Vandana Saini1* , Meenu Khurana1
, Meenu Khurana1 and Rama Krishna Challa2
and Rama Krishna Challa2
1Chitkara University School of Engineering and Technology, Chitkara University Himachal Pradesh, India.
2Department of Computer Science and Engineering, NITTTR, Chandigarh, India
Corresponding Author E-mail: vandana99.phd23@chitkarauniversity.edu.in
DOI : https://dx.doi.org/10.13005/bpj/3130
Abstract
Mammogram image segmentation is crucial for early detection and treatment of breast cancer. Timely detection can help in saving the patient’s life. By accurately identifying and isolating regions of interest in mammograms, we can improve diagnostic accuracy. In this paper a hybrid model for segmentation using Ostu thresholding with morphological operations and U-Net model is proposed for accurate segmentation of mammogram images. The incorporation of attention mechanisms and residual connections in U-Net helps in enhancing the model’s performance. The proposed model performs better than recent existing models, achieving high precision, recall, F1 score, accuracy, and area under curve (AUC). The proposed model is evaluated on the MIAS dataset and achieved an F1 score of 0.9764, precision of 0.9802, recall of 0.9980, accuracy of 0.9902, and an AUC of 0.99997. These results had shown significant improvements in comparison with existing models, making it a suitable and accurate model for the early detection and diagnosis of breast cancer.
Keywords
CAD; Otsu; Mammogram; MIAS; Segmentation; U-Net
Download this article as:| Copy the following to cite this article: Saini V, Khurana M, Challa R. K. A Hybrid Model for the Segmentation of Mammogram Images using Otsu Thresholding, Morphology and U-Net. Biomed Pharmacol J 2025;18(1). |
| Copy the following to cite this URL: Saini V, Khurana M, Challa R. K. A Hybrid Model for the Segmentation of Mammogram Images using Otsu Thresholding, Morphology and U-Net. Biomed Pharmacol J 2025;18(1). Available from: https://bit.ly/4aWOApY |
Introduction
Mammogram image segmentation plays a vital role in early and timely detection of breast cancer. The segmentation helps in the detection of lesions from the image, which is the major region of interest for a radiologist. There are many existing methods for image segmentation, such as thresholding, region-based, edge-based, and cluster-based methods. However, these are now classified as traditional methods since learning-based methods are more widely used due to their higher accuracy. Learning-based methods for image segmentation include U-Net, Fully Convolutional Network (FCN), and Mask R-CNN, among others. Due to their robust nature, high accuracy, and precision, these learning-based methods have become the first choice for researchers. Nevertheless, learning-based methods still suffer from issues such as a false positive rate due to complexities like large datasets, long training times with augmentations, and a lack of domain knowledge. This section further presents an overview of the strengths and weaknesses of each traditional and learning-based segmentation model, highlighting their applicability and potential limitations in the context of mammogram image segmentation.
Traditional Segmentation Models
In this section, various image segmentation models along with their positive as well as negative points are discussed in table 1.
Table 1: Summary of traditional image segmentation models
| Traditional Image Segmentation Models | ||
| Methods | Positive Points | Negative Points |
| Region-Based Segmentation2 | · Effective for segmenting homogeneous regions.· Good for images with distinct regions. | · Can be computationally intensive.· Sensitive to the selection of initial seed points. |
| Thresholding3 | · Simple and easy to implement· Fast processing time. | · Highly sensitive to noise and intensity variations.· Not suitable for complex images. |
| Watershed Segmentation13 | · Good for separating overlapping objects.· Effective for images with well-defined gradients | · Can lead to over-segmentation.· Sensitive to noise and requires pre-processing steps. |
| Cluster-Based Segmentation18 | · Can handle images with multiple regions.· Does not require prior knowledge of the image. | · May require a large number of iterations.· Sensitive to the initial choice of cluster centers. |
| Edge-Based Segmentation22 | · Effective for detecting boundaries.· Works well for images with high contrast edges. | · Sensitive to noise.· May produce fragmented edges.· Not effective for images with smooth boundaries. |
Image segmentation models discussed in table 1 are fundamental and still widely used for basic image analysis tasks. Among these methods most used models are discussed in detail below:
Otsu Thresholding Method
This is one of the most popular methods that separates the image foreground and background separately. In these methods an optimal threshold value is calculated every round using the maximum variance as parameter to separate foreground for background1,2.
Adaptive Thresholding Method
This method works on the pixel intensity value in an image. A threshold value is computed based on the image size, contrast and variance. The pixels that had intensity above the threshold are separated as foreground and left become background pixels3,4.
