Majid Moghbeli
Biology Department, Islamic Azad University, Damghn Branch, Damghan, Iran
DOI : https://dx.doi.org/10.13005/bpj/594
Abstract
Amylase is one of the most used enzymes in industry. It is an extracellular enzyme and has allocated 20% of the enzyme marketing to itself. Amylases are widely used in several industrial processes such as food, wood, paper, detergent, and pharmaceutical production, and fermentation. Due to their high potential of amylase overexpression, Bacilli are considered as the most important and prevalently used as the producers of this enzyme. Thus cloning an amylase expressing gene in a well-known host such as Escherichia coli and Bacillus subtilis can aid the process of identification and production of new amylases and their functional improvement. Bacillus licheniformis genome was extracted and the amylase-coding gene was isolated and amplified using PCR with genome as the template and specific primers. The amplified amylase gene was transferred to T.vector which subsequently was digested by HindIII and BamHI. The digested fragment was ligated to the shuttle vector pMR12 and transferred into E. coli TOP10 cells. After confirmation of successful transformation of E. coli TOP10 cells with the recombinant plasmid, it was extracted and directly transferred into B. subtilis WB600. Alpha-amylase gene was cloned in pMR12 vector and B subtilis as the host and the presence of the gene fragment in kanamycin-resistant colonies was confirmed using enzymatic digestion and PCR. Amylase gene was successfully cloned according to the stability of the recombinant vector harboring this gene after several consequent subcultures.
Keywords
Alpha-amylase gene; Cloning; Bacillus licheniformis; Bacillus subtilis; PCR
Download this article as:| Copy the following to cite this article: Moghbeli M. Cloning Alpha-Amylase Gene from Iranian Indigenous Bacillus Licheniformis in Bacillus Subtilis. Biomed Pharmacol J 2015;8(1) |
| Copy the following to cite this URL: Moghbeli M. Cloning Alpha-Amylase Gene from Iranian Indigenous Bacillus Licheniformis in Bacillus Subtilis. Biomed Pharmacol J 2015;8(1). Available from: http://biomedpharmajournal.org/?p=1240 |
Introduction
Starch and its substrates are widely spread throughout the nature and extensively used in industry. Starch is a polymer formed via glycosidic bonds. These bonds are stable in high pH and hydrolyzed in low pH. There are two type of glucose polymer chains in starch: 1) amylose, 2) amylopectin. Amylose is a linear polymer composed of more than 6000 glucose units which are bound together by glycosidic alpha 4 to 1 bonds. Decomposition and decay of starch is mainly achieved by the activity of soil microorganisms. The produced amylase by these organisms is usually secreted into their periphery or attached to the plasma membrane. However some microbial strains possess intracellular amylases. Starch is hydrolyzed by a variety of enzymes such as endoamylases, exoamylases, transferases, and gylcosidic bond-hydrolyzing enzymes [1] Alpha-amylases are classifies in endoamylases group which hydrolyze glycosidic alpha 4 to 1 bonds and found in a variety of microorganisms. Alpha-amylases are sub-categorized according to the substrate hydrolysis level. Saccharogenic type of this enzyme hydrolyzes 50 to 60% of the bonds [1, 2] The products of starch hydrolysis are implemented in food industry as additive for candy production, bakery products, conserving, and frozen food manufacturing. There are a variety of methods for production of hydrolyzed starch such as acid hydrolysis which has some drawbacks including high cost, low quality of products and the corrosion of systems. Enzymatic approach is conducted in 110 °C by Bacilli-derived amylases. This process is performed in 55 °C by Aspergillus niger and Aspergillus oryzae alpha-amylases. The main sources of alpha-amylase are plants, animals, and microorganisms (sorendra 2011). Bacteria and fungi secrete their amylase to the cell periphery. A. niger, A. oryzae, and Penicillium expansum produce a large amount of this enzyme (sorendra). Species of Bacilli such as B. cereus, B. circulans, B. subtilis, and B. licheniformis are important in industrial production of amylases [3] These bacteria are implemented in commercial production of thermostable alpha-amylases and are valuable in industry. Amylases are widely used in industrial operations including starch processing, textile starch removal, paper production, as additive in detergents, bread improvement, pharmaceutical industry, ethanol production, etc[4] . Alpha-amylase is the most used enzyme in starch industry and implemented in starch liquidification in which it is converted to glucose and fructose syrup [5].
