for industrial production due to microbial short growth period, low cost effective production, eco-friendly behavior, less handling issues for workers, productivity2 and easy manipulation of bacterial genes3. Bacteria and fungi tend to secrete amylases outside the cells to perform extra cellular digestion of starch into sugars. Increasing industrial demand for microbial amylases has been observed due to their specificity of reaction, mild conditions prerequisite for the reaction, and less energy consumption than the conventional non-enzymatic chemical methods.
Extensive application of amylase in food, starch liquefaction, saccharification, brewing, detergent, paper, textile and distilling industries, has brought about a greater attention for the increase in the indigenous production of α-amylase4. Bacillus is a common bacterial source for industrial amylase production. However, different strains have different optimal growth conditions and enzymatic production profile. Reportedly, Bacillus strains have been extensively used industrially to produce α-amylase including B. amyloliquefaciens, B. subtilis5, B. licheniformis6, B. stearothermophilus7, B. megaterium8 and B. circulans9.
To date, a number of amylase-producing bacteria has been documented, however, no study on amylase-producing bacteria from soil in the Nasinuan Community Forest, Kantharawichai District, Mahasarakham Province, Thailand has been reported. This forest seems to be rich in microbial biodiversity that can be useful in the production of industrial enzymes including amylase. This is the first report to identify amylase-producing bacteria isolated from Nasinuan Forest. These bacterial amylases can be locally applied to various industries including pulp and paper, textile, bioethanol, brewery, food processing, animal feed, and agriculture in Thailand in the future.
Materials and Methods
Soil samples
Soil samples were randomly collected below the soil surface 15 cm and kept in polystyrene bags from Nasinuan Community Forest, Kantarawichai District, Mahasarakham Province, Thailand (area of 9.6 hectare; coordinate of 16.340941, 103.210799).
Isolation of amylase-producing bacteria
Soil sample (10 g) was suspended in 90 mL of sterile 0.85% NaCl solution. The suspensions (100 µL) of serial dilutions were spread on starch agar (g/L); 3.0 beef extract, 10.0 soluble starch, 15.0 agar pH 7.0 and incubated at 37 ºC for 7 days. Next, the plates were flooded with Gram’s iodine. Any colonies with formation of clear zone around the colonies were subcultured in liquid broth and streaked at least five times to obtain pure isolates as confirmed by Gram staining and 1000X light microscopic observation. The pure isolates were point inoculated on starch agar and incubated at 37°C for 7 days. The diameters of the clear zones over the diameters of the colonies were measured using a ruler as the Halo : Colony ratio.
16S rRNA gene sequencing and phylogenetic analysis
Pure bacterial isolates were identified using genomic DNAs obtained from the above method and universal primers: forward primer 27F 5′-GAGAGTTTGATYCTGGCTCAG-3′ and reverse primer 1492R 5’AAGGAGGTGATCCARCCGCA -3′. In 25 µL PCR mixture, it was composed of genomic DNA 0.5 ng, 2X Master Mix (One PCR) of 100 mM Tris-HCl (pH 9.1), 0.1% TritonTMX-100, 200 mM dNTP, 1.5 mM MgCl2, 0.005 U Taq DNA Polymerase and 0.2 µM forward and reverse primer with volume adjustment with nuclease-free water. PCR thermocycler (Thermo Scientific Hybaid Px2) was programmed as follows: (1) initial denaturation for 2 min at 94 ºC for 1 cycle; (2) denaturation at 94 ºC for 45 s; annealing at 54 ºC for 45 s, and extension at 72 ºC for 1 min for 32 cycles; (3) final extension at 72 ºC for 7 min. Samples were held at 4 ºC till further analysis. The PCR products of 16S rRNAs (~ 1,500 bp) were detected on 0.8% agarose gel, purified using the PCR product purification kit (Vivantis, Malaysia), sent to First Base Co. Ltd. (Malaysia) for DNA sequencing. The 16S rRNA gene sequences were then compared with others available in GenBank using BLASTN program (Basic Local Alignment Search Tools)10. The Phylogenetic tree was constructed using Muscle method for sequence alignment and maximum likelihood method using MEGA X with 1,000 replicates of bootstrap values11. All 16S rRNA partial sequences of our amylase-producing isolates were deposited on NCBI database.
