Mansour Haddad
Department of Clinical Sciences, Faculty of Pharmacy, Philadelphia University, P.O box 3341, Irbid, Jordan P.C 211-10.
DOI : https://dx.doi.org/10.13005/bpj/455
Abstract
Rimonabant decreases body weight in man and improves metabolic parameters in animal models of obesity. The contribution of peripheral targets, particularly skeletal muscle, for CB1 cannabinoid receptor antagonism in these effects is, however, unclear. The purpose of this study was, therefore, to investigate the impact of CB1 receptors inhibition upon downstream signalling and to characterize the molecular mechanisms that mediate the direct effects of rimonabant in rat primary skeletal muscle cells. Skeletal muscle cells were cultured from vastus lateralis obtained from 180-200 g male Wistar rats according to Blau and Webster method with slight modification. mRNA expression, and downstream signalling genes were examined by gene expression microarray. QRT-PCR (Taqman) identified CB1 cannabinoid receptor mRNA expression with Ct values of 26.5-27.5 in myoblasts, myotubes and skeletal muscle tissue. The findings from the present study indicate, for the first time, that rimonabant up-regulates the mRNA content of neuropeptide Y (NPY), apelin (APLN) and lipocalin 2 (LCN2), and down-regulate the mRNA content of G protein alpha subunit (GNAI3) and nuclear receptor subfamily 4, group A (NR4A). These findings provide evidence for potential functional role of rimonabant in skeletal muscle in terms of insulin resistance, glucose and fat metabolism, inflammation and myogenesis. The impact of CB1 receptor expression in skeletal muscle will be the subject of further investigation
Keywords
Rimonabant; skeletal muscle; myotube; signalling
Download this article as:| Copy the following to cite this article: Haddad M. What Does Rimonabant Do in Rat Primary Skeletal Muscle Cells. Biomed Pharmacol J 2014;7(1) |
| Copy the following to cite this URL: Haddad M. What Does Rimonabant Do in Rat Primary Skeletal Muscle Cells. Biomed Pharmacol J 2014;7(1). Available from: http://biomedpharmajournal.org/?p=2851 |
Introduction
Obesity has grown in the United States and throughout the world at an unprecedented rate in recent decades (Ogden et al., 2006; Singh et al., 2011). Obesity, especially fat accumulation in the intra-abdominal region is linked to disease states such as: type 2 diabetes mellitus (Colditz et al., 1995), hypertension (Witteman et al., 1989), cardiovascular disease (Rimm et al., 1995), osteoarthritis, steatohepatitis, and cancer (Calle et al., 2004a; Calle et al., 2003; Calle et al., 2004b). Indeed, obesity has been linked to the development of insulin resistance and other metabolic abnormalities underlying the pathology of Type 2 diabetes mellitus. The pathogenesis of Type 2 diabetes mellitus is the failure of insulin action on metabolic tissues – known as insulin resistance. In other words, insulin resistance is the reduced ability of insulin to effectively stimulate glucose transport due to alteration of insulin receptor expression or insulin release in response to food ingestion (Del Prato et al., 2002; Ferrannini, 1998; Mosthaf et al., 1991). Moreover, insulin resistance can be associated with altered insulin receptor sensitivity which might be modulated by potential pharmacological agents such as RIM (Kahn, 1978).
Skeletal muscle is the largest tissue in the human body and represents ~40% of the human body mass and 35-40% of the total body weight in the rat (Delbono et al., 2007; Pedersen, 2011). Indeed, it plays a crucial role in maintaining body glucose homeostasis (James et al., 1985) and it clears out the majority (70-80%) of ingested glucose since it is the main site for insulin-dependent and non-insulin-dependent or contraction-mediated glucose uptake (Baron et al., 1988; Ferrannini et al., 1983; Toft et al., 1998). Therefore, skeletal muscle is generally considered as the most important site of insulin resistance. Insulin resistance in skeletal muscle participates in glucose intolerance and consequently in compensatory hyperinsulinemia (Nistala et al., 2006).
A novel therapeutic intervention in the treatment of obesity and hyperglycemia might occur through the antagonism of the endocannabinoid system. Indeed, studies from animals and humans have shown that the endocannabinoids are increased in the obese state. In addition, obese animal models showed that levels of endocannabinoids were elevated in the hypothalamus and peripheral tissues (Di Marzo et al., 2001; Matias et al., 2006; Osei-Hyiaman et al., 2005). Moreover, studies showed that circulating levels of AEA and 2-AG were raised, and 2-AG was also found to be elevated in visceral adipose tissue in obese and hyperglycaemic type 2 diabetic patients (Bluher et al., 2006; Engeli et al., 2005; Matias et al., 2006). Furthermore, CB1 knock-out mice were found to be resistant to diet-induced obesity (Osei-Hyiaman et al., 2005; Ravinet Trillou et al., 2004). Originally, CB1 receptor antagonism was investigated as a mediator of the hypophagic effect which leads to weight loss (Di Marzo et al., 2001; Vickers et al., 2003). However, independent to hypophagic weight loss attributed to CB1 receptor antagonism, CB1 receptor antagonism was also discovered to improve metabolic parameters, such as increased glucose uptake in skeletal muscle (Liu et al., 2005), increased glucose tolerance (Bermudez-Siva et al., 2006; Nogueiras et al., 2008) and decreased hyperinsulinemia (Doyon et al., 2006) as well as effects on lipids (increased HDL/LDL ratio and increased triglyceride) (Despres et al., 2005).