Materials and Methods
Isolation of the complete alpha-amylase gene (promoter and gene) from Iranian indigenous B. licheniformis by PCR
According to the alpha-amylase genes registered in Genbank (NCBI) two primers amyF:5ATTGGTAACTGTATCTCAGCTTGAAGA3 amyR:5TCCGTCCTCTCTGCTCTTCTATCTT3
were designed to amplify the entire gene from the beginning of the promoter to the terminator.
Genomic DNA was extracted from the indigenous strain isolated from Ardabil hot springs using phenol:chlorophorm protocol. In order to isolate and amplify the gene PCR was carried out in 25 µl volume. Each reaction consisted of 0.4 µmol of each primer, 0.2 µM dNTPs, 2 units of smarTaqDNA polymerase, 2.5 µl of 10X PCR buffer, MgCL2 with the final concentration of 2.5 mM, and 50 ng of the extracted DNA. PCR program included 5 minutes initial denaturation in 94 °C, 30 cycles of one minute denaturation in 94 °C, one minute annealing in 49 °C and 1 minute and 45 seconds elongation in 72 °C, and one cycle of 8 minutes final extention at 72 °C. The amplified fragment was visualized by electrophoresis and recovered from agarose gel.
Insertion of the complete alpha-amylase gene into T.vector and sequencing of the inserted fragment
The PCR product was cloned into T.vector. The recombinant plasmids were subsequently transferred to TOP10 competent cells via heat shock procedure. The bacterial cells were plated on culture media containing ampicillin antibiotic (80µg/ml), IPTG, and X-gal and incubated for 16 hours at 37 °C. Grown white colonies were subcultured and their recombinant plasmids were extracted using a plasmid extraction kit. The plasmids were sequenced with T.vector universal primers. The resulted sequence was aligned with other amylase genes registered in Genbank using Blast software and the homology levels were compared.
Isolation of alpha-amylase gene without signal peptide and its cloning in the shuttle vector pMR12
Two primers amyF1: GCAAGCTTGCAAATCTTAATGGGACGCTG and amyR1: GCGGATCCTCTGCTCTTCTATCTTTGAAC were designed with HindIII and BamHI restriction sites (underlined), respectively. amyF1 sequence was designed according to the downstream region of the signal peptide sequence in order to omit this part and implement the signal peptide of pMR12. According to pWB980 sequence map which is the source of pMR12 multiple cloning site, the usage of the above mentioned enzymes is appropriate for extracellular secretion of the exogenous expressed protein.
DNA extraction from bacteria was carried out according to Sambrook method and concentration and purity of the extracted DNA was determined using a spectrophotometer at the wavelength of 260 nm. In addition, 1% agarose gel electrophoresis method was implemented in order to visualize the quality and further purify the extracted DNA. PCR was performed in 25 µl volumes. Each reaction consisted of 0.4 µmol of each primer, 0.2 µM dNTPs, 2 units of smarTaqDNA polymerase, 2.5 µl of 10X PCR buffer, MgCL2 with the final conteration of 2.5 mM, and 50 ng of the extracted DNA. PCR program included one step of 5 minutes initial denaturation in 94 °C, 30 cycles of one minute denaturation in 94 °C, one minute annealing in 58 °C and 1 minute and 45 seconds elongation in 72 °C, and one cycle of 8 minutes final extension at 72 °C.
