Manuscript accepted on :
Published online on: 16-12-2015
Plagiarism Check: Yes
Zhizhen Dong1, Min Yao2, Li Wang3, Xing Gu4, Yun Shi4, and Hua Huang5
1Department of Diagnostics, Affiliated Hospital of Nantong University, Jiangsu Province, 226001, China. 2Department of Immunology, Medical School of Nantong University, Jiangsu Province, 226001, China. 3Department of Informatics, Medical School of Nantong University, Jiangsu Province, 226001, China. 4Department of Oncology, Affiliated Hospital of Nantong University, Jiangsu Province, 226001, China. 5Department of Pathology, Affiliated Hospital of Nantong University, Jiangsu Province, 226001, China.
DOI : https://dx.doi.org/10.13005/bpj/401
Abstract
Abnormal expression of liver insulin-like growth factor-II (IGF-II) regulating by the methylation of its gene promoter (P1~4) CpG islands through epigenetic silencing of fetal P2, P3, and P4, or adult P1 is associated with progression of HBV-related hepatocellular carcinoma (HCC). The aims of the present study were to investigate the alteration of methylational status in P3 region CpG islands, and analyze IGF-II expression and clinicopathological features in HBV-related HCC. The methylational status of P3 region CpG islands was observed in the matched parts of HCC tissues by methylation-specific PCR. Hepatic IGF-II expression was analyzed by immunohistochemistry. IGF-II mRNA was amplified by RT-PCR, and confirmed by sequencing. Serum IGF-II levels were quantitatively detected by ELISA assay. The frequencies of IGF-II mRNA positive fragment, IGF-II positive, and the P3 hypomethylated CpG islands were 100%, 87.5%, and 100% in HCC-, 53.3%, 47.5%, and 52.5% in paracancerous-, and none in noncancerous-tissues, respectively. Significant IGF-II expression in HCC tissues was related to differentiation degree (moderate or poor), tumor invasion, and positive-HBV DNA (P<0.05). An inverse correlation was found between P3 methylational degree and IGF-II expression. The levels of hepatic IGF-II and serum IGF-II expression were significantly elevated (P<0.001) in the HCC group more than any of the other groups. Abnormality of hepatic IGF-II expression is associated with hypomethylational status of its fetal promoter CpG sites, and circulating IGF-II abnormality is a useful biomarker for HCC diagnosis.
Keywords
Hepatocellular carcinoma; Insulin-like growth factor-II; Promoter; Methylation-specific PCR; Diagnosis; Biomarker
Download this article as:| Copy the following to cite this article: Dong Z, Yao M, Wang L, Gu X, Shi Y, Qiu L, Huang H, Yao D. Epigenetic Alterations of Hepatic IGF-II Gene Promoter and IGF-II Abnormal Expression in HBV-Related Hepatocellular Carcinoma. Biomed Pharmacol J 2013;6(2) |
| Copy the following to cite this URL: Dong Z, Yao M, Wang L, Gu X, Shi Y, Qiu L, Huang H, Yao D. Epigenetic Alterations of Hepatic IGF-II Gene Promoter and IGF-II Abnormal Expression in HBV-Related Hepatocellular Carcinoma. Biomed Pharmacol J 2013;6(2). Available from: http://biomedpharmajournal.org/?p=2673 |
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies in China, particularly in the eastern and southern areas, including the inshore area of the Yangtze River [1~3]. Major risk factors for HCC in these areas are exposure to aflatoxin B1 and infection by hepatitis B virus (HBV) [4]. Carcinogenesis of hepatocytes is a multi-factor, multi-step, and complex process [5]. Genetic and epigenetic changes including DNA methylation, histone modification, and DNA methylation are the core biological processes in HCC [6. 7]. DNA cytosine methylation is a central epigenetic modification that has essential roles in cellular processes including genome regulation, development and disease [8. 9]. The methylational status is closely associated with the HCC development and progression [10]. Insulin-like growth factor-II (IGF-II) is a kind of fetal growth factor and a mitogenic polypeptide closely related to insulin and highly expressed in rat hepatocyte carcinogenesis [11~13].
Identification of molecular abnormalities associated with an increased risk of HCC is particularly important to improve knowledge of both the pathways of liver carcinogenesis and the outcomes [14, 15]. IGF-II gene which contains 9 exons (E1~E9) and 4 promoters (P1~P4) has complex regulation of transcription, resulting in multiple mRNAs initiated by different promoters, which contribute to cell proliferation, differentiation, anti-apoptosis, and invasive behavior [16, 17]. Hepatic and circulating IGF-II is overexpressed during HCC occurrence and development [13]. As a hypoxia- inducible angiogenic factor stimulates the growth of HCC cells in vitro [18, 19]. However, little is known of the relationship between IGF-II gene’s promoter methylation status and hepatocarcinogenesis. In the present study, we investigated the alterations of IGF-II gene promoter methylational status, expression, and gene transcription in HCC tissues, and relationship between circulating IGF-II levels and clinical pathological features.