As previously stated, CB1 receptor mRNA and protein expression has been detected in skeletal muscle myotubes and tissues of rodents and humans (Cavuoto et al., 2007; Pagotto et al., 2006). In addition, in mice fed a high fat diet (HFD), the expression of CB1 in skeletal muscle was found to be up-regulated (Pagotto et al., 2006).
From agonist and antagonist studies, both in vitro and in vivo, it seems that the endocannabinoid system plays a role in glucose transport in skeletal muscle. In vitro, using cell culture models (L6 mouse myotube cell line and human primary skeletal muscle cells), Esposito et al and Eckardt et al, respectively showed that CB1 receptor antagonism using RIM enhanced basal and insulin-stimulated glucose transport activity (Eckardt et al., 2008; Esposito et al., 2008). In vivo, chronic CB1 receptor antagonism was found to increase insulin-stimulated glucose transport activity in obese mice (Liu et al., 2005). Furthermore, chronic CB1 receptor antagonism during euglycemic hyperinsulinemic clamp increased glucose uptake in diet-induced obese rats by several skeletal muscle groups (Nogueiras et al., 2008). These data suggest that the endocannabinoid system can play a role regarding glucose transport in skeletal muscle.
The glucose transport into the skeletal muscle is facilitated mainly by the GLUT4 isoform. The mechanism, through which CB1 antagonists affects glucose levels is unknown. Moreover, The effects of CB1 cannabinoid receptor antagonism in terms of glucose and fatty acid metabolism on peripheral targets, particularly skeletal muscle are unclear (Eckardt et al., 2009; Lindborg et al., 2011; Lipina et al., 2010). In an attempt to address this issue, the signalling events underlying the inhibition of the CB1 receptor in rat primary skeletal muscle cells were investigated. In other words, the effect of RIM on gene expression was also investigated in rat primary skeletal muscle cells.
Aims
The main aim of these experiments was to investigate the impact of CB1 inhibition upon signaling and to characterize the molecular mechanisms that mediate the direct effects of RIM on skeletal muscle. In other words, The main aim was to assess gene expression in rat primary skeletal muscle cells in response to Rimonabant.
Experimental Design and Methods
Primary skeletal muscle cells
Muscle culture was performed as Blau and Webster method with slight modification (Blau et al., 1981). Vastus lateralis muscles from Wistar rats were removed and immersed in phosphate buffered saline (PBS), washed to remove the remnants of blood, and minced finely with scissors and scalpel blades on a Petri dish.
Then, the minced muscle was transferred to a 50 ml flask containing a “flea‟ and 5-10 ml of 0.25% (W/V) trypsin/EDTA (1X) for incubation at 37ºC for 15 minutes. After that, the supernatant was transferred to a 50 ml flask and neutralised with an equal volume of medium (streptomycin, penicillin, foetal bovine serum and Ham’s F10), then centrifuged at 1700 rpm for 5 minutes. The collected cells were filtered through 100 μm nylon mesh (“cell strainers”) to purify the cells from the debris, and centrifuged for 10 minutes at 17,000 rpm (g=26) at room temperature.
The supernatant layer was removed and the cell pellet (satellite cells) was re-suspended in Ham‟s F10 growth medium, pre-plated on uncoated flasks for 10 minutes at 37 C° to purify these satellite cells from fibroblasts present in the extract, and then transferred to culture flasks coated with 0.2% (W/V) gelatin. The satellite cells were then grown to confluent myoblasts and differentiated into myotubes in growth medium; 20% (V/V) fetal bovine serum (FBS) and 5 ml of penicillin and streptomycin (10,000 units penicillin and 10 mg streptomycin/ml in 0.9% NaCl) were added to Ham‟s F10. After one day, the cells were fed with fresh medium, cells require fresh medium every 48 hours.
The cells were fed with 20% (V/V) FBS fresh medium for three weeks, then reduced to 10% (V/V) FBS fresh medium for two weeks and then changed to 6% horse serum and 10 mM glucose Ham‟s F10 for two to three days.
RNA Extraction and QRT-PCR (Taqman)
Myoblast and myotube cells were grown and differentiated as described above. The cells were collected in TriReagent, and processed according to the manufacturer’s directions. RNA was reverse transcribed into cDNA using Superscript III reverse transcriptase (Invitrogen). Then, QRT-PCR (Taqman) was performed as described according to the manufacturer’s directions.
Gene expression levels (in arbitrary units) were determined from the mean of triplicate determinants of each sample. Data from Taqman were only used if the slope of the standard curve for each plate was between -3.2 and -3.6 and R2 values of more than 0.99. In addition, Ct values of triplicate readings for an individual sample, which were more than 0.5 Ct apart, were excluded. The forward primer 5’CCAAAAGTGGAGAGCGACAAC3′, reverse primer 5’CGTCTCGAAGGTCCCAATGT3′ and probe 5’ATCCAGATCACCATGCCGTTCACA3′ were used for Cnr1.
Microarray
Experimental design
Myotubes were cultured in 25 cm2 flasks and incubated with RIM 100 nM for 24 hours (four flasks/condition). Ethanol 0.01% was used as a vehicle control. Fresh charcoal stripped fetal bovine serum 4% was replaced for four hours before performing the treatment.
Procedure for the microarray
The treated myotubes were lysed using Trizol and stored at -80°C, then total RNA was extracted and cleaned up with the Qiagen Rneasy kit according to the manufacturer’s instructions.