The PCR product was cloned in T.vector Recombinant plasmids were transferred into TOP10 competent cells with the same procedure as the previous step and cultivated on culture medium containing ampicillin, IPTG, and X-gal for 16 hours at 37 °C. White colonies were subcultured and their recombinant plasmids were extracted by a plasmid extraction kit. Enzymatic single and double digestion reactions were carried out simultaneously on recombinant T.vectors and the shuttle vector pMR12. The digested pMR12 vector and the separated fragment from T.vector were visualized and recovered from gel using electrophoresis and purification kit and prepared for ligation reaction.
The ligation reaction consisted of 6.5 µl vector, 2 µl gene, 4.5 µl ddH2O, 1.5 µl buffer, and 0.5 µl T4 Ligase in a 15-µl reaction which was incubated in 16 °C for 16 hours. The product of ligation reaction was transferred into calcium chloride treated TOP10 competent cells and plated on ampicillin containing medium. The grown colonies were screened and the recombinant plasmid was extracted.
Cloning the recombinant plasmid in Bacillus subtilis
Preparing B. subtilis WB600 competent cells: 20 ml of spc medium (tris-base, 50% w/v glucose, 1.2% w/v magnesium sulphate, 10% w/v bacto yeast extract, 1% w/v casaaminoacids) was inoculated with some fresh colonies grown on LB agar and incubated at 37 °C until it reaches OD = 0.5. Consequently, 2 ml of the culture was transferred to 200 ml SPII solution to be incubated at 37°C and 170 rpm shaking for 90 minutes. The mixture was centrifuged and the supernatant was separated and stored. 18 ml of the supernatant and 2 ml glycerol were added to the precipitated cells and transferred to -20 °C freezer.
25µl of bacterial competent cells was transferred into a 1.5ml microtube and equal volume of SPII+EGTA was added to the cells. Two volumes of 480 µl of the mixture were transferred into two sterile microtubes in which 20 µl of each the recombinant and empty plasmids (as the positive control) were also added separately. The microtubes were incubated and shook at 37 °C for an hour. 250 µl of tris-base solution containing 0.5% glucose was added to each mixture after the incubation. From each microtube, 200 µl was transferred onto kanamycin containing aga5r media and incubated at 37 °C for 24 hours. The grown colonies were screened by colony PCR.
Results
After confirmation of B. lichenisformis strain by cultivating on starch agar medium, its genomic DNA was extracted according to Sambrook procedure. The concentration and quality of the extracted DNA was determined by spectrophotometric methods (520 ng/µl, 280/260 ratio = 1.2) and 1% agarose gel electrophoresis.
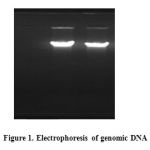 |
Figure 1 |
Amplification of amylase gene was carried out through PCR using amyF-amyR and amyF1-AmyR1 primer pairs. Five microliter of each reaction product was loaded on 1% agarose gel and electrophoresed for 45 minutes at 80 V.
 |
Figure 2: Electrophoresis of the PCR product with amyF-amyR as the primer pair
Lane 1: negative control. Lane 2: molecular weight marker. Lane 3: PCR product The length of the amplified complete alpha-amylase gene was approximately 1.7 kb. |
 |
Figure 3: Electrophoresis of the PCR product with amyF1-amyR1 primers
Lane 1: negative control. Lane 2: molecular weight marker. Lane 3: PCR product The length of the amplified fragments was found to be 1460 kb. |
After the cloning of amylase gene in T.vector and transferring into E. coli TOP10 competent cells and cultivating on X-gal containing medium, 45 white colonies were picked and examined. The extracted plasmids were cut in enzymatic single and double digestion reactions. Successful cloning was confirmed by the length of the produced fragments after digestion.
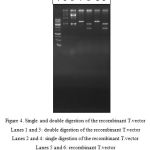 |
Figure 4: Single and double digestion of the recombinant T.vectorLanes 1 and 3: double digestion of the recombinant T.vector
Lanes 2 and 4: single digestion of the recombinant T.vector Lanes 5 and 6: recombinant T.vector Lane 7: molecular weight marker |
After the cloning of amylase gene in T.vector and transferring into E. coli TOP10 competent cells and cultivating on X-gal containing medium, 45 white colonies were picked and examined. The extracted plasmids were cut in enzymatic single and double digestion reactions. Successful cloning was confirmed by the length of the produced fragments after digestion.
