Manuscript accepted on :December 09, 2010
Published online on: 24-11-2015
Plagiarism Check: Yes
S. Kanimozhi1* and K. Perinbam2
1Sathyabama University, Jeppiaar Nagar, Old Mamallapuram Road, Chennai - 600 119 India.
2Faculty of Biomedical Engineering, Sathyabama University, Jeppiaar Nagar, Old Mamallapuram Road, Chennai - 600 119 India.
Abstract
Lipases (triacylglycerol acylhydrolases, EC 3.1.1.3) catalyze the hydrolysis and the synthesis of esters formed from glycerol and long-chain fatty acids. Lipases occur widely in nature, but only microbial lipases are commercially significant. In the present study, production of extracellular lipase in submerged fermentation of Pseudomonas sp.Lp1 isolated from oil contaminated soil has been investigated. The lipase production was optimized in shake flask experiments. The objective of the present investigation is to determine the activity of lipase was under varied physiochemical conditions of incubation time, pH, temperature, lipid carbon sources, nitrogen sources, surfactants and cations. The use of natural substrates in lipase production by Pseudomonas sp.Lp1 was also studied. The extracellular lipase production was maximum during the incubation time of 36 to 48 hours. The observed pH and temperature range optimum for maximum lipase production were 8.0-8.5 and 37-42 °C, respectively. Maximum lipase production occurred when olive oil (1%) was used when compared with Coconut oil, Sun flower oil Corn oil and Ground nut oil. Among the surfactants tested such as Triton X-100, Tween-20, Tween-80, Sodium dodecyl sulphate, all of them showed positive influence on lipase production,. Lipase activity was recorded to be the maximum when yeast extract and peptone were used as nitrogen source. About 45U/ml of enzyme activity was obtained in the medium prepared with whey as the natural substrate (2%).
Keywords
Lipase; Pseudomonas; Natural substrates; Production; Medium optimization
Download this article as:| Copy the following to cite this article: Kanimozhi S, Perinbam K. Optimization of Media Components and Growth Conditions to Enhance Lipase Production by Pseudomonas sp. Lp1. Biomed Pharmacol J 2010;3(2) |
| Copy the following to cite this URL: Kanimozhi S, Perinbam K. Optimization of Media Components and Growth Conditions to Enhance Lipase Production by Pseudomonas sp. Lp1. Biomed Pharmacol J 2010;3(2). Available from: http://biomedpharmajournal.org/?p=1562 |
Introduction
Lipases (triacylglycerol acylhydrolases, E.C. 3.1.1.3) are ubiquitous enzymes of considerable physiological significance and industrial potential. Lipases catalyze the hydrolysis of triacylglycerols to glycerol and free fatty acids. In contrast to esterases, lipases are activated only when adsorbed to an oil–water interface1 and do not hydrolyze dissolved substrates in the bulk fluid. A true lipase will split emulsified esters of glycerine and long-chain fatty acids such as triolein and tripalmitin. Lipases are serine hydrolases. In eukaryotes, lipases are involved in various stages of lipid metabolism including fat digestion, absorption, reconstitution, and lipoprotein metabolism. In plants, lipases are found in energy reserve tissues. How lipases and lipids interact at the interface is still not entirely clear and is a subject of intense investigation2. Lipases occur in animals, plants and microorganisms. Microbial lipases have a broad spectrum of industrial applications as they are more stable when compared with plant and animal lipases and they can be obtained cheaply3.
A relatively smaller number of bacterial lipases have been well studied compared to plant and fungal lipases. Amongst the lipase-producing organisms, Bacillus, Candida, Penicillium,Pseudomonas, Rhizomucor and Rhizopus spp. are outstanding ones4. Bacterial lipases are glycoprotein, but some extracellular bacterial lipases are lipoproteins. Most of the bacterial lipases reported so far are constitutive and are non–specific in their substrate specificity and a few bacterial lipases are thermostable. Among bacteria, Achromobacter,Alcaligenes, Arthrobacter, Pseudomonas, Staphylococcus and Chromobacterium and Serratia spp. have been exploited for the production of lipases5.