All RNA samples were examined using Agilent Bioanalyzer. Samples that had a RIN greater than 8 were included in the analysis. These values indicated that RNA degradation did not occur. Distinct bands of 28S and 18S RNA were visualized in all RNA samples isolated from the vehicle, ACEA, RIM and U0126-treated myotubes to confirm that the RNA was suitable for microarray procedure (Table 1).

|
Table 1: Quality of RNA isolated from myotubes.
|
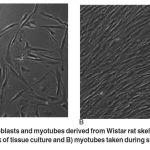 |
Figure 1: Representative myoblasts and myotubes derived from Wistar rat skeletal muscle. A) myoblasts taken during the third week of tissue culture and B) myotubes taken during sixth week of tissue culture
|
Briefly, synthesis of labelled cRNA, hybridization, and scanning of microarrays were carried out as described (Schafer et al.; Voss et al., 2005) according to established methods in the manufacturer’s protocols (Affymetrix, Santa Clara, CA).
Gene expression profiling, data processing and analysis
Global changes in gene expression induced by RIM were determined using Affymetrix Rat Genome 230 PM Array.
Pre-analysis data treatment
Before analysis took place, the raw microarray (Cel files) data was pre-processed through RMA (Robust Multichip Averaging) algorithm. RMA has the following components; background correction, normalization and probe summarization.
Background correction was based on the distribution of perfect match (PM) values amongst probes on an Affymetrix array. Plate and exon (PM only) arrays contain a set of antigenomic background probes that are not matched to any putative transcript region. By default, the Affymetrix software estimates probe background signal by the median response of all background probes with matching GC content to the probe in question. This background signal is then subtracted from the probe intensity to yield a background-corrected intensity.
Quantile normalization was performed within all arrays based on the raw intensities (Raymond et al., 2010). Normalization was performed to remove nonbiological effect among all arrays. This makes all arrays comparable. The RMA used quantile normalization. In this normalization, 1) probe intensities were ranked for each array, 2) the average across all arrays was taken and 3) the corresponding values of probe intensities were all set to the average. Consequently, these steps force the distribution of measurements on all arrays to be equal.
Probe summarization was performed through observing probe behavior [i.e., log transformed (PM) after background correction; any values attributed to background were eliminated] on the log scale as the sum of the actual expression value on the log scale (a probe specific term).
The summary of the final steps of data transformation were:
Log2 transformation of the intensities.
Tukey’s median polish was used to summarize the intensity values of individual probes into a single measurement for the corresponding gene.
After raw data was normalized, genespring GX 11 software was used to identify differentially expressed genes.
Initial characterisation of microarray gene expression data
The microarray data were summarized using Principal Components Analysis (PCA) based on gene expression patterns for each of the experimental conditions All four replicates treated with RIM were grouped in the scatter plot.
Correlation was performed between pairs of conditions using Affymetrix IDs common to all microarrays. In order to test the reproducibility of these data, the correlation co-efficient of technical replicates were calculated between conditions on the normalized data. All correlation coefficients were found to be higher than 0.98 across all conditions. The normalized data (not transformed to the median of all samples) was used in all subsequent analyses.
Statistical analysis
After preprocessing the microarray raw data, genespring GX 11 was used to identify differentially expressed genes. Using the normalized microarray data, conditions were compared using one-way ANOVA and Benjamini-Hochberg test. A statistically significant difference was accepted when the treatment effect yielded a P < 0.05 to correct for the likelihood of false positives. This p-value was used as a cut off for differentially expressed genes. Then, the 2 fold-change approach (increase or decrease) was utilized.
Data were further analysed using Ingenuity Pathways Analysis (IPA) http://www.ingenuity.com. The list of differentially regulated genes identified by the microarray analysis using genespring GX 11 was exported into IPA, which predicted biological functions of genes that are associated with particular biological processes.
The effect of the RIM on gene expression
The influence of Rimonabant on gene expression was investigated using Affymetrix microarray.
Interestingly, treatment with RIM altered mRNA expression of a number of genes. These genes represent transcription regulators (ANKRD57, BCL3, CEBPD, EGR2, GBX2, HIVEP1, ID4, IRX3), cytokines (CCL20, CXCL6), transporters (AQP1, LCN2, RBP1, SLC16A7), peptidases (C3, PRSS35), nuclear receptors (NR4A1, NR4A2, NR4A2, NR4A3, NR4A3), GPCR (PTGER4) and growth factor (BMP6). In particular, LCN2 and NPY were up-regulated by RIM compared to vehicle while NR4A were down-regulated by RIM compared to vehicle (Table 5 3).
Table 2: The fold change in the expression of genes influenced by RIM in rat primary skeletal muscle cells (+ means up-regulated and – means down-regulated in response to RIM).