Patients and Methods
Patients
There were 146 patients with HCC enrolled for this study at Affiliated Hospital of Nantong University, China. The patients’ ages ranged from 26 to 74 years old (median, 46 years). 134 patients (91.8%) had a history of cirrhosis, and 22 (15.1%) had a history of chronic hepatitis. All patients were diagnosed by blood biochemical tests, viral histology and B-ultrasonic examination. The incidence of hepatitis virus in these patients was 80.8% (118 of 146) in HBsAg, and 11.0 % (16 of 146) in antibody to HCV (ELISA, Beijing, China). Serum AFP level ranged from 33 to 2500 ng/mL (median, 425 ng/mL) and more than 50 ng/mL was taken as a positive result. Other patients included 39 with acute hepatitis, 72 with chronic hepatitis, 75 with decompensated cirrhosis, and 60 healthy subjects with negative-HBV markers (HBsAg, HBcAb, and HBV-DNA) and normal ALT levels from the Nantong Central Blood Bank. Bloods were collected in the morning. The diagnosis of HCC was based on the criteria proposed by Chinese National Collaborative Cancer Research Group [20].
HCC tissues
Human hepatoma-, para-cancerous- and distal cancerous- tissues in this study were obtained from 40 patients who underwent operations for liver cancers at the Affiliated Hospital of Nantong University, China. The livers were immediately frozen in liquid nitrogen and kept at -80℃ until required. The patients included 32 men and 8 women, ranging from 29 to 74 years old. The sizes of tumors were 24 cases with more than 5 cm and 16 with less than 5 cm. Serum AFP levels were 16 cases with more than 400 ng/mL, and 24 with less than 400 ng/mL. Histopathological analysis of all samples staining with Hematoxylin-Eosin (H&E) were evaluated and subjected to histological diagnosis by independent pathologists. All cancerous tissues were highly differentiated HCC; the para-cancerous tissues of the tumors were cirrhosis in 27 cases, chronic hepatitis in 13 cases, and atypical hyperplasia in 6 cases; and the noncancerous tissues were cirrhosis in 16 cases, chronic hepatitis in 15 cases, and atypical hyperplasia in 9 cases. Ethics Statement, this study was approved by the Institutional Review Board, Affiliated Hospital of Nantong University, and written informed consent was obtained.
Isolation of total RNA and synthesis of cDNA
Total RNAs were isolated from liver tissues by the guanidine thiocyanate method with RNAzole reagent (Promega) and purified as described elsewhere. RNAs were dissolved in tromethamine-HCl buffer (10 mmol/L, pH 8.0) containing EDTA 10 mmol/L. The level of total RNAs was measured by optical density at 260 nm in an ultraviolet spectrophotometer (Shimadzu UV-2201 type, Kyoto, Japan), and calculated μg/mg wet liver tissue, and it was stored at –85℃. For synthesis of cDNA, 2 μg of total RNAs was denatured in the presence of random hexamers (200 pmol/L, Promega, Madison, WI, USA) at 95℃ for 5 min and incubated with moloney murine leukemia virus reverse transcriptase (GIBCO, BRL) at 23℃ for 10 min, 42℃ for 60 min and 95℃ for 10 min, then on ice for 5 min, and stored at –20℃ for PCR analysis.
Amplification of nested PCR
The resulting cDNA was amplified by a nested PCR with two pairs of primers. The oligonucleotides were designed according to IGF-II sequence and synthesized with synthesizer (Model 381A, Applied Biosystems, Foster City, USA). The sequences of the 2 external primer pairs used for the initial PCR amplification were IGF-II-1 (sense), 5’-ATGGGAATGCCAATGGGGAAG-3’ (nt 251~271) and IGF-II-2 (anti- sense), 5’-CTTGCCCACGGGGTATCTGGG-3’ (nt 566~586), the size of amplified gene fragment was 336 bp. The sequences of the two internal primer pairs used for the second PCR amplification were IGF-II-3 (sense), 5’-TGCTGCATTGCTGCTTACC G-3’ (nt311~330) and IGF-II-4 (anti-sense), 5’-AGGTCACAGCTGCGGAAACA-3’ (nt 461~480). PCR amplification consisted of initial denaturation at 94℃ for 5 min, followed by 94℃ for 25 sec, 55℃ for 30 sec, and 72℃ for 90 sec for 30 cycles. The product was 170 bp. Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genome was used as a control. Primer sequence for GAPDH was GAPDH-1 (sense), 5’-ACCACAGTCCATGCCATCAC-3’(nt 601-620) and GAPDH-2 (anti-sense), 5’-TCCACCACCCTGTTGCTGTA-3’ (nt 1033-1052), the PCR product was 452 bp (GAPDH gene transcript, 40 pmol/L). The PCR products were electrophoresed on 2% agarose gels with ethidium bromide staining. The sizes were evaluated using PCR markers (Promega) as molecular weight standards.