Most of the bacterial lipases reported so far are constitutive and are non-specific in their substrate specificity and a few bacterial lipases are thermostable. Due to such attributes, lipases are used in detergents, manufacture of food ingredients, pitch control in pulp and paper industry6, production of aromas, production of insecticides and synthesis of drugs such as naxopren and ibuprofen and as a biocatalyst of stereo selective transformations. The exponential increase in the application of lipases in various fields in the past few years necessitated both qualitative and quantitative improvement in enzyme production. Bacterial lipases are mostly inducible enzymes and require some form of oil, fatty acid, fatty acid alcohol or fatty acid ester and surfactants for induction7. Increased productivity of lipase during the fermentation process is of great importance since lower costs of production could promote new industrial applications. The productivity of lipase is affected by different physio-chemical parameters such as temperature, pH, medium composition and presence of inducers among others. Factors affecting extracellular lipase production have been studied and reported by many investigators. Therefore, in the present investigation, a study was undertaken to optimize the lipase production by locally isolated Pseudomonas sp. Lp1 on relatively low cost media and natural substrate media.
Materials and Methods
Isolation and Screening
Lipolytic bacteria were isolated from edible oil contaminated soil samples collected from different locations of Chennai. For this, 1.0 g of soil was dissolved in 100 ml of sterile distilled water and serially diluted up to 10-5. The diluted samples were plated on sterile nutrient agar and incubated at 37°C for 24 hours. The isolates showing different colony morphological features were individually tested for its lipolytic potential by inoculating the isolates in Tween agar medium comprising of Peptone-10g-l; NaCl-5 g-l; CaCl2-0.01 g-l; Agar -20 g-l and Tween 20 -10 ml; pH -7.5). After 48 h of incubation at 37°C, the lipolytic activity was determined by the formation of precipitate zone around the colony 8. The most potent lipase producing isolate was selected and identified based on the biochemical and physiological characters according to the key of Bergey’s manual of Determinative Bacteriology.
Preparation of Inoculum and Lipase Production
The inoculums for lipase production and other studies was prepared by inoculating the isolate in Luria Bertani (LB) broth consists of Yeast extract – 5 g-l ; Peptone – 10 g-l , NaCl – 10 g-l ; pH – 8.0 and was incubated at 37°C in a rotary shaker for overnight. The fresh over night culture was used as inoculums for production of enzyme. The enzyme production was carried out by shake flask fermentation using production medium comprised of Peptone-5 g-l; Yeast extract-5 g-l; NaCl-0.5 g-l; CaCl2-0.05 g-l; Olive oil-10 ml (emulsified with gum acacia- 0.5%); pH-7.2 in which the olive oil was used as substrate and carbon source. 100 ml of sterile production broth in duplicates was prepared in 250 ml conical flasks and seeded with 3% inoculums aseptically. The flasks were incubated overnight at shaker incubator at 37°C, pH 7 and 150 rpm. Culture supernatant was harvested by centrifugation at 10,000 g for 10min at 4°C. The enzyme activity was determined in the cell free supernatant thereof.
Lipase assay
Extracellular lipase activity was measured by photometric method of Winkler9 by measuring the micromoles of 4-Nitrophenol released from 4-Nitrophenyl palmitate substrate. A stock solution (20mM) of 4-Nitrophenyl palmitate (4-NPP) was prepared in isopropanol. The reaction mixture contained 150 µl of 4-nitrophenyl palmitate, 100 µl enzyme solution and 0.1M Tris buffer (pH 8.5) was made a final volume of 3 ml was incubated at 55ºC for 10 min in water bath. The reaction was arrested by chilling at -20ºC for 10 min. The absorbance was measured at 410 nm using UV-Vis Spectrophotometer (Elico). The reaction mixture contained heat inactivated enzyme was used as control. The activity of lipase was determined by comparing the optical density value with 4-nitrophenol standard. One unit (U) was defined as the amount of enzyme catalyzing the liberation of 1 µM p-Nitrophenol/min under the assay conditions.
Assay of Total Protein Content
Protein analysis of the different supernatants was determined spectrophotometrically according to Braford10. The protein concentrations were determined through Bovine serum Albumin standard.
Optimization of Physico-Chemical parameters for Lipase Production
Various parameters like incubation time, pH, and temperature and media components were altered to obtain the maximum production of lipase. The lipase production was carried out by shake flask fermentation in duplicates at appropriate conditions with 3% fresh inoculum. The flasks were incubated under shaking conditions at 150 rpm in a shaker incubator. After fermentation, enzyme activity was determined.