Region Based Method
This method works based on pixel and the color intensity in the image. The method splits an image into multiple small windows based on the homogeneous set of pixels. The region-based partitioning is done to separate foreground and background. The regions with similar pixel values are snipped under one window and this process traverses the complete image. This method helps in effective segmentation due to grouping of similar pixels5.
Learning Based Segmentation Models
Various learning-based image segmentation models with their strengths and weakness are discussed in table 2. These models can capture complex patterns and details, making them suitable for advanced applications like medical imaging and autonomous driving.
Table 2: Summary of learning-based image segmentation methods
| Learning Based Segmentation Models | ||
| Methods | Positive Points | Negative Points |
| U-Net7 | · Focuses on relevant features.· Effective in complex medical imaging tasks. | · More complex architecture.· Increased computational and memory requirements. |
| Mask R-CNN8 | · Combines detection and segmentation.· High accuracy for instance segmentation.· Handles occlusions well. | · Computationally expensive.· Requires extensive training time and resources. |
| Fully Convolutional Networks (FCN)9 | · Improved gradient flow and feature reuse.· High performance on various segmentation tasks. | · May produce coarse segmentation boundaries.· Requires large, annotated datasets. |
| DeepLabV3+ 11 | · Handles multi-scale context.· Refines segmentation boundaries.· High performance on challenging datasets. | · Computationally intensive.· Requires significant computational resources for training and inference. |
| SegNet13 | · Efficient memory usage.· Suitable for real-time applications. | · May struggle with fine details in segmentation.· Requires large amounts of labeled data for training. |
| DenseNet23 | · Improved gradient flow and feature reuse.· High performance on various segmentation tasks. | · High memory usage.· May be prone to overfitting if not properly regularized. |
Learning-based Image segmentation models discussed in table 2 can improve with more data makes them powerful tools in modern image analysis. Among these methods most used models are discussed in detail below:
U-Net Model
This is one of the most effective and the popular image segmentation models. The model architecture is in U shape as the reason model is named as U-Net where net is network. It uses the convolution layer for the feature extraction from the image and pool layer for the image compression. But due to the image compression the image is down sampled, and the up sampling is done using skip connection, which is computationally expensive6,7.
Mask R-CNN
This is an extension of Fast R-CNN, that helps in predicting the segmentation mask. This model predicts the objects in the region, which are classified as ROI, as objects are the lesions that need to be segmented accurately. It uses the bounding boxes to mark the detected objects. This method is suitable for segmentation as well as object detection too from an image8.
Fully Convolution Network (FCN)
This model had a fully connected layer that is used for the replacement of convolution layers so that images of any size can be processed. Here each pixel in the image is assigned into a class. To up sample the image it has deconvolution layer, this up sampling is done using the created pixel class wise segmentation map9,10.
Traditional methods for image segmentation are simple, fast, and effective for specific tasks but may struggle with complex patterns whereas learning based methods provide high accuracy and can learn complex patterns from data but require large, labeled datasets and significant computational resources. So, there is a need for an accurate and robust model that combines the strengths of both methods. In this work we are proposing a hybrid model for mammogram image segmentation using the traditional and learning segmentation method. This helps in creating a less expensive and domain specific accurate model. The proposed model uses the Otsu threshold with morphological operations for better foreground and background separation and U-Net learning model for accurate detection of ROI. The combination of traditional methods helps in retaining the domain knowledge like lesion boundaries marking which helps in overall enhancement of the model.
Literature Review
Authors proposed a deep learning-based hybrid model for the mammogram image segmentation11. The proposed method is based on U-Net and BCDU net. The authors combine both models to form a Bi- directional Convolution LSTM U-Net. The proposed network had shown the good Jaccard and dice coefficient score of 78.72% and 83.76% respectively. Also, the accuracy of Region of Interest (ROI) segmentation is near 87%. The proposed model had reduced computational complexity by using LSTM with U-Net.
Authors used a Fuzzy C-Mean (FCM) method for mammogram image segmentation. The major goal of the paper is to enhance breast cancer diagnosis12. A hybrid method for segmentation is proposed using traditional methods like Contrast limited adaptive histogram equalization (CLAHE), morphological operations with FCM. The performance of the proposed method is evaluated on Mammographic Image Analysis Society (MIAS) dataset based on threshold, precision, sensitivity and specificity. The method had achieved the accuracy of more than 92% with 99% specificity.
Authors had discussed various existing segmentation methods based on traditional, machine and deep learning methodology13. The author had given emphasis on the fact that due to the cross disciplinary nature of this field, still there is no effective way to adopt the advancement in segmentation. The tumors are complex, and specialists could not understand the complex patterns and varying nature of tumors due to which developed models lack in varsity. The paper also concludes that the MIAS dataset is the most used dataset and U-Net is highest used model for the mammogram segmentation. The U-Net model does not need any annotated data as it is specifically developed for medical images segmentation only. Also, the models need high end GPUs for training making them complex for real time data processing.