Immunohiostochemistry
All immunohistochemistry kit was purchased from Fuzhou Maixin Biotechnology Development Company, China, and multiclonal anti-human IGF-II antibody was from Shanghai Jingmei Biotechnology Development Company, China. Sections (5 mm) were mounted on charged glass slides, deparaffinized with xylene for 2 × 10 min and rehydrated using a graded ethanol series. Antigen retrieval was performed by placing the samples in a microwave oven for 12 min, with occasional interruption to avoid tissue degradation by excessive heat. The slides were then treated with hydrogen peroxide, followed by incubation with the primary and secondary antibodies, a streptavidin-biotin complex, an amplification reagent, streptavidin-peroxidase and substrate-chromogen solution using the Envision system according to the manufacturers’ protocol (DAKO). The samples were then counterstained with hematoxylin, rinsed with ethanol, dried and visualized by light microscopy. Tissue samples to which no primary antibody had been added were used as negative controls. The antibody of IGF-II (1:10 dilution) was purchased from Santa Cruze (San Diego, CA). The slides were read by two pathologists and the percentage of the cytoplasmic staining was recorded.
DNA Extraction
Total DNA were purified by Wizard Cleanup DNA Purified kit. Ten mg of each liver tissue were homogenized with a homogenizer after addition of 600 μL of nuclear lysis buffer reagent for 10 sec, and incubated at 65 ℃ for 30 min. Then 3 μL of RNase to tubes, the mixture incubated at 37 ℃ for 30 min, put at room temperature for 5 min. And 200 μL of protein precipitation to tubes, mixed by vortex-mixing for 20 sec, put at 0 °C for 5 min, then centrifuged at 16000 rpm for 4 min at 4°C. The supernatants were collected, and added 600 μL isopropanol to a new tube and mixed gently; once obvious stripe like DNA was observed, then centrifuged at 16000 rpm for 1 min. The supernatants were removed, washed the DNA pellets twice with 600 μL of 70% ethanol, mixed and centrifuged at 16000 rpm for 1 min. The supernatants were removed again, the DNA pellets were air dried 15 min at room temperature and reconstituted in 100 μL of DNA Rehydration Solution and incubated at 65 °C for 1 h. The purity and concentration of DNA was measured by optical density at A260 and A280 nm in an ultraviolet spectrophotometer, and it was stored at 4 °C.
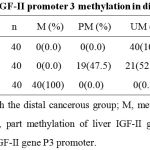 |
Table 1: The status of IGF-II promoter 3 methylation in different liver tissue
|
Methylation-specific PCR (MSP)
The modification and purified of DNA were performed using the DNA Modification Kit according to the protocol. The promoter methylation status of IGF-II gene was determined as described previously. The primers were designed using a software developed at the Johns Hopkins University (www. mspprimers.org) based on the accession number (X05331) of IGF-II gene promoter 3 (Fig.1A). The sequences of the 2 methylation primer pairs used for MSP amplification were P3M-F (sense), 5’-TTTTTAAATTATCGTGGTGGTTTTC-3’ and P3M-R (anti-sense), 5’-GTCTAAA TAACTCGCCTTTACGA-3’ (nt –889~-768, 122 bp). The sequences of the 2 demethylation primer pairs used for the MSP amplification were P3U-F (sense), 5’-TTTTTAAATTATTGTGGTGGTTTTTG-3’ and P3U-R (anti-sense), 5’-CATCTAA ATAACTCACTTTACAAC-3’ (nt –889~-767, 123 bp). The MSP conditions were as follow: HotStart Taq polymerase (Qiagen) used with initial activation and denaturation 94 ℃ for 5 min; 35 cycles (94 ℃ for 40 sec; 49 ℃ for 40 sec; 72 ℃ for 40 seconds) followed by final extension 72 ℃ for 10 min. In vitro methylated DNA and unmethylated lymphocytes DNA were used as controls, respectively. The MSP products were electrophoresed on 2% agarose gels with ethidium bromide staining. The sizes were evaluated using PCR markers as molecular weight standards.