Effect of Incubation time
The lipase production was carried out by liquid state fermentation at 37°C, 150 rpm. Culture was aseptically with drowned periodically at 12 hours intervals up to 72 hours. The lipase activity of the cells free culture filtrate was determined.
Effect of pH
The effect of medium pH on lipase production was assessed at different pH values of production medium such as 5.5, 6.5, 7.5, 8.5 and 9.5. The various pH were obtained by adjusting with 0.1N NaOH and 0.1N HCl. The flasks were incubated overnight in shaker incubator at 150 rpm at 37°C, pH 7. The culture filtrate was examined for lipase the activity.
Effect of Tmperature
The influence of temperature on lipase production was assessed by carrying out the fermentation at different temperatures such as 28ºC, 32ºC, 37ºC, 42ºC, 47ºC, 55ºC and 60ºC for 48 hours. The production medium with pH 8.5 were incubated overnight in shaker incubator at 150 rpm.The culture filtrate was examined for the lipase activity.
Effect of Lipid Carbon Sources
The effect of lipid carbon sources on lipase production was investigated by using different carbon sources namely Olive oil, Coconut oil, Sun flower oil, Corn oil and Ground nut oil at the concentration of 1%(v/v) and incubated at 40ºC, at pH 8.5, for 48 h under shaking conditions (150 rpm) and lipase activity was determined.
Effect of Nitrogen sources
In order to determine the effects of different nitrogen sources on lipase production the production medium was replaced with different nitrogen sources such as Yeast extract, peptone, potassium nitrate, ammonium nitrate and ammonium sulphate at the concentration of 0.5% (w/v). The samples were incubated at 40ºC, at pH 8.5, for 48 h under shaking conditions (150 rpm) and lipase activity was determined.
Effect of Surfactants
The following detergents viz.,(0.2%v/v or w/v) Sodium Dodecyl Sulphate, Triton X-100, Tween-20 and Tween-80 were added to the production medium in order to find out the effect of surfactants over the production of lipase. These mixtures were incubated at 40ºC, at pH 8.5 for 48 h under shaking conditions (150 rpm) and lipase content was determined.
Effect of Metal ions
The metal ions like Na2+, K+, Ca2+ ,Cu2+, Mg2+, Mn2+, Zn2+, Fe2+ and Fe3+ were added to the production medium at a concentration of 0.2%(w/v) and were incubated at 40ºC, at pH 8.5 for 48 h under shaking conditions(150 rpm) and the culture filtrate was analyzed for lipase activity.
Production of Lipase using natural substrates
Various natural oil cakes such as Coconut oil cake, Sesame oil cake, Ground nut oil cake, Cotton seed oil cake and Whey medium were used as substrates for effective enzyme production. The crude substrates were collected from edible oil industries. 2% (w/v) natural oil cakes were prepared by grinding them using distilled water and sterilized by autoclaving. The Whey medium 2% (v/v) was prepared from whey collected from milk industry and was steam sterilized. The pH was adjusted to 8.5 and fermentation was carried out in standard conditions. The amount of lipase produced in different substrates was estimated.
Results and Discussion
Isolation, Screening and Identification of Potential Strain
Microbiological analysis of soil samples from edible oil contaminated sites showed high bacterial count. The isolates obtained from the samples were screened for lipase production in Tween Agar. Among the total 52 isolates, 8 isolates showed positive towards lipase production by showing white precipitate around the colony. The isolate that showed comparatively more precipitation zone was considered as most potent strain and selected for the present study. Based on biochemical, cultural, and morphological characteristics, the isolate was identified as Pseudomonas sp. and designated as strain Lp1. (Figure.1).
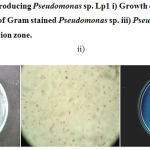 |
Figure 1: Lipase producing Pseudomonas sp. Lp1 i) Growth on Nutrient agar ii) Microscopic view of Gram stained Pseudomonas sp. iii) Pseudomonas sp.Lp1 showing precipitation zone.
|
Optimization of Physico-Chemical Parameters for Lipase Production
Effect of Incubation time on Lipase Production
The incubation time is an important factor for the production of extracellular lipase by the microorganisms11. The amount of lipase produced by Pseudomonas sp. Lp1 was observed after every 12 hours of incubation till 72 hours. The maximum lipase activity of about 68U/ml was observed after 48 h of fermentation .The maximum lipase production occurred during the late log phase of the organism. (Figure.2). After 48 h, the growth showed divergence from the exponential because in place of homogeneous growth, bacterial pellets began to form in which nutrients and oxygen supply became the growth limiting.