Authors had proposed a connected U-Net model for the mammogram image segmentation. The author worked on encoder and decoder-based architecture14. The proposed model uses attention and residual Net for enhancement of the model which is evaluated using DDSM dataset with generated synthetic data using GAN. The proposed network architecture has two convolution units of 3 x 3 with ReLU activation and decoder block is of 2 x 2 size which is connected with previous output. The model has achieved the dice score of more than 95% with intersection over union (IoU) score around 92% on the used dataset.
Authors had used a hybrid method using Haar transformation and U-Net together for the breast tissue image segmentation for the MRI image15. The proposed model takes the lying position image of a patient that is equivalent to standing position for the preservation of the natural shape of the breast. The dataset images are labelled as skin, tissues and fats. The Haar transformation is used to reduce the information loss from the image and U-Net to learn the accurate features from labelling. The U-Net architecture is designed to prevent the problem of overfitting by adding the early stopping mechanism. The proposed model had shown the higher accuracy of around 90% with IoU of 87.48%. The proposed method is helpful for the plastic surgeons for the breast reconstruction surgery.
Authors had reviewed the deep learning-based methods widely used for the breast cancer image segmentation16. The author had concluded that the deep learning models are preferred over traditional models due to their accuracy and robustness.
Authors had proposed a dual branch-based U-Net for the breast ultrasound image segmentation17. The success of U-Net for the segmentation of the medical image segmentation motivated the author to develop a dual branch variant. The proposed model architecture uses two distinct paths for the model encoding helps in better feature extractions. At encoder path one the original image is inputted and at second path the image is created using robert edge filtering, which helps in highlighting and preserving the edges. The encoder paths then combine at convolution layer followed by a pool layer. The weighted scheme is used for cross learning. The proposed model is evaluated on the Breast Ultrasound Images (BUSI) and UDIAT dataset. The proposed method had achieved an IoU of 77.46% and the dice coefficient of 87.28%.
Authors had proposed a fused architecture for segmentation using Attendseg and gravitational clustering algorithm by using the mathematical data modelling18. The proposed work is done on breast ultrasound (BUS) images dataset. Initially the author did the preprocessing before the segmentation which helped in better segmentation and retaining the important features of the image. Further the proposed model of segmentation is used with the data augmentation. Attendseg is used to separate the ROI for the input images whereas the gravitational and mathematical modelling is issued for efficient feature extraction. The proposed method had achieved an accuracy of 98.95%, IoU of 85.63% and DC 89.45%.
Some more existing segmentation models are discussed below in table 3.
Table 3: Existing Segmentation Models
| Ref. No. | Datasets Used | Models Used | Primary Contribution | Advantages | Limitations |
| [19] | MIAS | FF-CSO Algorithm | Proposed a novel FF-CSO (Firefly-Competitive Swarm Optimization) algorithm for breast cancer detection in mammograms. | Improved detection accuracy by combining FF and CSO algorithms. | Limited evaluation on different datasets; requires further validation on larger datasets. |
| [20] | MIAS | Chan-Vese technique | Introduced the Chan-Vese segmentation technique for breast tumor segmentation in mammograms. | Provides a more accurate segmentation for homogenous regions within breast tumors. | Struggles with non-homogenous and noisy regions in mammograms. |
| [21] | MIAS | Level set with Cuckoo search optimization | Developed a hybrid segmentation approach using level set method enhanced with Cuckoo search optimization for mammogram segmentation. | High accuracy and robustness in identifying breast masses. | Computationally intensive; may not generalize well to other datasets or segmentation tasks. |
| [22] | MIAS | Non-Convex border optimization, segmentation thresholding | Proposed a non-convex border optimization approach for early detection of mammography edges and boundaries. | Early detection of edges and boundaries, improving the accuracy of tumor detection in earlier stages. | Limited focus on the segmentation of complex structures, which might affect overall performance. |
| [23] | MIAS, DDSM, CBIS-DDSM | Deep learning (InceptionV3, DenseNet121, ResNet50, VGG16, MobileNetV2), U-Net | Enhanced breast cancer detection with deep learning models, using CNN architectures and U-Net for segmentation. | High accuracy and robustness across multiple datasets; demonstrated superior performance across metrics. | High computational cost due to the complexity of the models used. |
| [24] | MIAS | Transfer learning with pre-trained CNN architectures | Introduced a novel transfer learning technique for automatic detection and classification of breast cancer. | High sensitivity and specificity, leveraging the strengths of pre-trained CNN models. | Requires extensive computational resources, particularly for model training and tuning. |
| [25] | MIAS | Multi-objective Electromagnetism-Like Optimization Algorithm | Developed a multi-objective optimization algorithm for breast mass segmentation in mammograms. | Robust segmentation results with high accuracy and specificity. | Sensitivity could be higher; the model might be dataset-specific and less generalizable. |