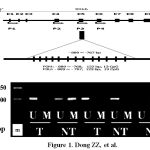 |
Figure 1: Primer design and methylation analysis of liver IGF-II gene promoter 3 (P3) in HCC. A, Exon-intron and 4 promoter structure of human IGF-II gene. The primers were designed based on the accession number (X05331) of P3. Exons are shown as numbered boxes (plain are coding). The 122 bp (P3M) or 123 bp (P3U) fragment of P3 amplified for methylation analysis is enlarged below. Vertical lines indicate 13 CpG positions; B, The analysis of P3 methylation status in different tissues. T, the HCC tissues; NT, the para-cancerous tissues; N, the distal-cancerous tissues; U,Demethylation; M,Methylation; m, DNA molecular marker.
|
Detection of IGF-II level
The levels of serum free IGF-II protein in patients with chronic diseases were detected by an enzymatically amplified two-step sandwich-type immunoassay (ACTIVETM IGF-II ELISA, TX). In this assay, standards, controls and serum samples were incubated in microtitration wells, which had been coated with anti-IGF-II antibody. After incubation and washing, the wells were treated with another anti-free IGF-II detection antibody labeled with the enzyme horseradish peroxidase (HRP). After a second incubation and washing step, the wells were incubated with the substrate tetramethyl benzidine. An acidic stopping solution was then added and the degree of enzymatic turnover of the substrate was determined by dual wavelength absorbance (A) measurement at 450 nm and 620 nm. The absorbance measured was directly proportional to the concentration of IGF-II present. A set of IGF-II standards was used to draw a standard curve of absorbance vs IGF-II concentration from which the IGF-II concentrations in the serum samples can be calculated according to the ELISA routine method.
Statistical analysis
All patients were divided into 4 groups: HCC, chronic hepatitis, cirrhosis, and normal subjects. Hepatoma tissues were divided into three groups: cancerous, para-cancerous, and distal cancerous-tissues. Results are expressed as mean ± standard deviation (SD). Differences between different groups were assessed by the Student’s t test or the χ2 test. P < 0.05 was considered to be significant.
Results
Methylation status of IGF-II gene P3 in HCC tissues
The promoter methylation status of hepatic IGF-II and the expression of hepatocyte IGF-II were analyzed in human HCC-, para-cancerous-, and distal cancerous- tissues, and the patterns by MPS or immunohistochemistry are shown in Figure 1. The incidence of IGF-II P3 methylation was 0% (0 of 40) in human HCC, 47.5% (19 of 40) in para-cancerous tissues, and 100% (40 of 40) in distal cancerous tissues, respectively. The summary of P3 methylation status analysis in different liver tissues is shown in Table 1. The incidence was increased gradually from cancerous to distal cancerous-parts of liver tissues, with significant differences among them (χ2 = 37.623, P < 0.001). The incidence of IGF-II P3 methylation was 0 % (0 of 40) in HCC tissues, and no significant difference was found between the methylation rates of IGF-II P3 in HCC and clinical parameters.
Expression of total RNA and IGF-II mRNA in HCC tissues
Different expression of hepatic total RNA (μg/mg wet liver tissue) was found in the different parts of 40 HCC tissues. The total RNA levels were significantly lower in HCC tissues (17.9 ± 27.7 μg/mg wet liver tissue) than in self-control surrounding- (32.9 ± 31.2 μg/mg wet liver tissue, P < 0.05) or non-cancerous liver tissues (41.4 ± 50.3 μg/mg wet liver tissue, P < 0.01), respectively. The amplified fragments of IGF-II mRNA from different liver tissues of HCC patients and the alignment of nucleotide sequences of the amplified fragments by sequence analysis are shown in Figure 2. The fragments (170 bp) of hepatic IGF-II genome were amplified by a RT-nested PCR assay (Figure 2C), the size of IGF-II DNA was identical to the original designed one, and confirmed by DNA sequencing analysis (Figure 2D). The incidence of positive IGF-II mRNA fragments was 100 % (40 of 40) in HCC tissues, and significantly higher (P < 0.01) than that in their surrounding (53.3 %, 21 of 40) or in their non-cancerous (0 %, 0 of 40) liver tissues, respectively.
 |
Figure 2: Amplification of IGF-II genomes from different liver tissues of HCC patients. IGF-II mRNAs were synthesized according to IGF-II cDNA with reverse- transcriptase, and amplified by nested PCR (170 bp) with different primers. The fragments of IGF-II gene were seen distinctly in HCC or their para-cancerous tissues. A, the detecting sensitive limitation (2 ng/L) according to total RNA with 10-2 ~ 10-8 fold dilution and then amplified by nested PCR; B, the amplified fragments (452 bp) of hepatic glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genome; C, the amplification of liver IGF-II genomes: No. 1~3, the positive fragments of IGF-II mRNA amplification in HCC; No. 4~5, no fragment in the para- or distal-cancerous tissues; No. 6, the positive fragments in the para-cancerous tissues. M: DNA molecular weight marker. D,Alignment of nucleotide sequences of the amplified IGF-II genome fragments in different livers by sequencing. Origin: the cited sequence (170 bp, nt 311-480) of IGF-II genome; Hepatoma: the amplification fragment of IGF-II genome in HCC tissue; A-HCC: the amplified fragment of IGF-II genome from the para-cancerous tissue in HCC patients.