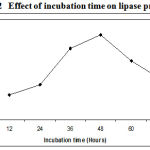 |
Figure 2: Effect of incubation time on lipase production.
|
After that lipase yield got reduced due to the depletion of nutrients, accumulation of toxic end products, and the change in pH of the medium. Several researchers have reported different incubation periods for optimal lipase production. Maximum lipase was produced after 72 h and 96 h of incubation, respectively, in the case of the Pseudomonas spp P. fragi and P. fluorescens BW 96CC12,13. The lipase production of lipase by Pseudomonas sp. was maximum after 48 hrs of incubation14. Pseudomonas fluorescens B68 catalyzed the transesterification reaction after 120 hours of incubation15. Bacillus sp. showed maximum lipase activity after 48 h of incubation16.
Effect of pH on Lipase Production
As pH is the important parameter required for the growth of bacterial culture in respective media lipase activity got affected with basic pH, which indicates that suitable pH is responsible for bacterial growth in the media. The data obtained clearly indicated that there was a strong influence of pH on lipase production (Figure.3) .The maximum lipase activity about 72U/ml by Pseudomonas sp. Lp1 was reported at pH 8.5. P. aeruginosa MB prefers neutral pH17. S. rubidaea requires alkaline pH (7). Lipase activity by Serratia grimesii was high at pH 8–95.
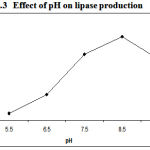 |
Figure 3: Effect of pH on lipase production.
|
Effect of Temperature on Lipase Production
Experiments on effect of temperature indicated that the lipase production by Pseudomonas sp. Lp1 was maximum between the temperatures of 37°C – 42°C at the optimum temperature of 42°C. But in low temperatures (25 to 35°C) as well as high above 47°C, the lipase production recorded was low (Figure. 4 ). Lipase production by S. marcescens was higher at the temperature of 25°C as compared to 30 and 35°C18. Similarly, lipase production observed in P. aeruginosa MB was higher at 30°C17. But in the present study, lipase activity showed gradual increase with the increase of temperature from 25 to 45°C and further increase of temperature, beyond 45°C the production decreased. Such type of result was also been reported in S. rubidaea7. The lipase activity of Bacillus strains was high (0.0029 μg/ml/min) when grown at the medium temperature of 37°C19. Lipase activity of Staphylococcus sp. Lp12 showed gradual increase with the increase of temperature from 25 to 45°C20.
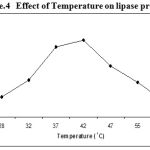 |
Figure 4: Effect of Temperature on lipase production.
|
Effects of Lipid Carbon Sources on Lipase Production
Carbon is a main component of cells and carbon sources are important substrates for energy production in microorganisms. Thus, bacterial lipases are generally produced in the presence of oil or any other lipid substrate viz., fatty acid esters, fatty acids, glycerol as carbon sources in the presence of any complex nitrogen source .In order to investigate the effects of carbon sources on lipase production various vegetable oils were tested as carbon source viz., Olive oil, Coconut oil, Sun flower oil, Corn oil and Ground nut oil. Maximum lipase production (58U/ml) occurred when olive oil was used and other oils like Corn oil and Coconut oil also have yielded comparatively considerable productions of about 38U/ml and 31U/ml respectively. (Figure.5). Triglycerides are important substrates for lipase production as they can act as an inducer as well as an inhibitor.
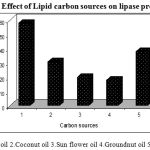 |
Figure 5: Effect of Lipid carbon sources on lipase production.
|
In the present study, all the tested triglycerides were found to induce the lipase synthesis by Pseudomonas sp. Lp1 with different level of enzyme production. This study is in agreement with the previous work on castor oil-induced lipase production by Pseudomonas aeruginosa KKA-521. Vegetable oil-induced lipase production was observed in Candida rugosa (DSM 203122 and sunflower oil and olive oil-induced extracellular lipase production by Yarrowia lipolytica23 and however, the thermostable Bacillus sp. has been reported to produce thermostable alkaline lipase with corn oil and olive oil (1%) as carbon sources24, 25. The lipase production was more when vegetable oil, olive oil, soya bean oil, sunflower oil and gingelly oil were used as the carbon source in the lipase producing strains26. Similar reports were shown in Pseudomonas sp. lipase production with various vegetable oils27.