| [26] | MIAS, DDSM, INbreast | Patchless Multi-Stage Transfer Learning | Introduced a patchless multi-stage transfer learning approach for improved classification of mammographic breast masses. | High accuracy and generalization across different datasets; eliminates the need for patch-based processing. | Patchless approach may not generalize well to all types of images, particularly those with irregular mass distributions. |
| [27] | MIAS, Custom Datasets | Fog Empowered Transfer Deep Learning | Proposed a fog computing empowered transfer learning model for enhanced breast cancer diagnosis. | High accuracy, sensitivity, and F1 score with efficient processing due to fog computing. | Requires specific fog computing infrastructure, which may not be universally available. |
| [28] | MIAS | Transfer Learning (AlexNet, Xception, ResNeXt, Channel Boosted CNN) | Proposed a transfer learning technique using various CNN architectures for breast cancer prognosis. | High accuracy, sensitivity, and specificity, with improved prognosis capability. | Focused primarily on classification; does not provide a detailed segmentation methodology. |
| [29] | MIAS | Atrous Pyramid Convolutional Deep Learning | Developed an Atrous Pyramid Convolutional deep learning approach for breast cancer detection and diagnosis. | Effective noise reduction and high detection accuracy, even in challenging cases. | Slightly lower sensitivity compared to other methods; may require fine-tuning for different datasets. |
| [30] | MIAS | Hybrid Model (EfficientNetB2, K-mean clustering, LSTM, CNN, Random Forest, Boosting) | Proposed a generic hybrid model for breast cancer classification using a combination of EfficientNetB2, K-mean clustering, LSTM, CNN, Random Forest, and Boosting. | High accuracy, sensitivity, and specificity; the hybrid model effectively leverages multiple techniques for improved classification. | Model complexity could pose challenges in implementation, requiring extensive computational resources and careful tuning. |
Materials and Methods
To improve the existing segmentation techniques, a novel hybrid method for segmentation is proposed using Otsu thresholding with morphological operations and Deep Convolutional Neural Network (DNN) architecture inspired by U-Net. U-Net with Attention Mechanisms and Residual Connections are used to improve segmentation accuracy and robustness. This method only focuses on the segmentation phase, as preprocessing is already done using CLAHE and a VGG-inspired model.
Model Architecture
The key components of the architecture are:
Otsu thresholding
This method calculates an optimal threshold value to separate the background and foreground regions of the image. The Otsu threshold, T, is determined by maximizing the between-class variance, as shown in the equation 1 below:
σ2B(T)=ω0(T)ω1(T)[μ0(T)−μ1(T)]
In our method, we modified the Otsu thresholding by incorporating local adaptive techniques to accommodate the varying illumination conditions across the mammogram image. This improvement enhances the overall segmentation performance.
 |
Algorithm 1: Otsu ThresholdingClick here to view Algorithm |
Morphological operations
Morphological operations are applied to the binary image obtained from the thresholding step. These operations, such as erosion, dilation, opening, and closing, can remove noise, fill gaps, and refine the segmented structures. We introduced a custom structuring element designed specifically for mammogram structures, which helps preserve the essential features and improve the segmentation quality.
U-Net
The U-Net architecture is a popular choice for segmentation tasks due to its encoder-decoder structure, which captures both high-level context and fine-grained details.
Attention Mechanisms
Attention gates are integrated to improve the focus on relevant features and reduce the influence of irrelevant regions. This helps in better delineation of the tumor boundaries.
Residual Connections
These connections help in mitigating the vanishing gradient problem, ensuring better gradient flow and making the network deeper and more robust.
The proposed model combines traditional and learning methods to take advantage of both methods.
The use of Otsu thresholding with morphological operations helps in making the model domain specific and to train model over smaller datasets. The Otsu thresholding with morphological operations is used for background and foreground separation, which helps in U-Net extracting accurate features from the images. The proposed U-Net follows the encoder decoder architecture based on convolutional layers as shown in figure 1, where encoder is used for the feature extraction and decoder is used to reconstruct the image to original resolution with minimum loss. The image is passed to the first two convolution layers of 3 x 3 kernel size and use 64 filters. A max pooling layer is present after every convolution layer that is nearly half the spatial dimensions of the image. This pattern follows up to Conv5 with the increasing filter from 128 to 1024. This helps the model in extracting the different features of the image. The high-level features from the image are captured using 16 x 16 x 1024. Further the decoder is used for the up sampling of the image, where created feature maps are enlarged to original input size of the image. After each up sampling a skip connection is used that helps in retaining the spatial information for better segmentation. These skip connections help in retaining the information that may be lost during encoding stage and results in poor segmentation. The model also uses an attention mechanism to better focus on the region of interest (ROI) and lesions. The residual connection helps in solving the issue of vanishing gradient, which helps training deeper networks. Also, In proposed model a total loss function is computed as combination of binary cross entropy and dice loss which helps in handling the issue of dataset classes imbalance.