| Fold Change | Symbol | Entrez Gene Name |
| 3.79 | LCN2 | lipocalin 2 |
| 3.71 | NPY | neuropeptide Y |
| 2.77 | CCL20 | chemokine (C-C motif) ligand 20 |
| 2.75 | C3 | complement component 3 |
| 2.58 | GBX2 | gastrulation brain homeobox 2 |
| 2.53 | RBP1 | retinol binding protein 1, cellular |
| 2.46 | TGM1 | transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase) |
| 2.42 | RASD1 | RAS, dexamethasone-induced 1 |
| 2.39 | BCL3 | B-cell CLL/lymphoma 3 |
| 2.31 | CEBPD | CCAAT/enhancer binding protein (C/EBP), delta |
| 2.30 | G0S2 | G0/G1switch 2 |
| 2.26 | CXCL6 | chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) |
| 2.17 | MGP | matrix Gla protein |
| 2.16 | C9orf16 | chromosome 9 open reading frame 16 |
| 2.14 | APLN | Apelin |
| 2.13 | RND1 | Rho family GTPase 1 |
| 2.10 | BMP6 | bone morphogenetic protein 6 |
| 2.08 | RASL12 | RAS-like, family 12 |
| 2.08 | KRT18 | keratin 18 |
| -2.01 | ANKRD57 | ankyrin repeat domain 57 |
| -2.02 | PRSS35 | protease, serine, 35 |
| -2.03 | RGS2 | regulator of G-protein signaling 2, 24kDa |
| -2.06 | RAD51 | RAD51 homolog (S. cerevisiae) |
| -2.07 | TRIO | triple functional domain (PTPRF interacting) |
| -2.09 | NR4A2 | nuclear receptor subfamily 4, group A, member 2 |
| -2.09 | BUB1 | budding uninhibited by benzimidazoles 1 homolog (yeast) |
| -2.09 | PTGER4 | prostaglandin E receptor 4 (subtype EP4) |
| -2.10 | STARD13 | StAR-related lipid transfer (START) domain containing 13 |
| -2.11 | KIF20B | kinesin family member 20B |
| -2.12 | GNAI3 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| -2.13 | SLC16A7 | solute carrier family 16, member 7 (monocarboxylic acid transporter 2) |
| -2.14 | EFEMP1 | EGF containing fibulin-like extracellular matrix protein 1 |
| -2.18 | LRRN4CL | LRRN4 C-terminal like |
| -2.21 | IRX3 | iroquois homeobox 3 |
| -2.21 | NR4A2 | nuclear receptor subfamily 4, group A, member 2 |
| -2.21 | ITGBL1 | integrin, beta-like 1 (with EGF-like repeat domains) |
| -2.24 | MKI67 | antigen identified by monoclonal antibody Ki-67 |
| -2.33 | HIVEP1 | human immunodeficiency virus type I enhancer binding protein 1 |
| -2.35 | KIF11 | kinesin family member 11 |
| -2.42 | ARL4C | ADP-ribosylation factor-like 4C |
| -2.56 | TRIB3 | tribbles homolog 3 (Drosophila) |
| -2.56 | AQP1 | aquaporin 1 (Colton blood group) |
| -2.58 | ECT2 | epithelial cell transforming sequence 2 oncogene |
| -2.72 | C1QTNF3 | C1q and tumor necrosis factor related protein 3 |
| -2.84 | NR4A3 | nuclear receptor subfamily 4, group A, member 3 |
| -3.23 | NR4A3 | nuclear receptor subfamily 4, group A, member 3 |
| -3.56 | EGR2 | early growth response 2 |
| -3.88 | HAS2 | hyaluronan synthase 2 |
| -4.23 | ID4 | inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| -5.22 | NR4A1 | nuclear receptor subfamily 4, group A, member 1 |
The treatment with RIM affected the expression of a number of genes involved in the activation of the following biological functions; adipogenesis of cells, inflammatory response, activation of phagocytes, proliferation of smooth muscle cells and impairment of tumorigenesis (Table 3).
Table 3: The biological functions ascribed to genes that were altered by treatment with RIM. For the gene abbreviation, see (Table 2).
| Category | Functions Annotation | Predicted Activation State | Molecules |
| Cellular Development | adipogenesis of cells | Increased | C3,CEBPD,NPY,NR4A1,NR4A2, NR4A3 |
| Connective Tissue Development and Function | adipogenesis of cells | Increased | C3,CEBPD,NPY,NR4A1,NR4A2, NR4A3 |
| Inflammatory Response | inflammatory response | Increased | C3,CCL20,CXCL6,GNAI3,LCN2,NPY,NR4A2,PTGER4 |
| Inflammatory Response | activation of phagocytes | Increased | C3,CXCL6,LCN2,NPY |
| Cell-To-Cell Signaling and Interaction | activation of phagocytes | Increased | C3,CXCL6,LCN2,NPY |
| Hematological System Development and Function | activation of phagocytes | Increased | C3,CXCL6,LCN2,NPY |
| Immune Cell Trafficking | activation of phagocytes | Increased | C3,CXCL6,LCN2,NPY |
| Antigen Presentation | activation of phagocytes | Increased | C3,CXCL6,LCN2,NPY |
| Cell-To-Cell Signaling and Interaction | activation of cells | Increased | BCL3,C3,CXCL6,EGR2,KRT18, LCN2,NPY |
| Cellular Growth and Proliferation | proliferation of smooth muscle cells | Increased | CEBPD,NR4A1,NR4A2,NR4A3 |
| Skeletal and Muscular System Development and Function | proliferation of smooth muscle cells | Increased | CEBPD,NR4A1,NR4A2,NR4A3 |
| Cancer | tumorigenesis | Decreased | AQP1,ARL4C,BCL3,BMP6,BUB1,C3,CEBPD,CXCL6,ECT2,EFEMP1,HAS2,ID4,ITGBL1,KIF11,KIF20B,KRT18,LCN2,MGP,MKI67,NR4A1,NR4A2,NR4A3,PTGER4,RAD51,RASL12,RBP1,RGS2,STARD13,TRIO |
Discussion
Skeletal muscle is a main site of fatty acid and glucose metabolism and involved in energy balance (Zurlo et al., 1990). Skeletal muscle also produces skeletal movement through its contraction and maintains body glucose homeostasis, and so pharmacological tools that target the molecular mechanisms controlling skeletal muscle metabolism, functions and physiological roles may be therapeutically useful for metabolically related disorders.