Effects of Surfactants on Lipase Production
In order to determine the effects of surfactants on lipase production, Sodium Dodecyl Sulphate (SDS), Triton X-100, Tween 20 and Tween 80 were tested. All the surfactants tested showed positive influence on lipase production by Pseudomonas sp. Lp1 compared to the control. Medium containing Tween 20 showed highest lipase production after 48 h of incubation (Figure. 6). Very low lipase production was observed when SDS was used. From the present study it is also evident that the lipase production by microorganisms could be induced by the addition of surfactants. Similarly, the studies of lipase production by P. aeruginosa EF2 indicated the positive influence of Tween 8028. Higher levels of lipase production were observed when the substrate formed an emulsion, thereby presenting an interfacial area to the enzyme29. The maximum lipase production by S. rubidaea was induced by Tween 20 ((27.10±0.01) U/ml), followed by polyethylene glycol 300 ((26.00±0.06) U/ml) (7). Tween 80 in the medium of Staphylococcus sp.Lp12 was shown to enhance lipase production after 48 h of incubation20.
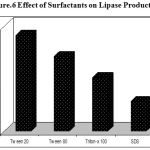 |
Figure 6: Effect of Surfactants on Lipase Production.
|
Effects of Nitrogen sources on Lipase Production
Figure.7 shows the effect of nitrogen sources on lipase production. Both organic and inorganic nitrogen sources play an important role in the synthesis of the enzyme by Pseudomonas sp. Lp1. Different nitrogen sources were incorporated in the production medium viz., Yeast extract, and peptone, potassium nitrate, ammonium nitrate and ammonium sulphate at the concentration of 0.5% (w/v)). Lipase activity was recorded to be the maximum when yeast extract and peptone were used, whereas in the medium with the inorganic nitrogen salts, the lipase production was comparatively less. Peptone contains cofactors and amino acids which match strains physiological requirements of lipase production30. Complex nitrogen sources such as yeast extract, peptone and corn steep liquor have traditionally been used for lipase production31, 32. Peptone has also been shown to support lipase production in the case of Aspergillus sp.33 and Fusarium sp.34. Pseudomonas sp. strain 5 evidenced higher degrees of lipase production (0.335 U/ml) in cases in which peptone was added to the basal medium14. Bacillus strain produced a maximal lipase activity in a medium that contained yeast extract (3.0%) as the nitrogen source16. Peptone was found to be the best organic nitrogen source for lipase production in Staphylococcus sp.Lp1220. The presence of organic nitrogen sources increased the lipase production in the cultures of Pseudomonas and Bacillus species27.
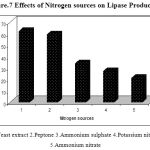 |
Figure 7: Effects of Nitrogen sources on Lipase Production.
|
Effects of Metal ions on Lipase Production
Divalent cations stimulate or inhibit enzyme production in microorganisms The effect of a variety of inorganic salts Na2+, K+, Ca2+ ,Cu2+, Mg2+, Mn2+, Zn2+, Fe2+ and Fe3+ on lipase production was evaluated. The production of extracellular lipase by Pseudomonas sp. Lp1 was enhanced in the medium supplemented with Ca2+ K+ and Mg2+ at the concentration of 0.2% (Fig. 8).The metal ions like Mg2+ and Na2+ also significantly increased the extracellular lipase production in Pseudomonas sp. Lp1. The medium with the ions like Cu2+ and Zn2+ there was reduction in the lipase production than the control medium. The metal ions like Fe2+ and Fe3+ inhibited the lipase production completely. Ca2+ was determined to stimulate lipase, thereby suggesting that Pseudomonas sp. Lp1 lipase was a metal-activated enzyme, in which the ions often function in a structural, rather than a catalytic, and role. The ions bind to the enzyme and alter the conformation of the protein to counter greater enzyme stability .The metal ions function as electrophiles, which seek the opportunity to share electron pairs with other atoms, such that a bond or charge-charge interaction might be formed35. This effect was attributable to the interaction between salt ions and the enzyme surface charge, which might markedly affect the ionization of some amino acid residues, thus changing the enzyme conformation and altering enzyme activity13. The preference for metal ions of the crude enzyme differed from that of the purified lipase36. The stimulation of lipase production occurred in Burkholderia sp. in the presence of Ca2+ and Mg2+37. The stimulation of lipase production was observed in Bacillus sp. RSJ1 in the presence of calcium chloride38. However, most other metal ion salts were inhibitory to lipase production. Iron was found to play a critical role in the production of lipase by Pseudomonas sp. G639. In addition to the various chemical constituents the production of extracellular lipase by P. aeruginosa S5 was enhanced in cases in which the peptone medium was supplemented with Na2+. Lipase production by Bacillus sp. (Pa2) was increased when Mg2+ (0.25%) was added in the production medium16. Lipase production by P. pseudoalcaligenes F-111 was enhanced when a phosphate containing medium was provided with Mg2+40. Maximal lipase production by P. pseudoalcaligenes KKA-5 occurred at Mg2+ concentration of 0.8 M21.