|
Immunohistochemical analysis of IGF-II expression in HCC tissues
The IGF-II expressions in HCC-, para-cancerous-, and distal-cancerous-tissues by immunohistochemical analysis are shown in Figure 3. The positive staining of hepatic IGF-II showed yellow-brown particles, located in cytoplasm with only a few in cellular nuclei but none in cell membrane. The frequency of positive IGF-II expression in the HCC (87.5 %, 35/40) or para-cancerous (47.5 %, 19/40) group was significantly higher than that in the non-cancerous group (0 %, 0/40, P < 0.05), respectively.
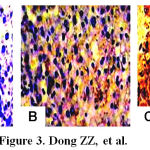 |
Figure 3: Immunohistochemical analyses with anti-human IGF-II in the different liver tissues. A, the absence of cytoplasmic staining for IGF-II from non-cancerous tissues of HCC (S-P, original magnification × 100); B, And the IGF-II weakly positive staining in cytoplasm and cell membrane from the surrounding tissues of HCC (S-P, original magnification × 100); C, the IGF-II strongly positive staining in cytoplasm and cell membrane from HCC tissues (S-P, original magnification ×100).
|
Clinicopathological features of IGF-II expression
The clinicopathological features of IGF-II expression in HCC tissues are shown in Table 2. The expression of IGF-II was significantly higher in HCC with moderate or low differentiation than that with well differentiation (92.9 %, 100 % vs 50.0 %, P < 0.001 or P < 0.01). Hepatic IGF-II expression was markedly lower in HCC without serosa invasion than that with serosa invasion (61.5 % vs 100 %, P < 0.05). The level of IGF-II expression in HBV DNA-positive HCC was significantly higher than that in HBV DNA-negative ones (100 % vs 58.3 %, P < 0.05). No significant difference was found between it with the patients’ sex, age, tumor size, tumor number, and serum AFP concentration.
 |
Table 2: The clinical pathological features of IGF-II expression in HCC tissues
|
Circulating IGF-II level in diagnostic value of HCC
The circulating IGF-II expression and diagnostic evaluation for HCC in patients with liver diseases are shown in Figure 4. Of these patients, the circulating IGF-II level was significantly higher (P < 0.001) in HCC than in liver cirrhosis, chronic hepatitis, and acute hepatitis (Figure 4A). The pathologic characteristics of circulating IGF-II expression showed that the higher expression of hepatic IGF-II in HCC was associated with HBV infection (P < 0.001, Figure 4B). However, no significant difference was found between IGF-II expression and HCC patients’ sex, age, tumor size, or AFP level (Figure 4C). The analysis of two markers for the whole range of sensitivities and specificities using the area (0.823 for AFP and 0.771 for IGF-II, Figure 4D) under ROC curves indicated that the abnormality of serum IGF-II level could be a useful molecular marker for HCC diagnosis.
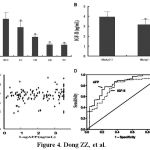 |
Figure 4 Levels of serum IGF-II expression and relationship between IGF-II and HBV infection in hepatocellular carcinoma. A, Serum IGF-II levels in different liver diseases, *P < 0.001 vs. the HCC group; HCC, hepatocellular carcinoma; LC, liver cirrhosis; CH, Chronic hepatitis; AH, Acute hepatitis; NC, Normal control. B, Relationship between IGF-II expression and HBV infection in hepatocellular carcinoma, HBsAg (+), the HBsAg positive HCC patients (n = 110, IGF-II = 3.93 ± 0.50 ng/mL), HBsAg (-), the HBsAg negative HCC patients (n = 36, IGF-II = 3.16 ± 0.80 ng/mL, t = 5.390, P < 0.001). C, Relationship between serum IGF-II and AFP level; D, Receiver operating characteristic (ROC) curves for hepatocellular carcinoma, and the area under the ROC curves was 0.823 for AFP and 0.771 for IGF-II.
|
Discussion
Epigenetic analysis of HCC plays a major role in the understanding of HCC processes and targeted therapies [21, 22]. Hepatocarcinogenesis is a multi-factor, multi-step, and complex process. Genetic and epigenetic changes regulate the expression of cancer- related genes, including DNA methylation, histone modification, and chromosome remodeling. DNA methylation within CpG islands in carcinogenesis is very important. The mechanism of carcinogenesis by DNA methylation include: oncogenes were activated by DNA methylation, for example, c-Jun, c-Myc and c-Ha-ras; and tumor suppressor genes were inactivated, for example, P16. DNA methylation has negative relationship with genes expression [6, 7, 23, 24]. This study was to investigate the changes of IGF-II gene promoter methylation status in different human liver tissues, and its correlations with hepatic IGF-II expression and clinical pathological features.