Architecture Details
Encoder
The encoder part of the U-Net captures context through a series of convolutional layers followed by max-pooling. Each convolutional block in the encoder consists of:
Convolutional Layer (3×3 kernel, padding, ReLU activation)
Batch Normalization
Convolutional Layer (3×3 kernel, padding, ReLU activation)
Batch Normalization
Max Pooling (2×2)
Attention Mechanisms
Attention gates are placed after each convolutional block in the encoder to refine the feature maps before passing them to the decoder. The attention mechanism calculates attention coefficients, which are used to weigh the importance of features.
Residual Blocks
Residual connections are added to each convolutional block to ensure better gradient flow and to improve the learning capability of the network. Each residual block consists of:
Convolutional Layer (1×1 kernel, to match dimensions)
Addition of input features and the convolutional output
ReLU activation
Decoder
The decoder part of the U-Net upsamples the feature maps to the original input size, progressively combining them with the corresponding encoder feature maps through skip connections. Each upsampling block in the decoder consists of:
Transpose Convolutional Layer (2×2 kernel, stride 2)
Concatenation with the corresponding encoder feature map
Convolutional Layer (3×3 kernel, padding, ReLU activation)
Batch Normalization
Convolutional Layer (3×3 kernel, padding, ReLU activation)
Batch Normalization
Output Layer
The final layer consists of a 1×1 convolution to map the features to the desired number of classes (e.g., tumor and non-tumor), followed by a softmax activation function for pixel-wise classification.
| Algorithm: Novel Attention-Enhanced Residual U-Net for Mammogram Image Segmentation |
| Input: Otsu + Morphological Image |
| Output: Segmented image S with pixel-wise classification (tumor, non-tumor) |
| 1. Initialize the U-Net architecture parameters: |
| – Encoder layers: L_enc = {l_enc1, l_enc2, …, l_encN} |
| – Decoder layers: L_dec = {l_dec1, l_dec2, …, l_decN} |
| – Attention gates: A = {a1, a2, …, aN} |
| – Residual connections: R = {r1, r2, …, rN} |
| – Number of classes C = 2 (tumor, non-tumor) |
| – Learning rate α, Adam optimizer parameters, Dice loss weight λ_D, Cross-Entropy loss weight λ_CE |
| 2. Define convolutional block ConvBlock: |
| Input: Feature map F_in |
| Output: Feature map F_out |
| ConvBlock(F_in): |
| F1 = Conv2D(F_in, filters=f, kernel_size=(3,3), padding=’same’) |
| F1 = ReLU(BatchNorm(F1)) |
| F2 = Conv2D(F1, filters=f, kernel_size=(3,3), padding=’same’) |
| F_out = ReLU(BatchNorm(F2)) |
| return F_out |
| 3. Define attention gate AttnGate: |
| Input: Encoder feature map F_enc, Decoder feature map F_dec |
| Output: Refined feature map F_ref |
| AttnGate(F_enc, F_dec): |
| F_enc_att = Conv2D(F_enc, filters=f, kernel_size=(1,1), padding=’same’) |
| F_dec_att = Conv2D(F_dec, filters=f, kernel_size=(1,1), padding=’same’) |
| F_att = ReLU(F_enc_att + F_dec_att) |
| α_att = Sigmoid(Conv2D(F_att, filters=1, kernel_size=(1,1), padding=’same’)) |
| F_ref = α_att * F_dec |
| return F_ref |
| 4. Define residual block ResBlock: |
| Input: Feature map F_in |
| Output: Feature map F_out |
| ResBlock(F_in): |
| F_res = Conv2D(F_in, filters=f, kernel_size=(1,1), padding=’same’) |
| F_out = ConvBlock(F_in) + F_res |
| return F_out |
| 5. Encoder: |
| F_enc = I |
| for l in L_enc: |
| F_enc = ConvBlock(F_enc) |
| F_enc = MaxPool2D(F_enc, pool_size=(2,2)) |
| F_enc = ResBlock(F_enc) |
| end for |
| 6. Bottleneck: |
| F_bottle = ConvBlock(F_enc) |
| 7. Decoder: |
| F_dec = F_bottle |
| for l in reverse(L_dec): |
| F_dec = UpSampling2D(F_dec, size=(2,2)) |
| F_enc = corresponding F_enc from encoder |
| F_dec = Concatenate([F_dec, F_enc]) |
| F_dec = AttnGate(F_enc, F_dec) |
| F_dec = ConvBlock(F_dec) |
| end for |
| 8. Output layer: |
| S = Conv2D(F_dec, filters=C, kernel_size=(1,1), padding=’same’) |
| S = Softmax(S) |
| 9. Loss function: |
| Define Dice Loss L_D: |
| L_D = 1 – (2 * Σ(y_true * y_pred) + ε) / (Σ(y_true) + Σ(y_pred) + ε) |
| Define Cross-Entropy Loss L_CE: |
| L_CE = -Σ(y_true * log(y_pred)) |
| Total Loss L: |
| L = λ_D * L_D + λ_CE * L_CE |
| 10. Training: |
| Initialize Adam optimizer with learning rate α |