In the present study, CB1 receptor mRNA was detected in both skeletal muscle tissue and rat primary cells using QRT-PCR (Taqman). In previous studies, CB1 receptor was found to be expressed in human and rodent skeletal muscle (Cavuoto et al., 2007). Interestingly, CB1 receptor protein expression was found to be significantly decreased in soleus muscle from obese compared to lean Zucker rats (Lindborg et al., 2011). However, CB1 receptor mRNA expression in soleus muscle was found to be increased after high fat feeding in C56BL/6 mice (Pagotto et al., 2006).
In a previous study investigating the effect of RIM in overweight or obese patients with type 2 diabetes, RIM was found to reduce bodyweight and cause a clinically significant reduction in HbA1c levels (Scheen et al., 2006). Gene expression changes were studied in peripheral tissues such as liver and adipose from diet-induced obese mice treated with AM251 (Zhao et al., 2010). They found down-regulation of genes within fatty acid and cholesterol synthetic pathways such as sterol regulatory element binding proteins 1 and 2 in both liver and adipose tissues. However, these gene expression changes have not been studied in skeletal muscle. Therefore, in the present study a comprehensive analysis of differential gene expression in response to RIM treatments in rat primary skeletal muscle cells was achieved using Affymetrix Rat Genome 230 PM Array. The four technical replicates used for this analysis were found to be reproducible since the Pearson correlations for normalized intensity data for all replicates were above 0.98.
In this study, RIM up-regulated the mRNA content of NPY, APLN and LCN2, and down-regulated GNAI3 and NR4A1. These genes were suggested to be related to insulin resistance although the exact mechanisms are not known. There are limited studies on these genes in skeletal muscle; acute administration of apelin in chow-fed mice was associated with enhanced utilization of glucose in skeletal muscle (Dray et al., 2008). Similarly, administration of NPY to rats was associated with increased glucose utilization in skeletal muscle (Vettor et al., 1998). Although the cross-talk between cannabinoids and GNAI3 has not been studied in skeletal muscle, CB1 receptor activation was suggested to hinder insulin-stimulated IR autophosphorylation dependent on the association between GNAI3 and IR in pancreatic beta-cells (Kim et al., 2011). It is worth noting that GNAI2 in skeletal muscle was suggested to have a role in insulin sensitivity through the suppression of protein-tyrosine phosphatase 1B (PTP1B) (Tao et al., 2001).
LCN2 knockout mice exhibit significant decrease in fasting glucose levels and insulin sensitivity (Law et al., 2010). In addition, LCN2 concentrations correlated with hyperglycemia and insulin resistance in humans (Wang et al., 2007), whereas mRNA content of LCN2 was increased in liver and adipose tissue of diabetic/obese mice. Moreover, it was reported that cAMP can affect the mRNA content of NR4A1 in skeletal muscle (Kawasaki et al., 2011; Pearen et al., 2008; Pearen et al., 2006). NR4A1 might modulate fat and glucose metabolism through regulating the expression of genes related to oxidative metabolism in skeletal muscle (Pearen et al., 2008). It is worth mentioning that all of the above genes (NR4A1, NPY, APLN, LCN2 and GNAI3) were detected in rat skeletal muscle tissue using Agilent microarray at the following ranking (506, 12597, 1827, 12118 and 15033) out of 41000. However, more research is needed to investigate the role of RIM in skeletal muscle metabolism. Moreover, measurement of NPY and apelin in response to RIM should be recommended to be investigated in skeletal muscle cells.
There is little information in the literature about the roles of these genes in skeletal muscle. Overall, RIM might have a role in skeletal myogenesis through regulating the expression of those genes (CEBPD, NR4A1, NR4A2, NR4A3). Therefore, further research is needed to address this issue.
Moreover, RIM might have a role in inflammatory role in skeletal muscle through regulating of those genes (C3, CCL20, CXCL6, GNAI3, LCN2, NPY, NR4A2, PTGER4 ). In fact, the level of complement C3 was shown to be improved by treatment with antidiabetic agents such as thiazolidinediones (Ebeling et al., 1999). In addition to that, markers of inflammation including the proinflammatory cytokines and acute phase proteins are associated with developing and severity of type 2 diabetes (Donath et al., 2011; Spranger et al., 2003; Xie et al., 2011). Furthermore, type 2 diabetes has been associated recently with subclinical chronic inflammation (Donath et al., 2011; Spranger et al., 2003; Xie et al., 2011). Therefore, RIM might play a role with type 2 diabetes through improving the inflammation.
There is also a suggestion that RIM might give a response as an agonist through other receptors such as GPR55 (Godlewski et al., 2009). It is also worth noting that the serum used in this experiment might also contain low endocannabinoid level that might interact with signalling of RIM. Therefore, it is very hard to explain the response of RIM. From this data, no conclusion can also be made as to whether the RIM effects depend on the inhibition of the CB1 receptor (a CB1 receptor dependent manner). Further work should be performed to get a clear comprehensive image in these issues, such as repeating this experiment from different animals using either microarray or QRT-PCR (Taqman) or using delipidated serum instead of charcoal stripped serum. Further work is also required to understand these responses such as using siRNA for CB1 receptor or using GPR55 antagonist.
In summary, the microarray findings revealed treatment myotubes with RIM influenced the mRNA content of genes (NPY, APLN, LCN2, NR4A1 and GNAI3) related to insulin resistance, glucose and fat metabolism. Further research is warranted to establish the precise role of endocannabinnoids in the regulation of gene expression in skeletal muscle and the importance of this role in the development of insulin resistance and obesity.