Many studies showed that the methylation status of many genes was closely related to occurrence and development of HCC [23, 25]. The methylation status frequencies of tumor suppressor gene–APC promoter in HCC tissues were significantly higher than para-cancerous liver tissues [26]. Simultaneously, the expression of APC in HCC tissues was also higher than para-cancerous liver tissues, suggesting the correlation between APC gene expression and promoter methylation status. The hyper-methylation frequency of glutathione S-transferase pi (GSTP1) promoter showed significant difference between HCC and liver cirrhosis with or without HBV infection (P < 0.05), suggesting hyper-methylation of GSTP1 may be involved in hepatocarcinogenesis [27, 28]. The frequencies of IGF-II gene P3 methylation status were 100 % in non-cancerous tissues, 47.5 % in para-cancerous tissues, and 100% in HCC, respectively, suggesting the correlation between IGF-II promoter methylation status and hepatocarcinogenesis and is an early event in the development of HCC.
IGF-II is highly expressed in the fetal liver and early after birth, which is mainly based on activation of P2~P4. But its expression is strongly reduced in adulthood, mainly based on activation of P1 [4, 16]. Several studies have shown elevated expression levels of IGF-II in pre-neoplastic lesions and very high levels in HCC, and so was the IGF-II gene P2~P4, indicating the correlation between IGF-II gene expression and promoter. The methylation rates of IGF-II gene P3 in the adjacent tissues in the poor differentiated HCC were lower than the well-differentiated ones (P < 0.01). However, IGF-II was highly expressed in HCC tissues, while the IGF-II promoter methylation status was low [23, 24, 9], showing that IGF-II methylation is a central epigenetic modification that has essential roles in cellular processes including genome regulation, development and disease.
HCC is strongly associated with HBV infection, especially HBV X gene (HBX) which is closely related to hepatocarcinogenesis [4, 29]. Many studies showed HBV infection contributed to methylation of tumor suppressor genes [26, 27]. However, the data in the present study have shown that the methylation rates of IGF-II gene P3 in HCC tissues with positive HBV DNA was significantly lower than those with negative HBV DNA, indicating that the demethylation of IGF-II gene P3 could relate to HCC patients with HBV infection, suggesting the positive correlation between demethylation status of IGF-II gene P3 and hepatocarcinogenesis, and epigenetic analysis of HCC related genes plays a major role in the understanding of carcinogenic processes and targeted therapies [30, 31].
HCC prognosis is poor, and early detection is of the utmost importance [32, 33]. The alteration of IGF-II gene P3 methylation status and HCC development was strengthened by the fact that IGF-II over-expression in tissues and sera of HCC patients involved in this study. This alteration was in agreement with the presence of an increasing methylation gradient from the cancer to the distal, with a higher level of methylation from well to poorly differentiated HCC. IGF-II gene was suspected to be involved in early carcinogenic processes [13]. The demethylating process strengthened by the fact that the gene activation and transcription result in hepatic IGF-II production and then secrete into blood. The circulating IGF-II level was significantly higher (P < 0.001) in HCC than in liver cirrhosis, chronic hepatitis, and acute hepatitis, and its abnormality should be a useful biomarker for HCC diagnosis.
In conclusion, the present data indicate that elevated expression of IGF-II in HCC tissue was the result of IGF-II P3 demethylation, and help to explore the molecular mechanisms responsible for reactivation of IGF-II in development and therapy of HCC [34~36]. DNA methylation may be the molecular-targeted therapies of HCC. Further studies will allow us to investigate the changes of IGF-II gene promoter methylation status in patients’ peripheral blood with HCC, and will elevate early diagnosis and monitor metastasis of HCC. These reference epigenomes provide a foundation for future studies exploring this key epigenetic modification in human HCC and development.