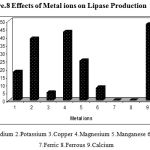 |
Figure 8: Effects of Metal ions on Lipase Production.
|
Production of Lipase using Crude Substrates
The real and beneficial production of enzyme is the production from the natural sources and industrial wastes. In this study, several natural oil cakes after de oiling and whey were used as substrates for lipase production by Pseudomonas sp. Lp1. The wastes like Coconut oil cake, Sesame oil cake, Ground nut oil cake, Cotton seed oil cake and Whey were used. The results revealed that the maximum production was observed in whey medium of about (45 U/ml) (Figure.9). Cultural conditions for the production of lipase by Aspergillus niger strain MTCC 2594 by solid state fermentation using gingly oil cake were standardized. A lipase activity of 363.U/g of dry substrate was obtained after 72h under optimal conditions41. Similar result was reported42.
The almond meal is the best crude medium for the higher production of lipase by Bacillus sp which provides all required carbon, nitrogen and contained sucrose, gum, asparagines and proteins43. Similarly in the present study whey medium also supplied the nutrients needed for the growth and lipase production by Pseudomonas sp. Lp1. (Fig.9).
 |
Figure 9: Production of Lipase using Crude Substrates.
|
Conclusion
The present study revealed that extracellular lipase production by Pseudomonas sp. Lp1 isolated from oil contaminated soil samples was found to be enhanced at optimized culture conditions such as medium incubation time, pH, temperature and various substrate concentrations. From the results, it could be concluded that the medium pH of 8.0 and temperature range of 37-42 °C when incubated up to 48 hours was optimum for maximizing lipase production by Pseudomonas sp. Lp1. The assessment of various substrates for optimizing the production of lipase by Pseudomonas sp. Lp1 revealed that the medium components contained Corn oil – 1%(emulsified with gum acacia- 0.5%) ,Peptone -0.5%,Yeast extracts 0.5%,Tween 20 – 0.2% Nacl – 0.2% and CaCl2 – 0.2%. The natural substrate medium [Whey medium (Whey – 2.0%v/v)] also supported the growth of Pseudomonas sp. Lp1and induced the production of lipase significantly. Thus Pseudomonas sp. Lp1is a potential strain to produce lipase in both chemically defined medium and natural medium extracellularly.
References
- Martinelle, M., Holmquist, M., Hult, K. On the interfacial activation of Candida antarctica lipase A and B as compared with Humicola lanuginosa lipase. Biochim. Biophys. Acta., 1995; 1258:272- 6.
- Balashev, K., Jensen, T.R., Kjaer, K., Bjornholm, T. Novel methods for studying lipids and lipases and their mutualinteraction at interfaces: Part I. Atomic force microscopy. Biochimie., 2001; 83:387–97
- Ghosh, R.K., Saxena, R.K., Gupta, R., Yadav, R.P., Davidson, W.S. Microbial lipases: production and applications. Sci. Prog., 1996; 79:119–57.
- Rapp,P., Backhaus, S. Formation of extracellular lipases by filamentous fungi, yeast and bacteria. Enzyme. Microb. Technol., 1992; 14: 938–943.
- Abdou, A.M. Purification and partial characterization of psychrotrophic Serratia marcescens lipase. J. Dairy. Sci., 2003; 86: 127–132.
