Sanjay Telang
Government Science and Commerce College, Benazir, Bhopal, India.
DOI : https://dx.doi.org/10.13005/bpj/430
Abstract
Proteases have been isolated from A. niger isolates originally isolated from common carp (Cyprinus carpio). The three isolates showed slight difference in their productivity of the enzyme. The characteristics of the enzyme from three isolates however was same i.e. alkaline in nature. The alkaline proteases are industrially very important.
Keywords
Fungal pathogen; Cyprinus carpio; Proteases Activity
Download this article as:| Copy the following to cite this article: Telang S. Aspergillus niger from Common Carp (Cyprinus carpio) with Proteases Activity. Biomed Pharmacol J 2013;6(2) |
| Copy the following to cite this URL: Telang S. Aspergillus niger from Common Carp (Cyprinus carpio) with Proteases Activity. Biomed Pharmacol J 2013;6(2). Available from: http://biomedpharmajournal.org/?p=2768 |
Introduction
Proteases constitute one of the most important groups of industrial enzymes and have applications in different industries for example in detergent, food, feed, pharmaceutical, leather, silk and for recovery of silver from used X-ray films (Anisworth, 1994; Fujiwara, 1993). This enzyme accounts for 30% of the total world enzyme production (Horikoshi, 1996). A variety of microorganisms such as bacteria, fungi, yeasts and actinomycetes are known to produce this enzyme (Reese et al., 1950; Taguchi et al., 1983; Kim et al., 1993; Manjeet et al., 1998).
The objective of this work was to isolate protease from the fungus Aspergillus niger originally isolated from the skin of common carp.
Materials and Methods
Fungal strain and maintenance
The fungal strains A. niger BEN1, A. niger BEN3 and A. niger BEN7 were used to isolate protease. The strains were maintained in refrigerator at 4oC on PDA slant.
Liquid culture
Minimal synthetic medium supplemented with casein (1% w/v) was used to raise liquid culture in flask of size 25 ml. A 100 ml medium was inoculated with about 107 spores of the fungus and the medium was incubated at 37oC on a shaking incubator.
Preparation of enzyme source
After three, four and five days of growth, the broth was centrifuged at 10000 rpm and the supernatant was used as source of enzyme.
Assay of enzyme
The protease activity was assayed by casein digestion method (Kunitz, 1947). The reaction mixture contained suitably diluted enzyme and casein in 0.1M sodium carbonate buffer pH 10. The reaction mixture was incubated at 40°C for 10 min. The reaction was terminated by the addition of 3 ml of 10% trichloroacetic acid. The terminated reaction mixture was incubated at room temperature for 30 min. The precipitate formed was filtered through Whatman No. 1 filter paper. The absorbance of the filtrate was measured at 280 nm. Tyrosine was used as standard. One unit of protease activity is defined as the amount of enzyme which liberates one micromoles of tyrosine per minute per gram dry substrate under experimental conditions. Protein was estimated by the method of Lowry et al. (1951).
Effect of pH
The pH optimum of the enzyme was determined by varying the pH of the reaction mixtures using the following buffers (100 mM): sodium acetate (pH 3.0-5.5), sodium phosphate (pH 6.0-7.0) and Tris-HCl (pH 7.5-8.0).
The protease activity at different period of incubation is given in table 1.
Table 1: Protease activity shown by the fungal isolates in different period of growth.
| Fungal isolates | Proteases activity (unit/ml) in the broth of different period of growth | |||
| Third day | Fourth Day | Fifth day | Sixth day | |
| A. niger BEN1 | 2 | 3 | 4 | 4 |
| A. niger BEN3 | 2 | 4 | 5 | 5 |
| A. niger BEN7 | 2 | 3 | 4 | 4 |
The enzyme activity in the broth was found to increase from third day and became constant with effect from fifth day. The activity was found to be a bit better (5 unit/ml) in case of the strain A. niger BEN3.
The effect of pH on enzyme activity is shown by the figure 1. The enzyme showed maximum activity at alkaline pH, although residual activity continued to be seen upto a pH of 3.
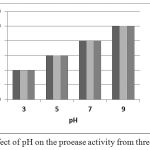 |
Figure 1: Effect of pH on the proease activity from three isolates.
|
The alkaline protease is very important industrially. Earlier alkaline protease has been reported from fungi and bacteria (Veloorvalappil et al., 2013).
Conclusions
niger isolated originally from carps produced proteases under in vitro condition.
Of the three strains BEN3 produced more unit of enzymes under parallel conditions All the three isolates produced alkaline proteases.
References
- Anisworth, J. Soap and detergents. Chem. Eng. News., 72: 34-59 (1994)..
- Fujiwara, N. Production of thermophilic alkaline protease from Bacillus B18. J. Biotechnol., 30:245-256 (1993).
- Horikoshi, Alkalophils from an industrial point of view. FEMS Microbiol. Rev., 18: 259-270 (1996).
- Reese, E.T., Sin, R.G.H. and Levinson, H.S. The biological degradation of cellulose derivatives and its relationship to the mechanism of cellu-lose hydrolysis. J. Bacteriol., 59: 480 – 485 (1950).
- Taguchi, H., Hamoki, M., Matsuzava, H. and Ohta, J. Heat stable extra-cellular proteolytic enzyme produced by Thermus caldophilous strain GK24 an extremely thermophilic bacterium. Biochem., 93: 7 – 13 (1983).
- Kim, , Dhillon, J., Chaudhary, S. and Singh, R. Properties of alkaline protease isolated from Nocardiopsis dassonvillei. Korean Biochem. J., 26: 81- 85 (1993).
- Manjeet, K., Dhillon, S., Chaudhary, S. and Singh, R. production purification and characterization of a thermostable alkaline protease from Bacillus polymyxa. Indian J. Microbiol., 38: 63 – 67 (1998).
- Kunitz, M. Crystalline soyabean trypsin inhibitor General pro-perties. J. Gen. Physiol., 30: 291–310 (1947).
- Lowry, O.H., Rosebrough, N.J., Farr and Randall, R.J. Protein measurement with Folin phenol reagent. Biol. Chem., 193: 265 – 275. (1951).
- Veloorvalappil, N. J., Robinson, B. S., Selvanesan, P., Sasidharan, S., Kizhakkepawothail, N. U., Sreedharan, S., Prakasan, P., Moolakkariyil,S.J. and Sailas, B. Versatility of microbial proteases. Advances in Enzyme Research., 1: 39-51 (2013).