References
- Baron, AD, Brechtel, G, Wallace, P, Edelman, SV (1988) Rates and tissue sites of non-insulin- and insulin-mediated glucose uptake in humans. Am J Physiol 255(6 Pt 1): E769-774.
- Bermudez-Siva, FJ, Serrano, A, Diaz-Molina, FJ, Sanchez Vera, I, Juan-Pico, P, Nadal, A, Fuentes, E, Rodriguez de Fonseca, F (2006) Activation of cannabinoid CB1 receptors induces glucose intolerance in rats. Eur J Pharmacol 531(1-3): 282-284.
- Bluher, M, Engeli, S, Kloting, N, Berndt, J, Fasshauer, M, Batkai, S, Pacher, P, Schon, MR, Jordan, J, Stumvoll, M (2006) Dysregulation of the peripheral and adipose tissue endocannabinoid system in human abdominal obesity. Diabetes 55(11): 3053-3060.
- Calle, EE, Kaaks, R (2004a) Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer 4(8): 579-591.
- Calle, EE, Rodriguez, C, Walker-Thurmond, K, Thun, MJ (2003) Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med 348(17): 1625-1638.
- Calle, EE, Thun, MJ (2004b) Obesity and cancer. Oncogene 23(38): 6365-6378.
- Cavuoto, P, McAinch, AJ, Hatzinikolas, G, Janovska, A, Game, P, Wittert, GA (2007) The expression of receptors for endocannabinoids in human and rodent skeletal muscle. Biochem Biophys Res Commun 364(1): 105-110.
- Colditz, GA, Willett, WC, Rotnitzky, A, Manson, JE (1995) Weight gain as a risk factor for clinical diabetes mellitus in women. Ann Intern Med 122(7): 481-486.
- Del Prato, S, Marchetti, P, Bonadonna, RC (2002) Phasic insulin release and metabolic regulation in type 2 diabetes. Diabetes 51 Suppl 1: S109-116.
- Delbono, O, Xia, J, Treves, S, Wang, ZM, Jimenez-Moreno, R, Payne, AM, Messi, ML, Briguet, A, Schaerer, F, Nishi, M, Takeshima, H, Zorzato, F (2007) Loss of skeletal muscle strength by ablation of the sarcoplasmic reticulum protein JP45. Proceedings of the National Academy of Sciences of the United States of America 104(50): 20108-20113.
- Despres, JP, Golay, A, Sjostrom, L (2005) Effects of rimonabant on metabolic risk factors in overweight patients with dyslipidemia. N Engl J Med 353(20): 2121-2134.
- Di Marzo, V, Goparaju, SK, Wang, L, Liu, J, Batkai, S, Jarai, Z, Fezza, F, Miura, GI, Palmiter, RD, Sugiura, T, Kunos, G (2001) Leptin-regulated endocannabinoids are involved in maintaining food intake. Nature 410(6830): 822-825.
- Donath, MY, Shoelson, SE (2011) Type 2 diabetes as an inflammatory disease. Nat Rev Immunol 11(2): 98-107.
- Doyon, C, Denis, RG, Baraboi, ED, Samson, P, Lalonde, J, Deshaies, Y, Richard, D (2006) Effects of rimonabant (SR141716) on fasting-induced hypothalamic-pituitary-adrenal axis and neuronal activation in lean and obese Zucker rats. Diabetes 55(12): 3403-3410.
- Dray, C, Knauf, C, Daviaud, D, Waget, A, Boucher, J, Buleon, M, Cani, PD, Attane, C, Guigne, C, Carpene, C, Burcelin, R, Castan-Laurell, I, Valet, P (2008) Apelin stimulates glucose utilization in normal and obese insulin-resistant mice. Cell Metab 8(5): 437-445.
- Ebeling, P, Teppo, AM, Koistinen, HA, Viikari, J, Ronnemaa, T, Nissen, M, Bergkulla, S, Salmela, P, Saltevo, J, Koivisto, VA (1999) Troglitazone reduces hyperglycaemia and selectively acute-phase serum proteins in patients with Type II diabetes. Diabetologia 42(12): 1433-1438.
- Eckardt, K, Sell, H, Taube, A, Koenen, M, Platzbecker, B, Cramer, A, Horrighs, A, Lehtonen, M, Tennagels, N, Eckel, J (2008) Cannabinoid type 1 receptors in human skeletal muscle cells participate in the negative crosstalk between fat and muscle. Diabetologia.
- Eckardt, K, Sell, H, Taube, A, Koenen, M, Platzbecker, B, Cramer, A, Horrighs, A, Lehtonen, M, Tennagels, N, Eckel, J (2009) Cannabinoid type 1 receptors in human skeletal muscle cells participate in the negative crosstalk between fat and muscle. Diabetologia 52(4): 664-674.
- Engeli, S, Bohnke, J, Feldpausch, M, Gorzelniak, K, Janke, J, Batkai, S, Pacher, P, Harvey-White, J, Luft, FC, Sharma, AM, Jordan, J (2005) Activation of the peripheral endocannabinoid system in human obesity. Diabetes 54(10): 2838-2843.
- Esposito, I, Proto, MC, Gazzerro, P, Laezza, C, Miele, C, Alberobello, AT, D’Esposito, V, Beguinot, F, Formisano, P, Bifulco, M (2008) The cannabinoid CB1 receptor antagonist rimonabant stimulates 2-deoxyglucose uptake in skeletal muscle cells by regulating the expression of phosphatidylinositol-3-kinase. Mol Pharmacol 74(6): 1678-1686.