- Jaeger, K.E., Reetz, M.T. Microbial lipases form versatile tools for biotechnology. Trends. Biotechnol., 1998; 16: 369-403.
- Immanuel, G., Esakkiraj, P., Jeladhas, A., Iyapparaj, P., Arunachalam, P. Investigation of lipase production by milk isolate Serratiarubidaea. Food Technol. Biotechnol., 2008 4; 6(1): 60-65
- Rua Luisa, M., Schmidt-Dannert, C., Wahl, S., Sprauer, A. Schmid, R. D. Thermoalkalophilic lipase of Bacillus thermocatenulatus large-scale production, purification and properties: aggregation behaviour and its effect on activity. J. Biotechnol., 1997; 56:89-102.
- Winkler, U.K., Stuckmann, M. Glycogen, hyaluronate and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J. Bacteriol., 1979;38: 663–670.
- Bradford. M. A rapid and sensitive method for the quantification of µg quantities of protein. Anal. Chem., 1976; 72: 248-254.
- Shirazi, S.H., Rahman, S.R., Rahman, M.M. Production of Extra cellular lipases by . Saccharomyces cerevisiae. World J. Microbiol. Biotechnol., 1998; 14(4): 595-597.
- Pabai, F., Kermasha, S., Morin, A. Use of continuous culture to screen for lipase-producing microorganisms and interesterification of butterfat by lipase isolates. Can. J. Microbiol., 1996; 42:446–452
- Dong, H., Gao, S., Han, S., Cao, S. Purification and characterization of a Pseudomonas sp. lipase and its properties in non-aqueous media. Appl. Microbiol. Biotechnol., 1999; 30:251–256
- Abdul Rahman, R.N.Z.R., Syarul Nataqain Baharum., Abu Bakar Salleh., Mahiran Basri. S5 Lipase : An Organic Solvent Tolerant Enzyme. J. Microbiol., 2006; 44(6):583-590.
- Yu Luo., Yitao Zheng., Zhengbing Jiang., Yushu Ma., Dongzhi Wei. A novel psychrophilic lipase from Pseudomonas fluorescens with unique property in chiral resolution and biodiesel production via transesterification. Appl. Microbiol. Biotechnol., 2006; 73:349–355
- Shah, K. R., Patel, P. M., Bhatt, S. A. Lipase Production By Bacillus Sp. Under different Physio-Chemical Conditions. J. Cell and Tissue Res., 2007; 7(1): 913-916.
- Marcin, C., Kat.,L., Greasham, R., Chartrain, M. Optimization of lipase production by Pseudomonas aeruginosa MB 5001 in batch cultivation. J. Ind. Microbiol. Biotechnol., 1993; 12: 29-34.
- Gao, L., Xu, J.H., Li, X.J., Liu, Z.Z. Optimization of Serratia marcescens .Lipase production for enantio selective hydrolysis of 3- phenyl glycidic acid ester. J. Ind. Microbiol. Biotechnol., 2004; 31: 525-530
- Selva Mohan, T., Palavesam, A., Immanvel, G. Isolation and characterization of lipase-producing Bacillus strains from oil mill waste. Afr. J. Biotechnol., 2008; 7 (15): 2728-2735.
- Pogaku, P., Suresh, A., Srinivas,P., Ram Reddy, S. Optimization of lipase production by Staphylococcus sp. Lp12. Afr. J. Biotechnol., 2010; 9(6): 882-886.
- Sharon, C., Furugoh, S., Yamakido, T., Ogawa, H.I., Kato, Y. Purification and characterization of a lipase from Pseudomonas aeruginosa KKA-5 and its role in castor oil hydrolysis. J. Ind. Microbiol. Biotechnol., 1998; 20: 304–307.
- Lakshmi, B., Kangueane, P., Abraham, B., Pennatheu, G. Effect of vegetable oils in secretion of lipase from candida rugosa 9DSM2031) Lett. Appl. Microbiol., 1999; 29: 66-70.
- Dominguez, A., Deive, F.J., Sanroman, M.A.., Longo, M.A. Effect of lipids and surfactants on extracellular lipase by Yarrowia lipolytica. J. Chem. Technol. Biotechnol., 2003; 78:1166–1170.
- Sugihara, A., Tani, T., Tominaga,Y. Purification and Characterization of a Novel Thermostable Lipase from Bacillus sp. J. Biochemi., 1991; 109: 211-216.