| for each epoch: |
| for each batch (I_batch, y_batch): |
| F_enc = Encoder(I_batch) |
| F_bottle = Bottleneck(F_enc) |
| F_dec = Decoder(F_bottle, F_enc) |
| S_pred = OutputLayer(F_dec) |
| L_batch = Loss(y_batch, S_pred) |
| Backpropagate and update weights using Adam optimizer |
| end for |
| end for |
| 11. Return segmented image S |
Results and Discussion
The performance of the proposed hybrid model is evaluated on the MIAS dataset. The various performance parameters are calculated to evaluate the model like F1, recall, precision, accuracy and area under curve (AUC). The proposed model had achieved a high F1 score of 0.97 that is balancing the recall and precision, which shows that the proposed model is performing well. As the recall and precision of the proposed model is around 0.98, it indicates that the model has less or almost no false alarms. This shows that the proposed model can segment the lesions very accurately. Also, with the accuracy and AUC of segmentation over 0.99, the proposed model very accurately detects positive and negative cases. Below in Table 4 the existing models that use MIAS dataset are compared with our proposed model on the basis of various evaluation metrics like F1, precision, recall, accuracy, AUC.
Table 4: Evaluation of Proposed Model
| Ref. No | Model Used | Accuracy | Sensitivity | F1 Score | Precision | AUC |
| [23] | U-Net based segmentation | 98.87% | 98.98% | 97.99% | 98.79% | 0.9888 |
| [24] | Transfer learning with pre-trained CNN architectures | 98.96% | 97.83% | 97.66% | 97.35% | 0.995 |
| [25] | Multilevel multi-objective electromagnetism like optimization technique | 98.93% | 92.11% | — | — | — |
| [26] | Patchless approach eliminating need for patch separation | 99.92% | 99.87% | 99.89% | — | 1.00 |
| [27] | Pre-trained deep learning models using Fog computing | 99.1% | 99.86% | 99.37% | 98.89% | — |
| [28] | Transfer learning-based segmentation approach | 98.96% | 98.5% | 98.5% | 98.78% | 0.997 |
| [29] | Quantum Wavelet Transform (QWT) filtering, Atrous pyramid CNN | 98.57% | 92% | — | 98.57% | 0.8877 |
| [30] | Efficient NetB2 +MGSVM | 99.47% | 99.31% | 99.44% | 99.44% | 0.9944 |
| Proposed hybridModel | Otsu thresholding + Attention ResUNet | 99.02% | 99.80% | 97.64% | 98.02% | 0.99997 |
As shown in Table 4, the proposed model outperforms in contrast with existing models. The proposed model initially uses the Otsu thresholding method which helps in accurately classification of background and foreground pixels, which helps in calculating the optimal threshold. Further morphological operations are applied that help in refining the binary image. Then the image is fed to U-Net architecture where encoder and decoder architecture helps in extracting fine details and features. The integration of attention mechanism helps the model only to focus on ROI instead of other irrelevant areas which ensure accurate marking of tumor boundaries. The model is robust as it also uses residual connection to solve gradient problem.
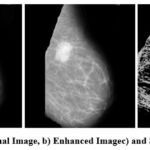 |
Figure 1: a) Orignal Image, b) Enhanced Image and c) Segmented ImageClick here to view Figure |
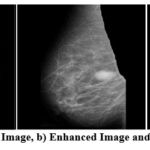 |
Figure 2: a) Orignal Image, b) Enhanced Image and c) Segmented ImageClick here to view Figure |
Figure 1 and 2 above shows the original image in pgm format as in MIAS dataset. There is a total of 322 such images in MIAS dataset. The next image is an enhanced image which is denoised using the CLAHE and VGG inspired network. CLAHE is used for contrast correction and VGG inspired is proposed learning model that is trained over noisy images to understand the complex pattern of noise and various artifacts in images. The noise is generated in images using noise synthesis, and the images are used to train models to understand noisy image patterns and to denoise the image. The model is evaluated based on PSNR value. Further the segmented image is shown. The segmented image clearly shows the lesions or ROI part removing all the irrelevant area. This helps the radiologist and a learning model to classify the segmented images correctly as benign or malignant. The proposed methodology is quite accurate and useful for future classification.