- Ferrannini, E (1998) Insulin resistance versus insulin deficiency in non-insulin-dependent diabetes mellitus: problems and prospects. Endocr Rev 19(4): 477-490.
- Ferrannini, E, Barrett, EJ, Bevilacqua, S, DeFronzo, RA (1983) Effect of fatty acids on glucose production and utilization in man. J Clin Invest 72(5): 1737-1747.
- Godlewski, G, Offertaler, L, Wagner, JA, Kunos, G (2009) Receptors for acylethanolamides-GPR55 and GPR119. Prostaglandins Other Lipid Mediat 89(3-4): 105-111.
- James, DE, Jenkins, AB, Kraegen, EW (1985) Heterogeneity of insulin action in individual muscles in vivo: euglycemic clamp studies in rats. Am J Physiol 248(5 Pt 1): E567-574.
- Kahn, CR (1978) Insulin resistance, insulin insensitivity, and insulin unresponsiveness: a necessary distinction. Metabolism: clinical and experimental 27(12 Suppl 2): 1893-1902.
- Kawasaki, E, Hokari, F, Sasaki, M, Sakai, A, Koshinaka, K, Kawanaka, K (2011) The effects of beta-adrenergic stimulation and exercise on NR4A3 protein expression in rat skeletal muscle. J Physiol Sci 61(1): 1-11.
- Kim, W, Doyle, ME, Liu, Z, Lao, Q, Shin, YK, Carlson, OD, Kim, HS, Thomas, S, Napora, JK, Lee, EK, Moaddel, R, Wang, Y, Maudsley, S, Martin, B, Kulkarni, RN, Egan, JM (2011) Cannabinoids inhibit insulin receptor signaling in pancreatic beta-cells. Diabetes 60(4): 1198-1209.
- Law, IK, Xu, A, Lam, KS, Berger, T, Mak, TW, Vanhoutte, PM, Liu, JT, Sweeney, G, Zhou, M, Yang, B, Wang, Y (2010) Lipocalin-2 deficiency attenuates insulin resistance associated with aging and obesity. Diabetes 59(4): 872-882.
- Lindborg, KA, Teachey, MK, Jacob, S, Henriksen, EJ (2011) Effects of in vitro antagonism of endocannabinoid-1 receptors on the glucose transport system in normal and insulin-resistant rat skeletal muscle. Diabetes Obes Metab 12(8): 722-730.
- Lipina, C, Stretton, C, Hastings, S, Hundal, JS, Mackie, K, Irving, AJ, Hundal, HS (2010) Regulation of MAP kinase-directed mitogenic and protein kinase B-mediated signaling by cannabinoid receptor type 1 in skeletal muscle cells. Diabetes 59(2): 375-385.
- Liu, YL, Connoley, IP, Wilson, CA, Stock, MJ (2005) Effects of the cannabinoid CB1 receptor antagonist SR141716 on oxygen consumption and soleus muscle glucose uptake in Lep(ob)/Lep(ob) mice. Int J Obes (Lond) 29(2): 183-187.
- Matias, I, Gonthier, MP, Orlando, P, Martiadis, V, De Petrocellis, L, Cervino, C, Petrosino, S, Hoareau, L, Festy, F, Pasquali, R, Roche, R, Maj, M, Pagotto, U, Monteleone, P, Di Marzo, V (2006) Regulation, function, and dysregulation of endocannabinoids in models of adipose and beta-pancreatic cells and in obesity and hyperglycemia. J Clin Endocrinol Metab 91(8): 3171-3180.
- Mosthaf, L, Vogt, B, Haring, HU, Ullrich, A (1991) Altered expression of insulin receptor types A and B in the skeletal muscle of non-insulin-dependent diabetes mellitus patients. Proceedings of the National Academy of Sciences of the United States of America 88(11): 4728-4730.
- Nistala, R, Stump, CS (2006) Skeletal muscle insulin resistance is fundamental to the cardiometabolic syndrome. J Cardiometab Syndr 1(1): 47-52.
- Nogueiras, R, Veyrat-Durebex, C, Suchanek, PM, Klein, M, Tschop, J, Caldwell, C, Woods, SC, Wittmann, G, Watanabe, M, Liposits, Z, Fekete, C, Reizes, O, Rohner-Jeanrenaud, F, Tschop, MH (2008) Peripheral, but not central, CB1 antagonism provides food intake-independent metabolic benefits in diet-induced obese rats. Diabetes 57(11): 2977-2991.
- Ogden, CL, Carroll, MD, Curtin, LR, McDowell, MA, Tabak, CJ, Flegal, KM (2006) Prevalence of overweight and obesity in the United States, 1999-2004. JAMA 295(13): 1549-1555.
- Osei-Hyiaman, D, DePetrillo, M, Pacher, P, Liu, J, Radaeva, S, Batkai, S, Harvey-White, J, Mackie, K, Offertaler, L, Wang, L, Kunos, G (2005) Endocannabinoid activation at hepatic CB1 receptors stimulates fatty acid synthesis and contributes to diet-induced obesity. J Clin Invest 115(5): 1298-1305.
- Pagotto, U, Marsicano, G, Cota, D, Lutz, B, Pasquali, R (2006) The emerging role of the endocannabinoid system in endocrine regulation and energy balance. Endocr Rev 27(1): 73-100.
- Pearen, MA, Myers, SA, Raichur, S, Ryall, JG, Lynch, GS, Muscat, GE (2008) The orphan nuclear receptor, NOR-1, a target of beta-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinology 149(6): 2853-2865.