After the amplification of alpha amylase gene with amyF1 and amyF2 primers and the genome of B. licheniformis as the template, the PCR product was cloned in T.vector and transferred into E. coli cells. This time the digestion reactions were prepared for the recombinant T.vector harboring the alpha-amylase gene with BamHI and HindIII restriction sites. The shuttle vector pMR12 was also cut by the same emzymes in single and double digestion reactions. Both digested plasmid and the separated gene fragment from T.vector were recovered from agarose gel and used in a ligation reaction. The ligation product was transferred into E. coli TOP10 cells. pET21 was used in order to control the transformation process. Colony PCR as implemented to confirm the presence of alpha-amylase gene in the transformed plasmid in E. coli TOP10 cells.
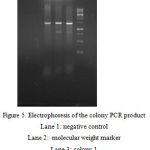 |
Figure 5: Electrophoresis of the colony PCR product
Lane 1: negative control Lane 2: molecular weight marker Lane 3: colony 1 Lane 4: colony 2 Lane 5: colony 3 |
Despite the confirmation by colony PCR, plasmid extraction and digestion were also carried out for further reassurance.
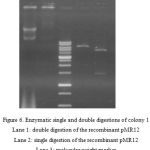 |
Figure 6: Enzymatic single and double digestions of colony 1
Lane 1: double digestion of the recombinant pMR12 Lane 2: single digestion of the recombinant pMR12 Lane 3: molecular weight marker Lane 5: the recombinant pMR12 |
Transformation of the recombinant pMR12 harboring alpha-amylase gene was carried out according to the described procedure in materials and methods. Approximately 23 positive colonies were observed which were cultivated on kanamycin containing medium and used for plasmid extraction afterwards.
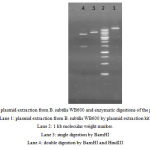 |
Figure 7: plasmid extraction from B. subtilis WB600 and enzymatic digestions of the plasmids. Lane 1: plasmid extraction from B. subtilis WB600 by plasmid extraction kit. Lane 2: 1 kb molecular weight marker. Lane 3: single digestion by BamHI Lane 4: double digestion by BamHI and HindIII |
Discussion
Alpha-amylase is one of the most important industrial enzymes which acts on starch, glycogen and oligosaccharides as the substrate. It is an extracellular enzyme and has allocated 20% of the enzyme marketing to itself[6] The microbial alpha-amylase has several industrial applications and is widely used in pharmaceutical and medical processes, analytical chemistry, starch processing, fermentation, and carbohydrates, detergent, and animal feed manufacturing [7,8]. According to the industrial importance of this enzyme considerable efforts have been focused on isolation of new producer bacterial species in order to address the newly emerged industrial strategies; e.g. alkaline amylases which are appropriate for detergent industry.
Thermostabilty is considered a desirable feature in manufacturing. The advantages of thermostable amylases include reducing the contamination risk and external cooling costs, higher solubility of reagents, and reduced viscosity which assists rapid mixing and more efficient pumping[8]. There are a variety of methods for bacterial genomic DNA extraction. In order to extract the genomic DNA, Sambrook and manual methods using lysis buffer were implemented in this study. Both methods are simple, rapid and efficient. Wang et al. (2009) used Xu method to extract genomic DNA from samples[9].