References
- El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology, 2012; 142(6): 1264-73.e1[PMID: 22537432]
- El-Serag HB, Marrero JA, Rudolph L, Reddy KR. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology, 2008; 134 (6): 1752-63 [PMID: 18471552]
- Yang JD, Roberts Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol, 2010; 7(8): 449-58 [PMID: 20628345]
- Kim DG. Differentially expressed genes associated with hepatitis B virus HBx and MHBs protein function in hepatocellular carcinoma. Methods Mol Biol. 2006; 317(1): 141-55.[PMID: 16264227]
- Jain S, Singhal S, Lee P, Xu R. Molecular genetics of hepatocellular neoplasia. Am J Transl Res 2010; 2(1): 105-18 [PMID: 20182587]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet, 2007, 8(4): 286–98. [PMID:17339880]
- Morisawa T, Marusawa H, Ueda Y, Iwai A, Okazaki IM, Honjo T, Chiba T. Organ-specific profiles of genetic changes in cancers caused by activation-induced cytidine deaminase expression. Int J Cancer. 2008; 123(12): 2735-40. [PMID: 18781563]
- Fuchs BC, Fujii T, Dorfman JD, Goodwin JM, Zhu AX, Lanuti M, Tanabe KK. Epithelial -to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res. 2008; 68(7): 2391-9. [PMID: 18381447]
- Wu J, Qin Y, Li B, He WZ, Sun ZL. Hypomethylated and hypermethylated profiles of H19DMR are associated with the aberrant imprinting of IGF2 and H19 in human hepatocellular carcinoma. Genomics. 2008; 91(5): 443-50. [PMID: 18358696]
- Tang SH, Yang DH, Huang W, Zhou HK, Lu XH, Ye G. Hypomethylated P4 promoter induces expression of the insulin-like growth factor-II gene in hepatocellular carcinoma in a Chinese population. Clin Cancer Res. 2006; 12(14 Pt 1): 4171-7. [PMID: 16857788]
- Breuhahn K, Schirmacher P. Reactivation of the insulin-like growth factor-II signaling pathway in human hepatocellular carcinoma. World J Gastroenterol. 2008; 14(11): 1690-8. .PMID: 18350600
- Ubagai T, Kikuchi T, Fukusato T, Ono Y. Aflatoxin B1 modulates the insulin-like growth factor-2 dependent signaling axis. Toxicol In Vitro. 2010; 24(3): 783-9. [PMID: 20036727]
- Qiu LW, Yao DF, Zong L, Lu YY, Huang H, Wu W, Wu XH. Abnormal expression of insulin-like growth factor-II and its dynamic quantitative analysis at different stages of hepatocellular carcinoma development. Hepatobiliary Pancreat Dis Int. 2008;7(4): 406-11.[PMID: 18693177]
- Couvert P, Carrié A, Pariès J, Vaysse J, Miroglio A, Kerjean A, Nahon P, Chelly J, Trinchet JC, Beaugrand M, Ganne-Carrié N. Liver insulin-like growth factor 2 methylation in hepatitis C virus cirrhosis and further occurrence of hepatocellular carcinoma. World J Gastroenterol. 2008; 14(35): 5419-27. [PMID: 18803353]
- Mukherjee B, Ghosh S, Das T, Doloi M. Characterization of insulin-like- growth factor II (IGF II) mRNA positive hepatic altered foci and IGF II expression in hepatocellular carcinoma during diethylnitrosamine-induced hepatocarcinogenesis in rats. J Carcinog, 2005; 4(1): 1-12. [PMID: 16092956]
- Scharf JG, Braulke T. The role of the IGF axis in hepatocarcinogenesis. Horm Metab Res. 2003; 35(11-12): 685-93. [PMID: 14710347]
- Dong ZZ, Yao DF, Yao DB, Wu XH, Wu W, Qiu LW, Jiang DR, Zhu JH, Meng XY. Expression and alteration of insulin-like growth factor II-messenger RNA in hepatoma tissues and peripheral blood of patients with hepatocellular carcinoma. World J Gastroenterol, 2005; 11(30): 4655-60. [PMID: 16094705]
- Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology 2008; 48(6): 2047-63 [PMID: 19003900]
- Li S, Yao D, Wang L, Wu W, Qiu LW, Yao M, Yao N, Zhang H, Yu D, Ni Q. Expression characteristics of HIF-1α and its clinical values in diagnosis and prognosis of hepatocellular carcinoma. Hepat Mon. 2011: 11(10): 821-8 [PMID: 22224081]
- Chinese Society of Liver Cancer, Chinese Anti-cancer Association. The criteria of diagnosis and stage for primary liver cancer. Zhonghua Gangzangbing Zazhi, 2001, 9(3): 324. In Chinese