Determination of amylase enzyme activity
The method followed Bhaskara et al. (2011)12. The three bacteria with the highest Halo: Colony ratios were chosen for determination of amylase enzyme activity. Each isolate was subcultured in starch liquid broth (g/L): 10.0 soluble starch, 10.0 peptone, 20.0 yeast extract, 0.05 KH2PO4, 0.015 MnCl2.4H2O, 0.25 MgSO4.7H2O, 0.05 CaCl2.2H2O, 0.01 FeSO4.7H2O and incubated at 37 ºC, 150 rpm for 1, 2 and 3 days. The clear supernatant (crude extracellular amylase enzyme) was obtained after centrifugation at 10,000g for 15 min at 4 ºC. The crude extract was concentrated using MWCO 10 kDa ultracentrifuge protein concentrator (Vivaspin, Sartorius, UK).
The DNS method13 was used to determine the amylase activity of each bacterial isolate at each enzyme induction days. One mL of crude enzyme was mixed with 1 mL of 1% starch solution in 1 mL 0.05 M sodium phosphate buffer pH 7.0. The samples were incubated at 37°C for 30 min. After incubation, 0.5 mL DNS solution was added to each sample to stop the reaction and then boiled at 100°C for 5 min in the water bath. The colour intensity of the solution was observed by measuring the optical density (OD) using a spectrophotometer at 575 nm. The reading was compared to a prepared blank solution (without crude enzyme). The OD values of samples at T30min were subtracted from those of samples at T0min since glucose still remained in the T0min samples after enzyme induction process at 1, 2, 3 days. The process was carried out in triplicates. The concentration of glucose produced for each solution was obtained from the glucose standard curve. The activity of amylase was calculated. One unit of amylase activity is defined as the amount of amylase required to catalyze the formation of reducing sugar which is equal to 1 µmol of glucose per min under assay conditions14.
The crude enzyme of the bacterial isolate having the highest activity was chosen for further work.
In order to determine the specific enzyme activity of the selected isolates, the Folin-Lowry method for total protein estimation was used15. The specific enzyme activity was measured using the following formula.
Specific activity (U/mg) = Enzyme activity (U/ml)/Extracellular protein concentration (mg/ml)
Optimal pH and temperature of amylase enzyme activity
Soluble starch solutions (1%) in different pH from 3 to 10 were tested. One mL of different 1% substrate solution was added along with 1 mL of the respective buffers; 0.05 M citrate buffer (pH 3 to 5), 0.05 M sodium phosphate buffer (pH 6 and 7), 0.05 M Tris-HCl (pH 8 and 9) and 0.05 M glycine NaOH (pH 10). One mL of crude enzyme was added to these buffers as well. The samples were incubated at 37°C for 30 min. The specific amylase activity was calculated. The pH at which the highest activity was observed was noted. Likewise, different substrate solutions were made by dissolving 1% soluble starch in pH 7.0 solutions One mL of 1% soluble starch was added along with 1 mL of 0.05 M sodium phosphate buffer (pH 7). One mL of crude enzyme was added to the buffers as well. The samples were incubated at 20, 30, 40, 50, 60, 70, and 80°C for 30 min. The specific amylase activity was calculated. The temperature at which the highest activity was observed was noted. Both optimal pH and temperature were used to determine the final specific amylase enzyme activity.
Statistical analysis
One-Way Analysis Of Variance (One-way ANOVA) was used with Duncan Multiple Range’s Test on SPSS Statistics Ver. 17.0. Results were expressed as means ± SD with statistical difference when p < 0.05.
Results
Isolation of amylase-producing bacteria
In this study, 13 amylase-positive isolates showed clear zones on 1% starch agar with different Halo : Colony ratios. The colonies showing clear zones of iodine solution were taken as positive starch-degrading bacterial colonies. All bacterial isolates showed similar colony morphologies and appeared as Gram-positive and rod-shaped bacteria (Table 1). The result showed Halo : Colony ratios ranging from 1.18 to 1.71 (Table 1). Bacterial isolates were then used to determine the optimal amylase enzyme induction duration and only the 3 isolates with highest specific activities were shown in 3.4.
Strain identification of amylase-producing bacteria
All 13 amylase-positive bacterial strains were subjected to 16S rRNA gene sequencing for strain identification. The BLAST results displayed that all amylase-positive isolates belong to the genus Bacillus, except for one isolate belonging to Enterobacter (Table 2). Their closest relatives were found in India, China, Korea, Indonesia, Argentina, Italy, Israel, USA, Argentina and South Africa with a range of 96-100% identity.