Yamabhai et al. (2008) designed their primers according to B. licheniformis DSM13 alpha-amylase gene which was registered in Genbank in order to isolate and amplify its homologues gene in B. licheniformis 8785[10] Mobini et al. (2013) used a pair of primers with AscI and NotI restriction sites which amplified the alpha amylase gene from Iranian indigenous B. licheniformis without its signal peptide for cloning and transferring this gene in Saccharomyces cerevisiae[5]. Primers amyF1 and amyR1 were designed in this study to isolate and amplify amylase gene from B. licheniformis and cloning in shuttle expression vector. Since we aimed for expression of this enzyme with the strong signal peptide of pMR12 vector, the primers were designed in a way to omit amylase signal peptide upon the amplification in PCR.
Zhang et al. (2011) isolated a strain of B. amyloliquefaciens from high temperature soils of hot springs in China and cultivated the strain on a medium containing starch (2 g/L), peptone (1 g/L), NaCl (5.0 g/L), and meat extract (0.3 g/L) in order to produce alpha-amylase [9] Abd-el fateh et al. (2013) used 250-ml flasks filled with 50 ml of a medium containing starch (5 g/L), yeast extract (5 g/L), (NH2)4SO4 (5.2 g/L), MgSO4.7H2O (2.0 g/L), KH2PO4 (3 g/L), and CaCl2.2H2O (25.0 g/L) in order to produce alpha-amylase by B. licheniformis AI20 [11]
Sulivan et al. (1984) implemented LB broth medium for E. coli and Penassay broth for B. subtilis cultivation[13]. In another study Doekel used YTx2 medium for E. coli and B. subtilis cultivation Doekel [14]. In this study LB medium (liquid and solid) was used for both E. coli and B. subtilis growth. Various concentrations have been implemented in previous studies for screening the recombinants. For instance Brückner (1984) screened the recombinants by 100 µg/ml ampicillin in E. coli and 5 µg/ml kanamycin in B. subtilis [12]. Sulivan (1984) used 30 mg/ml ampicillin and kanamycin for the selection of transformed E. coli cells[13] . In the present study transformed E. coli and B. subtilis cells were screened using 100 µg/ml ampicillin and 10 µg/ml kanamycin, respectively.
Sulivan (1984) used CsCl-EtBr gradient for bulk extraction and boiling (according to Holms, 1981) for small scale extraction of plasmids[13]. Trieu-Cout (1991) used the following solution in the presence of lysozyme to extract plasmid DNA from gram positive bacteria: 25% Sucrose, 25mM Tris-HCl, 10mM EDTA, pH 8.0. Shuichi et al. (1983) transferred the thermostable alpha-amylase gene from B. stearothermophilus to plasmids pTB53 and pTB90 by which they transformed B. subtilis and B. stearothermophilus cells[16]. Transformed B. stearothermophilus harboring the recombinant plasmid showed a fivefold enzyme production (9.20 U/mgCDW) in comparison with the wild strain[16] .
subtilis WB600 was implemented in this study as the main host for the recombinant pMR12 harboring alpha-amylase gene. This bacterium has a strong ability in secreting proteins directly into the medium which facilitates the downstream processes. Sulivan (1984) used the method described by Davis (1980) for E. coli transformation[13]. They also used liquid medium for the selection of transformants (Sulivan, 1984). However in the present study heat shock and CaCl2 method was implemented for transformation and solid medium for selection which was found to be efficient. Differently, Nagami (1988) used calcium shock technique[17].
Yasbin et al. (1975) used two synthetic media called GMI (CaCl2, MgCl2, glucose) and GMII (GMI without CaCl2 and MgCl2 with concentrations of 5.0 mM and 5.2 mM) in addition to tryptose blood agar and Penassay broth media for B. subtilis transformation[18]. Transformation of B. subtilis was carried out in the present study through two procedures, being a two-step method described by Harwood in “Molecular biological methods for Bacillus” (Harwood, 1990) and using OTB solution. The results of B. subtilis transformation and confirmation of the recombinant pMR12 presence after subsequent subcultures suggested that the constructed shuttle vector by usS had the appropriate structural stability. Cloning alpha-amylase gene from B. licheniformis in pMR12 and B. subtilis WB600 can be an important step towards the production of this enzyme with higher efficiency
References
- El-Fallal, A., et al., Starch and Microbial α-Amylases: From Concepts to Biotechnological Applications. Carbohydrates–Comprehensive Studies on Glycobiology and Glycotechnology. InTech, 2012: p. 459-488.