- Pearen, MA, Ryall, JG, Maxwell, MA, Ohkura, N, Lynch, GS, Muscat, GE (2006) The orphan nuclear receptor, NOR-1, is a target of beta-adrenergic signaling in skeletal muscle. Endocrinology 147(11): 5217-5227.
- Pedersen, BK (2011) Muscles and their myokines. J Exp Biol 214(Pt 2): 337-346.
- Ravinet Trillou, C, Delgorge, C, Menet, C, Arnone, M, Soubrie, P (2004) CB1 cannabinoid receptor knockout in mice leads to leanness, resistance to diet-induced obesity and enhanced leptin sensitivity. Int J Obes Relat Metab Disord 28(4): 640-648.
- Raymond, F, Metairon, S, Kussmann, M, Colomer, J, Nascimento, A, Mormeneo, E, Garcia-Martinez, C, Gomez-Foix, AM (2010) Comparative gene expression profiling between human cultured myotubes and skeletal muscle tissue. BMC Genomics 11
- Rimm, EB, Stampfer, MJ, Giovannucci, E, Ascherio, A, Spiegelman, D, Colditz, GA, Willett, WC (1995) Body size and fat distribution as predictors of coronary heart disease among middle-aged and older US men. Am J Epidemiol 141(12): 1117-1127.
- Schafer, HL, Linz, W, Falk, E, Glien, M, Glombik, H, Korn, M, Wendler, W, Herling, AW, Rutten, H AVE8134, a novel potent PPARalpha agonist, improves lipid profile and glucose metabolism in dyslipidemic mice and type 2 diabetic rats. Acta Pharmacol Sin 33(1): 82-90.
- Scheen, AJ, Finer, N, Hollander, P, Jensen, MD, Van Gaal, LF (2006) Efficacy and tolerability of rimonabant in overweight or obese patients with type 2 diabetes: a randomised controlled study. Lancet 368(9548): 1660-1672.
- Singh, GK, Siahpush, M, Hiatt, RA, Timsina, LR (2011) Dramatic increases in obesity and overweight prevalence and body mass index among ethnic-immigrant and social class groups in the United States, 1976-2008. J Community Health 36(1): 94-110.
- Spranger, J, Kroke, A, Mohlig, M, Hoffmann, K, Bergmann, MM, Ristow, M, Boeing, H, Pfeiffer, AF (2003) Inflammatory cytokines and the risk to develop type 2 diabetes: results of the prospective population-based European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam Study. Diabetes 52(3): 812-817.
- Tao, J, Malbon, CC, Wang, HY (2001) Galpha(i2) enhances insulin signaling via suppression of protein-tyrosine phosphatase 1B. The Journal of biological chemistry 276(43): 39705-39712.
- Toft, I, Bonaa, KH, Lindal, S, Jenssen, T (1998) Insulin kinetics, insulin action, and muscle morphology in lean or slightly overweight persons with impaired glucose tolerance. Metabolism: clinical and experimental 47(7): 848-854.
- Vettor, R, Pagano, C, Granzotto, M, Englaro, P, Angeli, P, Blum, WF, Federspil, G, Rohner-Jeanrenaud, F, Jeanrenaud, B (1998) Effects of intravenous neuropeptide Y on insulin secretion and insulin sensitivity in skeletal muscle in normal rats. Diabetologia 41(11): 1361-1367.
- Vickers, SP, Webster, LJ, Wyatt, A, Dourish, CT, Kennett, GA (2003) Preferential effects of the cannabinoid CB1 receptor antagonist, SR 141716, on food intake and body weight gain of obese (fa/fa) compared to lean Zucker rats. Psychopharmacology (Berl) 167(1): 103-111.
- Voss, MD, Beha, A, Tennagels, N, Tschank, G, Herling, AW, Quint, M, Gerl, M, Metz-Weidmann, C, Haun, G, Korn, M (2005) Gene expression profiling in skeletal muscle of Zucker diabetic fatty rats: implications for a role of stearoyl-CoA desaturase 1 in insulin resistance. Diabetologia 48(12): 2622-2630.
- Wang, Y, Lam, KS, Kraegen, EW, Sweeney, G, Zhang, J, Tso, AW, Chow, WS, Wat, NM, Xu, JY, Hoo, RL, Xu, A (2007) Lipocalin-2 is an inflammatory marker closely associated with obesity, insulin resistance, and hyperglycemia in humans. Clin Chem 53(1): 34-41.
- Witteman, JC, Willett, WC, Stampfer, MJ, Colditz, GA, Sacks, FM, Speizer, FE, Rosner, B, Hennekens, CH (1989) A prospective study of nutritional factors and hypertension among US women. Circulation 80(5): 1320-1327.
- Xie, W, Du, L (2011) Diabetes is an inflammatory disease: evidence from traditional Chinese medicines. Diabetes Obes Metab 13(4): 289-301.
- Zhao, W, Fong, O, Muise, ES, Thompson, JR, Weingarth, D, Qian, S, Fong, TM (2010) Genome-wide expression profiling revealed peripheral effects of cannabinoid receptor 1 inverse agonists in improving insulin sensitivity and metabolic parameters. Mol Pharmacol 78(3): 350-359.
- Zurlo, F, Larson, K, Bogardus, C, Ravussin, E (1990) Skeletal muscle metabolism is a major determinant of resting energy expenditure. J Clin Invest 86(5): 1423-1427.