Phylogenetic analysis
The phylogenetic tree of 13 amylase-positive bacterial strains and 2 reference strains with putative amylase enzymes using MEGA 7.0 showed that E. cloacae 2.1AL2 was evolutionarily different from the other Bacillus strains (Fig. 1). B. thuringiensis serovar konkukian (NCBI accession no. AB617494.1) reference strain evolved differently to our Bacillus bacteria. However, another reference strain B. cereus ATCC 14579 (NCBI accession no. MG708176.1) isolated from safflower leaf, Eqypt showed similar evolution to our bacteria (Fig. 1).
 |
Table 1: Characteristics and Halo : Colony ratios of 13 amylase-producing bacterial strains |
Optimal amylase enzyme induction duration
It was shown that the incubation duration that induced amylase production with the highest specific activity was 2-3 days among all 3 isolates without statistic differences. Bacillus sp. 3.5AL2 seemed to produce the highest specific activity at 0.91 ± 0.12 U/mg at day 2 at 37°C (Table 3). Thus, this strain and this condition was used for inducing amylase production for further analysis.
Optimal pH and temperature of amylase enzyme activity
Bacillus sp. 3.5AL2 showed the highest specific activity at pH 7.0 when 37°C was fixed and at 60°C when pH 7.0 was fixed (Fig. 2). Thus, both optimal conditions were used to determine specific amylase activity and 1.97 ± 0.41 U/mg (Table 4) was obtained.
Table 2: Thirteen amylase-positive bacterial strains identified by 16S rRNA analysis
| Isolate | Accession
no. a |
Closest relativeb | Accession no.c | % Identityd | Origine |
| 1.1AL1 | MK578206 | Bacillus pseudomycoides FJAT-hcl-17 | KY653098.1 | 99% | Host Paris polyphylla var. chinensis, China |
| 1.1AL2 | MK578207 | Bacillus sp. 188Cu-As | KM349197.1 | 99% | Biofilms, Argentina |
| 1.2AL3 | MK578208 | Bacillus anthracis SAK4 | MG706137.1 | 100% | Soil, Republic of Korea |
| 1.4AL1 | MK578209 | Bacillus sp. CNJ732 PL04 | DQ448749.1 | 99% | Marine sediment, USA |
| 1.4AL3 | MK578210 | Bacillus cereus B19 | MK229038.1 | 99% | Wheat grain, Isarael |
| 2.1AL2 | MK578211 | Enterobacter cloacae HNXY160623 | KX431213.1 | 98% | Avian embryo, China |
| 2.3AL1 | MK578212 | Bacillus thuringiensis BD17-E12 | HF584771.1 | 99% | Grapevine root system, Italy |
| 2.3AL8 | MK578213 | Bacillus cereus DFT-1 | KY750685.1 | 98% | Seawater of industrial area, Indonesia |
| 2.3AL9 | MK578214 | Bacillus sp. PTP1 | KY910137.1 | 96% | Papaya mealybug gut, India |
| 2.4AL2 | MK578215 | Bacillus pseudomycoides 74 | MH910178.1 | 99% | India |
| 3.2AL1 | MK578216 | Bacillus thuringiensis
IARI-IIWP-38 |
KF054891.1 | 99% | Wheat rhizospere, India |
| 3.4AL1 | MK578217 | Bacillus cereus F4a | MK088302.1 | 99% | Tea rhizosphere soil, India |
| 3.5AL2 | MK578218 | Bacillus cereus SP1-AB4 | MH013307.1 | 99% | Marine sponge, South Africa |
aGenBank accession no. of our strains deposited on NCBI website (http://www.ncbi.nlm.nih.gov/pubmed)
bClosest species with highest % identity and highest Max score on BLAST search
cGenBank accession no. of closest relative strains on NCBI website
dBased on BLAST search results, identity (%) of strains compared to the
closest relatives.
eBased on BLAST search results, origin of the closest relatives.
 |
Figure 1: Phylogenetic tree of 13 amylase-positive bacterial strains and 2 Bacillus reference strains |
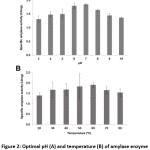 |
Figure 2: Optimal pH (A) and temperature (B) of amylase enzyme activity from Bacillus sp. 3.5AL2 |
Table 3: Optimal amylase enzyme induction duration
|
Isolate |
Specific amylase activity
(U/mg protein) |
||
| 24 h | 48 h | 72 h | |
| Bacillus sp. 1.1AL2 | 0.21 ± 0.04Ab | 0.40 ± 0.04Ca | 0.41 ± 0.04Ba |
| B. cereus 2.3AL8 | 0.45 ± 0.06Ab | 0.62 ± 0.04Ba | 0.65 ± 0.06Aa |
| Bacillus sp. 3.5AL2 | 0.32 ± 0.11Ab | 0.91 ± 0.12Aa | 0.86 ± 0.11Aa |
Values are means ± SD in triplicate.