Conclusion
In this paper a hybrid model for segmentation using mammogram images is proposed based on Otsu thresholding with morphological operations and U-Net. The integration of traditional and learning methods lays a strong foundation to develop an efficient segmentation architecture. The goal of the segmentation model is to decrease the false detection rate for the accurate diagnosis. The adaptive filtering in Otsu thresholding helps in better classification of similar pixels and create accurate homogeneous pixel sets for foreground and background and morphological operations helps in prevention of the edge’s structures. The preservation of edges is very important for further U-Net to identify the tumor boundaries pattern. U-Net will learn the boundaries patterns and help in accurately segmenting the tumor. The incorporation of attention mechanism, residual and skip connection also helps in enhancing the model performance. The model had attained accuracy, AUC and precision around 99%, which makes the proposed model accurate. In future, the segmented data will be further used for the classification task.
Acknowledgement
I would like to express my sincere gratitude to my supervisors for their valuable guidance and support throughout my research. Special thanks to the Department of Computer Science and Engineering for providing the resources and facilities necessary to complete this study. I am also grateful to my family and friends for their constant encouragement and support.
Funding Sources
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
The author(s) do not have any conflict of interest.
Data Availability Statement
This statement does not apply to this article.
Ethics Statement
This research did not involve human participants, animal subjects, or any material that requires ethical approval.
Informed Consent Statement
This study did not involve human participants, and therefore, informed consent was not required.
Clinical Trial Registration
This research does not involve any clinical trials
Author Contributions
- Vandana Saini: Design of segmentation model, Implementation and manuscript writing, Manuscript communication.
- Meenu Khurana: Suggestion regarding design of segmentation model, Visualization, Supervision, manuscript writing – review & editing, approval of manuscript.
- Rama Krishna Challa: Suggestion regarding design of segmentation model, visualization, approval of manuscript and Supervision.
References
- Ait lbachir I, Es-salhi R, Daoudi I, Tallal S, Medromi H. A survey on segmentation techniques of mammogram images. In: Azouzi RE, Menasche DS, Sabir E, Pellegrini FD, Benjillali M, eds. UNet. Singapore: Springer;2016: 545-556.
CrossRef - Priya DS, Sarojini DB. Breast cancer detection in mammogram images using region-growing and contour-based segmentation techniques. J Comput Sci Appl. 2013;1(1):23-29.
- Saleck MM, Ould Mohamed Dyla MH, Benany MM, Mohamed Rmili M. Otsu’s thresholding for semi-automatic segmentation of breast lesions in digital mammograms. Int J Adv Comput Sci Appl. 2022;13(10):45-52.
CrossRef - Mohamed Saleck M, Ould Taleb N, El Moustapha El Arby Chrif M, Benany Mohamed Mahmoud E. A comparison study between Otsu’s thresholding, fuzzy C-means, and K-means for breast tumor segmentation in mammograms. In: Shakya S, Tavares JMRS, Fernández-Caballero A, Papakostas G, eds. Fourth International Conference on Image Processing and Capsule Networks. Singapore: Springer; 2023:725-734.
CrossRef - Yu X, Wang SH, Zhang YD. Multiple-level thresholding for breast mass detection. J King Saud Univ Comput Inf Sci. 2023;35(1):115-130.
CrossRef - Khairnar S, Thepade SD, Gite S. Effect of image binarization thresholds on breast cancer identification in mammography images using OTSU, Niblack, Burnsen, Thepade’s SBTC. Intell Syst Appl. 2021;10-11(1).
CrossRef - Hossain MS. Microcalcification segmentation using modified U-net segmentation network from mammogram images. J King Saud Univ Comput Inf Sci. 2022;34(2):86-94.
CrossRef - Shaaban SM, Gaber Z, Semary S, Dewidar AM. Impact of Vitamin B12 on outcome of Early Stage Luminal A and B Breast Cancer, single center experience. Medical and Pharmaceutical Journal. 2023 ;2(1):17-27. DOI:10.55940/medphar202227
CrossRef - Xu S, Adeli E, Cheng J-Z, Xiang L, Li Y, Lee S-W, Shen D. Mammographic mass segmentation using multichannel and multiscale fully convolutional networks. Int J Imaging Syst Technol. 2020;30(4):1095-1107.
CrossRef - Tammineedi VSV, Raju C, Girish Kumar D, Yalla V. Improvement of segmentation efficiency in mammogram images using dual-ROI method. Int J Health Inf Syst 2022;17(2):1-14.