- Wang,Y., Srivastava, K.C., Shen,G..J., Wang, H.Y. Thermostable alkaline lipase from a newly isolated thermophilic Bacillus, strain A30-1 (ATCC 53841). J. Fermen.Bioeng., 1995; 79: 433-438.
- Rohit, S., Yusuf, C., Ullamchand, B. Production, purification, characterization and application of lipases. Biotechnol. Adv., 2001; 19: 627- 662.
- Shivareddy, D.M., Raghavendra, L., Ksheerasagar, Abhilash, M. Isolation, optimization, production and molecular characterization of lipase from Bacillus and Pseudomonas species and protein profiling. Res. J. Pharm. Bio.l Chem. Sci., 2010; 1(1): 27-38.
- Gilbert, E.J., Drozd, J.W., Jones, C.W. Physiological regulation and optimization of lipase activity by Pseudomonas aeruginosa. J. Gen. Microbiol., 1991; 137:2215–2221.
- Wu, H.S., Tsai, M.J., Kinetics of tributyrin hydrolysis by lipase. Enzyme Microbiol. Technol., 2004; 35: 488-493
- Freire, D.M.G., Teles, E.M.F., Bon, E.P.S., Sant Anna, G.L., Lipase production by Penicillium restrictum in a bench-scale fermentor: effect of carbon and nitrogen nutrition, agitation and aeration. Appl. Bacteriol. Biotechnol., 1997; 63/64: 409-421.
- Chahinian, H., Vanot, G., Ibrik, A., Rugani, N., Sarda, L., Comeau, C. Production of extracellular lipases by Penicillium cyclopium. Biosci. Biotech. Biochem., 2000; 64: 215-222.
- Sharma, R., Chisti, Y., Banerjee, U.C. Production, purification, characterization and application of lipases. Biotechnol. Adv., 2001; 19: 627-662.
- Cihangir, N., Sarikaya, E., Investigation of lipase producing by a new isolate of Aspergillus sp. World. J. Microbiol. Biotechnol., 2004; 20: 193-197.
- Gulati, R., Isar, J., Kumar, V., Prasad, A.K., Parmar, V.S., Saxena, R.K. Production and a noval alkaline lipase by Fusarium globulosum using neem oil and its applictions. Pure. App. Chem., 2005;77: 251-262.
- Glusker, J.P., Katz, A.K., Bock. C.W. Metal ions in biological systems. J. Rigaku., 1999;16: 8-16.
- Rahman, R.N.Z.R.A., Baharum, S.N., Salleh, A.B., Basri, M. High yield purification of an organic solvent tolerant lipase from Pseudomonas sp. strain S5. Anal. Biochem., 2005; 341: 267-274.
- Rathi, P., Saxena, R.K., Gupta, R. A novel alkaline lipase from Burkholderia cepacia for detergent formulation. Process. Biochem., 2001; 37:187–192
- Sharma, R., Soni, S.K., Vohra, R.M., Jolly, R.S., Gupta, L.K., Gupta, J.K. Production of extracellular alkaline lipase from a Bacillus sp. RSJ1 and its application in ester hydrolysis. Ind. J. Microbiol., 2002; 42:49–54
- Kanwar, L., Gogoi, B.K., Goswami, P. Production of a Pseudomonas lipase in n-alkane substrate and its isolation using an improved ammonium sulfate precipitation technique. Bioresour. Technol., 2002; 84:207–211.
- Lin, S.F., Chiou, C.M., Yeh, C., Tsai, Y.C. Purification and partial characterization of an alkaline lipase from Pseudomonas pseudoalcaligenes F-111. Appl. Environ. Microbiol ., 1996; 62:1093–5.
- Kamini, N.R., Mala, J.G.S., Puvanakrishnan, R. Lipase production from Aspergillus niger, by.solid-state fermentation using gingili oil cake. Process. Bioche., 1997; 33: 505-511.
- Catherine Schuepp., Selim Kermasha, A., Marie-Caroline Michalsk, P., Andre Morin. Production, partial purification and characterisation of lipases from Pseudomonas fragi CRDA 037.Process. Bioche., 1996; 32: 225-232.
- Ikram ul-Haq., Shumaila Idrees., Ibrahim Rajoka, M. Production of lipases by Rhizopus oligosporous by solid-state fermentation. Process. Biochem., 2001; 37: 637–641.