Capital and small letters indicate statistic differences (p < 0.05) in the column and row, respectively.
Table 4: Specific amylase enzyme activity at optimal pH and temperature
| Strain | Activity (U/mg) | Optimal Temp. (°C) | Optimal pH |
| Bacillus sp. 3.5AL2 | 1.97 ± 0.41 | 60 | 7.0 |
Discussion
Thai customs (2017) data showed that Thailand has imported industrial enzymes from other countries with the value of 2,625 million Bahts in 2016 and the import rate was increased by 9.40% per year. One of the most widely used enzymes in Thailand, amylase, has been applied in garments, textile and food industries. However, there is a lack of local production of commercial amylase and thus this work aimed to isolate soil bacteria from local forest with the capacity to produce amylase for industrial uses.
Starch-rich residues may be a better potential source where amylase-positive bacteria can be isolated16. In addition, these could be isolated from places such as cassava farms, soil, and processing flour factories17. In this work, 13 amylase-producing bacteria were isolated and identified as Bacillus spp. and Enterobacter sp. which is similar to the previous reports18, 19, 20 where Bacillus spp. were mostly found as amylase producers.
Our results showed that Bacillus sp. 3.5AL2, the isolate with the highest specific activity among 13 isolates, produced the highest specific activity at 0.91 ± 0.12 U/mg at 2-day incubation at 37°C. The optimal pH and temperature for amylase activity were 7.0 and 60°C, respectively. Both optimal conditions gave the specific amylase activity from Bacillus sp. 3.5AL2 of 1.97 ± 0.41 U/mg. It is known that the enzymes that can work at high temperature are considered as having an advantage in industrial processes such as starch liquefaction21. Thus, amylase from Bacillus sp. 3.5AL2 has a potential use in such industries.
Similarly, Bacillus sp. GM890 showed the optimal temperature for amylase at 60°C20, Bacillus licheniformis AI20 show the highest activity between the range of 60-80°C22 and Bacillus sp. WA21 had a lower optimal temperature of 55°C for amylase19.
Most of the starch – degrading bacterial strain revealed a pH range between 6.0 and 7.0 for normal growth and enzyme activity4. Likewise, in this work, Bacillus sp. 3.5AL2 had an optimal pH at 7.0 which does not require any addition of acid or alkali. Our finding was in accordance with Bacillus sp. isolated from dhal industry waste exhibiting an optimal temperature of 60°C23 and B. amyloliquefaciens with an optimal pH of 7.024. Nevertheless, Bacillus sp. WA21 showed the optimal pH of the amylase at 6.0 which is less than that found in this study19.
This kind of a study is very important for the starting point of commercial amylase production in Thailand. However, additional research investigation is essential to make amylase production cost-effective. There is a need for thorough enzyme characterization, further determination of the thermostability, pH stability, influence of different metal ions and different substrates, especially agricultural wastes in Thailand, and their concentrations.
After obtaining all the data needed as mentioned above, it is thought that the production of commercial amylase in Thailand enzyme industries will be increasing and widespread in every region and in turn will be benefiting the country’s economy due to self-reliance on its own resources to produce amylase enzyme.
Conclusion
This is the first report of identifying 13 amylase-producing bacterial isolates from soil in Nasinuan Community Forest, Maha Sarakham. All bacteria were identified as Bacillus spp. except for one from Enterobacter genus. These bacteria can be used for amylase production and applied locally and nationally in agriculture, food processing and textile industries in the future. Thus, this will reduce the cost of industrial enzyme import from other countries, offer sustainability of local enzyme production and enhance the economy of the nation.
Acknowledgments
The authors would like to thank Mahasarakham University (Grant Year 2018) for financial support awarded to SD, and Faculty of Technology, Mahasarakham University (Grant Year 2017) awarded to VL.
Conflict of Interest
Authors declare no conflict of interest.
References
- Pandey A. Glucoamylase research: An overview. 47:439-445 (1995).