CrossRef - Zhu, R., Xin, B., Deng, N., & Fan, M. (2022). Semantic segmentation using DeepLabv3+ model for fabric defect detection. Wuhan University Journal of Natural Sciences, 27(6), 539–549.
CrossRef - Sharma R, Kamra A. Enhancing diagnosis of breast cancer through mammographic image segmentation using fuzzy C-means. Int J Sustain Build Technol Urban Dev. 2023;14(4):488-499.
- Michael E, Ma H, Li H, Kulwa F, Li J. Breast cancer segmentation methods: current status and future potentials. Biomed Res Int. 2021: 1-29
CrossRef - Baccouche A, Garcia-Zapirain B, Castillo Olea C, Elmaghraby AS. Connected-UNets: a deep learning architecture for breast mass segmentation. npj Breast Cancer. 2021;7(1):151: 1-12.
CrossRef - Yang KB, Lee J, Yang J. Multi-class semantic segmentation of breast tissues from MRI images using U-Net based on Haar wavelet pooling. Sci Rep. 2023;13(1): 1-12
CrossRef - Carriero A, Groenhoff L, Vologina E, Basile P, Albera M. Deep learning in breast cancer imaging: state of the art and recent advancements. 2024;14(848):1-36
CrossRef - Pramanik P, Pramanik R, Schwenker F, Sarkar R. DBU-Net: Dual branch U-Net for tumor segmentation in breast ultrasound images. PLoS ONE. 2023;18(11): 1-20.
CrossRef - Yu L, Shaheema SB, Sunil J, Govindan V, Mahimiraj P, Li Y, Jamshed W, Hassan AM. Breast cancer segmentation using a hybrid AttendSeg architecture combined with a gravitational clustering optimization algorithm using mathematical modelling. Open Phys. 2023;21(1): 1-13.
CrossRef - Pawar RV, Saraf S, Dixit U, Jadhav AS. Diagnosis of mammographic images for breast cancer detection using FF-CSO algorithm. Paper presented at: Advanced Computing and Communication Technologies for High Performance Applications (ACCTHPA) Meetings; 20-21 January,2023; Ernakulam, India: 1-5.
CrossRef - Kamil MY, Radhi EA. Breast tumor segmentation in mammography image via Chan-Vese technique. Indones J Electr Eng Comput Sci. 2021;22(2):809-817.
CrossRef - Reddy BRG, Kumar HP. Segmentation of mammogram images using level set with cuckoo search optimization. Comput Methods Biomech Biomed Eng Imaging Vis. 2022;11(3):914-921.
CrossRef - Nalini N, Jagadeesh P, Bharathi PS, Amudha V, Ramkumar G, Nagalakshmi TJ. Edges and boundary detection of mammography images in earlier stages through non-convex border optimization of segmentation thresholding algorithm. Paper presented at: International Conference on Advances in Computing, Communication and Applied Informatics (ACCAI) Meetings; 28-29 January,2022; Chennai, India: 1-7.
CrossRef - Salama WM, Aly MH. Deep learning in mammography images segmentation and classification: automated CNN approach. Alexandria Eng J. 2021;60(5):4701-4709.
CrossRef - Saber A, Sakr M, Abo-Seida O, Keshk A, Chen H. A novel deep-learning model for automatic detection and classification of breast cancer using the transfer-learning technique. IEEE Access. 2021;9:71194-71209.
CrossRef - Ittannavar SS, Havaldar RH. Segmentation of breast masses in mammogram image using multilevel multiobjective electromagnetism-like optimization algorithm. Biomed Res Int. 2022: 1-15.
CrossRef - Ayana G, Park J, Choe S. Patchless multi-stage transfer learning for improved mammographic breast mass classification. 2022;14(5) 1280: 1-23.
CrossRef - Pati A, Parhi M, Pattanayak BK, Sahu B, Khasim S. CanDiag: Fog empowered transfer deep learning-based approach for cancer diagnosis. 2023;7(3) 57: 1-18.
CrossRef - Diwakaran M, Surendran D. Breast cancer prognosis based on transfer learning techniques in deep neural networks. Inf Technol Control. 2023;52(2):381-396.
CrossRef - Pour ES, Esmaeili M, Romoozi M. Employing atrous pyramid convolutional deep learning approach for detection to diagnose breast cancer tumors. Comput Intell Neurosci. 2023:7201479: 1-17
CrossRef - Abioye OA, Thomas S, Odimba CR, Olalekan AJ. Generic hybrid model for breast cancer mammography image classification using EfficientNetB2. Dutse J Pure Appl Sci. 2023;9(3b):456-462.
CrossRef