- Luo JH, Ren B, Keryanov S, Tseng GC, Rao UN, Monga SP, Strom S, Demetris AJ, Nalesnik M, Yu YP, Ranganathan S, Michalopoulos GK. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006; 44(4): 1012-24. [PMID: 17006932]
- Cantarini MC, de la Monte SM, Pang M, Tong M, D’Errico A, Trevisani F, Wands JR. Aspartyl-asparagyl beta hydroxylase over-expression in human hepatoma is linked to activation of insulin-like growth factor and notch signaling mechanisms. Hepatology. 2006; 44(2):446-57. [PMID: 16871543]
- Liu JB, Zhang YX, Zhou SH, Shi MX, Cai J, Liu Y, Chen KP, Qiang FL. CpG island methylator phenotype in plasma is associated with hepatocellular carcinoma prognosis. World J Gastroenterol. 2011; 17(42): 4718-24. [PMID: 22180715]
- Feng Y, Xue WJ, Li P, Sha ZY, Huang H, Rui L, Li HX, Mao QS. RASSF1A hypermethylation is associated with aflatioxin B1 and polycyclic aromatic hydrocarbon exposure in hepatocellular carcinoma. Hepatogastroenterology. 2011; 59(118):1883-8. [PMID: 22172412]
- Yasen M, Obulhasim G, Kajino K, Mogushi K, Mizushima H, Tanaka S, Tanaka H, Hino O, Arii S. DNA binding protein A expression and methylation status in hepatocellular carcinoma and the adjacent tissue. Int J Oncol. 2012; 40(3): 789-97. [PMID: 22159460]
- Jain S, Chang TT, Hamilton JP, Lin SY, Lin YJ, Evans AA, Selaru FM, Lin PW, Chen SH, Block TM, Hu CT, Song W, Meltzer SJ, Su YH. Methylation of the CpG sites only on the sense strand of the APC gene is specific for hepatocellular carcinoma. PLoS One. 2011; 6(11): e26799. [PMID: 22073196]
- Jain S, Chen S, Chang KC, Lin YJ, Hu CT, Boldbaatar B, Hamilton JP, Lin SY, Chang TT, Chen SH, Song W, Meltzer SJ, Block TM, Su YH. Impact of the location of CpG methylation within the GSTP1 gene on its specificity as a DNA marker for hepatocellular carcinoma. PLoS One. 2012; 7(4): e35789. [PMID: 22536438]
- Huang ZH, Hu Y, Hua D, Wu YY, Song MX, Cheng ZH. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol, 2011; 91(3):702-7.[PMID:21884695]
- Tang SH, Yang DH, Huang W, Zhou M, Zhou HK, Lu XH, Ye G. Differential promoter usage for insulin-like growth factor-II gene in Chinese hepatocellular carcinoma with hepatitis B virus infection. Cancer Detect Prev. 2006; 30(2): 192-203. [PMID: 16697535]
- Tovar V, Alsinet C, Villanueva A, Hoshida Y, Chiang DY, Solé M, Thung S, Moyano S, Toffanin S, Mínguez B, Cabellos L, Peix J, Schwartz M, Mazzaferro V, Bruix J, Llovet JM.. IGF activation in a molecular subclass of hepatocellular carcinoma and pre-clinical efficacy of IGF-1R blockage. J Hepatol. 2010; 52(4): 550-9.[PMID: 20206398]
- Kim YJ, Yoon JH, Kim CY, Kim LH, Park BL, Shin HD, Lee HS. IGF2 polymorphisms are associated with hepatitis B virus clearance and hepatocellular carcinoma. Biochem Biophys Res Commun.2006;346(1):38-44.[PMID: 16750516]
- Yao DF, Dong ZZ, Yao M. Specific molecular markers in hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2007; 6(3):241-7.[PMID: 17548245]
- el-Houseini ME, Mohammed MS, Elshemey WM, Hussein TD, Desouky OS, Elsayed AA. Enhanced detection of hepatocellular carcinoma. Cancer Control, 2005; 12(4):248-53[PMID: 16258497]
- Jeng YM, Chang CC, Hu FC, Chou HY, Kao HL, Wang TH, Hsu HC. RNA- binding protein insulin-like growth factor II mRNA-binding protein 3 expression promotes tumor invasion and predicts early recurrence and poor prognosis in hepatocellular carcinoma. Hepatology. 2008; 48(4): 1118-27[PMID: 18802962]
- Nussbaum T, Samarin J, Ehemann V, Bissinger M, Ryschich E, Khamidjanov A, Yu X, Gretz N, Schirmacher P, Breuhahn K. Autocrine insulin-like growth factor-II stimulation of tumor cell migration is a progression step in human hepatocarcinogenesis. Hepatology, 2008; 48(1): 146-56[PMID: 18537183]
- Cheng W, Tseng CJ, Lin TT, Cheng I, Pan HW, Hsu HC, Lee YM. Glypican- 3-mediated oncogenesis involves the Insulin-like growth factor-signaling pathway. 2008; 29(7): 1319-26[PMID: 18413366]