- Burhan A.I., Nisa U., Gokhan C., Omer C., Ashabil A., and Osman G. Enzymatic properties of a novel thermostable, thermophilic, alkaline and chelator resistant amylase from an alkaliphilic Bacillus isolate ANT-6. Process Biochem. 38: 1397-1403 (2003).
- Gurung N., Ray S., Bose S., and Rai, V.A. Broader view: Microbial enzymes and their relevance in industries, medicine and beyond. Biomed Res Internat. 1-18. doi:10.1155/2013/ 329121 (2013).
- Gupta R., Gigras P., Mohapatra H., Goswami V.K., and Chauhan B. Microbial α-amylases: a biotechnological perspective. Process Biochem. 38:1599–1616 (2003).
- Takasaki Y. An amylase producing maltotetrose and from maltopentose from circulans. Agric Biol Chem. 47:2193–2199 (1983).
- Fogarty W.M., and Kelly C.T. Amylase, amyloglucosidase and related glucanases. Rose A.H. Economic Microbiology, Microbial Enzymes and Bioconversion, New York Academic Press Inc, vol. 5. Pp. 115–170 (1980).
- Wind R.D., Buitelaar R.M.G., Huizing H.J., and Dijkhuizen L. Characterization of a new Bacillus stearothermophilus isolate: a highly thermostable α-amylase producing strain. Appl Microbiol Biotechnol. 41:155-162 (1994).
- Brumm P.J., Hebeda R.E., and Teague W.M. Purification & characterization of commercialized, cloned megaterium α-amylase. Part I: purification & hydrolytic properties. Starch. 43:319–323 (1991).
- Takasaki Y. An amylase producing maltotriose from subtilis. Agric Bio. Chem. 49:1091–1097 (1985).
- Lowry O.H., Rosebrough N.J., and Randall R.J. Protein measurement with the folin phenol reagents. J Biol Chem. 193: 265-275 (1951).
- Altschul S.F., Gish W., Miller W., Myers E.W., and Lipman D.J. Basic local alignment search tool. J Mol 215:403–410 (1990).
- Bhaskara R.K.V., Ashwini K., Gaurav K., and Karthik L. Optimization, production and partial purification of extracellular α-amylase from Bacillus marini. Arc Appl Sci Res. 3:33-42 (2011).
- GhoseK. Measurement of cellulase activities. Pure Appl Chem. 59: 257-268 (1987).
- Haq U.I., Ashraf H., Iqbal J., and Qadeer M.A. Production of α-amylase by Bacillus licheniformis using an economical medium. Biores Technol. 87:57-61 (2003).
- Mishra S., and Behera N. Amylase activity of starch degrading bacteria is isolated from soil receiving kitchen wastes. Afr J Biotechnol. 7: 3326–3331 (2008).
- Fossi B.T., Taveaand F., and Ndjonenkeu T. Production and Partial Characterization of a Themostable amylase from Ascomycetes yeast strain isolated from starchy soils. Afr J Biotechnol. 4:14–18 (2005).
- Cordeiro C.A.M. Production and propertiesof α–amylase from thermophilic Bacillus sp. Brazil J Microbiol. 33:57-61 (2002).
- Asad W. Extracellular enzyme production by indigenous thermophilic bacteria: Partial purification and characterization of α–amylase by Bacillus sp. WA21. Pakistan J Botany. 43:1045-1052 (2011).
- Kim T.U. Purification and characterization of a maltotetraose-forming alkaline α-amylase from an alkalophilic Bacillus strain, GM8901. Appl Envin Microbiol. 61: 83105–3112 (1995).
- Sivaramakrishnan S., Gangadharan D., Madhavan K.N., and Pandey A. Solid culturing of Bacillus amyloliquefaciens for alpha amylase production. Food Technol 44:269-274 (2006).
- Abdel-Fattah Y.R., Soliman N.A., El-Toukhy N.M., El-Gendi H., and Ahmed R.S. Purification, and characterization of thermostable α-amylase produced by Bacillus licheniformis isolate AI20. J Chem. 2013:1-11 (2013).
- Thippeswamy S., Girigowda K., and Mulimani V.H. Isolation and identification of α-amylase producing Bacillus from Dhal
industry waste. Indian J Biochem Biophysics. 43: 295-298 (2006). - Abd-ElhalemT., El-Sawy M., Rawia F.G., and Khadiga A.A.T. Production of amylase from Bacillus amyloliquefaciens under submerged fermentation using some agro–industrialby-products. Ann Agri Sci. 60:193-202 (2015).
- Tamura K., and Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512-526 (1993).