- Thippeswamy, S., K. Girigowda, and V. Mulimani, Isolation and identification of alpha-amylase producing Bacillus sp. from dhal industry waste. Indian journal of Biochemistry and biophysics, 2006. 43(5): p. 295.
- Souza, P.M.d., Application of microbial α-amylase in industry-A review. Brazilian Journal of Microbiology, 2010. 41(4): p. 850-861.
- Mojsov, K., Microbial alpha-amylases and their industrial applications: a review. International Journal of Management, IT and Engineering (IJMIE), 2012. 2(10): p. 583-609.
- Javan, F. and M. Mobini-Dehkordi, Application of alpha-amylase in biotechnology. 2012.
- Demirkan, E., Production, purification, and characterization of\ alpha-amylase by Bacillus subtilis and its mutant derivates. Turkish Journal of Biology, 2011. 35(6): p. 705-712.
- Akcan, N., High level production of extracellular β-galactosidase from Bacillus licheniformis ATCC 12759 in submerged fermentation. African Journal of Microbiology Research, 2011. 5(26): p. 4615-4621.
- Özcan, N., A. Altinalan, and M.S. EKİNCİ, Molecular Cloning of an\ alpha-Amylase Gene from Bacillus subtilis RSKK246 and Its Expression in Escherichia coli and in Bacillus subtilis. Turkish Journal of Veterinary and Animal Sciences, 2001. 25(2): p. 197-201.
- Li, W., et al., Cloning of the thermostable cellulase gene from newly isolated Bacillus subtilis and its expression in Escherichia coli. Molecular biotechnology, 2008. 40(2): p. 195-201.
- Yamabhai, M., et al., Secretion of recombinant Bacillus hydrolytic enzymes using Escherichia coli expression systems. Journal of Biotechnology, 2008. 133(1): p. 50-57.
- Abdel-Fattah, Y.R., et al., Production, purification, and characterization of thermostable α-amylase produced by Bacillus licheniformis Isolate AI20. Journal of Chemistry, 2012. 2013.
- Brückner, R., O. Shoseyov, and R.H. Doi, Multiple active forms of a novel serine protease from Bacillus subtilis. Molecular and General Genetics MGG, 1990. 221(3): p. 486-490.
- Sullivan, M.A., R.E. Yasbin, and F.E. Young, New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene, 1984. 29(1): p. 21-26.
- Doekel, S., K. Eppelmann, and M.A. Marahiel, Heterologous expression of nonribosomal peptide synthetases in B. subtilis: construction of a bi-functional B. subtilis/E. coli shuttle vector system. FEMS microbiology letters, 2002. 216(2): p. 185-191.
- Aiba, S., K. Kitai, and T. Imanaka, Cloning and expression of thermostable α-amylase gene from Bacillus stearothermophilus in Bacillus stearothermophilus and Bacillus subtilis. Applied and environmental microbiology, 1983. 46(5): p. 1059-1065.
- Aiba, S., et al., Stabilisation in bacillus stearothermophilus of a recombinant plasmid carrying the homologous α‐amylase gene. Journal of Chemical Technology and Biotechnology, 1986. 36(12): p. 599-609.
- Henner, D., M. Yang, and E. Ferrari, Localization of Bacillus subtilis sacU (Hy) mutations to two linked genes with similarities to the conserved procaryotic family of two-component signalling systems. Journal of bacteriology, 1988. 170(11): p. 5102-5109.
- Yasbin, R.E., G. Wilson, and F.E. Young, Transformation and transfection in lysogenic strains of Bacillus subtilis: evidence for selective induction of prophage in competent cells. Journal of bacteriology, 1975. 121(1): p. 296-304.








