Anak Agung Gede Indraningrat1* , Pande Putu Christine Putri Purnami1
, Pande Putu Christine Putri Purnami1 , Ema Damayanti2
, Ema Damayanti2 , Made Dharmesti Wijaya3
, Made Dharmesti Wijaya3 , Dewa Ayu Putri Sri Masyeni4
, Dewa Ayu Putri Sri Masyeni4 , Ni Luh Putu Eka Kartika Sari5
, Ni Luh Putu Eka Kartika Sari5
1Department of Microbiology and Parasitology, Faculty of Medicine and Health Sciences, Warmadewa University, Denpasar-Bali, Indonesia.
2Research Center for Food Technology and Processing, National Research and Innovation Agency, Gunungkidul, Indonesia.
3Department of Pharmacology, Faculty of Medicine and Health Sciences, Warmadewa University, Denpasar-Bali, Indonesia,
4Department of Internal Medicine, Faculty of Medicine and Health Sciences, Warmadewa University, Denpasar-Bali, Indonesia.
5Department of Biochemistry and Physiology, Faculty of Medicine and Health Sciences, Warmadewa University, Denpasar-Bali, Indonesia.
Corresponding Author E-mail:indraningrat@warmadewa.ac.id
DOI : https://dx.doi.org/10.13005/bpj/3029
Abstract
Eucheuma cottonii is a type of seaweed that are commonly found in Indonesia. As many other marine organisms, E. cottonii builds a strong bonding with its endophytic bacteria. These bacteria are well known to synthesize various of bioactive compounds including antibacterial compounds to protect its host from bacterial infections and pathogenic bacteria. Previous study has successfully isolated bacterial encoded ISP1RL4 with antibacterial potential against nonresistant Gram-positive and Gram-negative bacterial target. This research aimed to identify the ISP1RL4 isolate based on DNA sequencing, to evaluate antibacterial activity of the crude extract of ISP1RL4 isolate against multidrug-resistant bacterial target and to analysis chemical profiling of the extracts. Briefly, the cell mass of ISP1RL4 bacterial isolate was fermented for 2 weeks in 100 mL sterile liquid ISP-2 medium and then filtered. Extraction was carried out using ethyl acetate with an extraction ratio of 1:1 twice. Ethyl acetate extracts of ISP1RL4 were tested against multidrug-resistant bacteria Methicilin-resistant Staphylococcus aureus (MRSA), Escherichia coli ESBL, Klebsiella pneumoniae ESBL, and Acinetobacter baumanii. Our findings revealed that the ISP1RL4 phylogenetically related to Pseudomonas aeruginosa strain M4 with 100% of sequence similarity. The crude extract of P. aeruginosa ISP1RL4 showed diameter zone of inhibition of 9.0±1.0 mm, 10.3±2.0 mm and 9.4±0.1 mm against MRSA, E. coli ESBL, and K. pneumoniae ESBL respectively. No antibacterial activity of the crude extract was observed against A. baumanii. The liquid chromatography-high resolution mass spectrometry (LC-HRMS) analysis detected 381 compounds with 2-Amino-1,3,4-octadecanetriol (11.2%) identified as the major antibacterial compound present in ethyl acetate extracts of P. aeruginosa ISP1RL4. In addition, gas chromatography/mass spectrometry (GC/MS) analysis identified 39 compounds and 11 of them have been associated as antibacterial molecules. Among these 11 molecules, four prominent antibacterial compounds (> 8%) were 2-hexanol, 3-hexanol, 3-Pentanol, 2-methyl- and 2-hexanone. Overall, the ability of P. aeruginosa ISP1RL4 crude extract to inhibit selected multidrug-resistant bacterial target and the presence antibacterial compounds in the extract provided a promising result that the isolate could potentially be a promising antibacterial producer.
Keywords
Anti-bacterial; Bioprospecting; Marine; Multidrug resistant bacteria; Secondary metabolites; Pseudomonas aeruginosa
Download this article as:| Copy the following to cite this article: Indraningrat A. A. G, Purnami P. P. C. P, Damayanti E, Wijaya M. D, Masyeni D. A. P. S, Sari N. L. P. E. K. Antibacterial Potential of Pseudomonas aeruginosa ISP1RL4 Isolated from Seaweed Eucheuma cottonii Against Multidrug-resistant Bacteria. Biomed Pharmacol J 2024;17(4). |
| Copy the following to cite this URL: Indraningrat A. A. G, Purnami P. P. C. P, Damayanti E, Wijaya M. D, Masyeni D. A. P. S, Sari N. L. P. E. K. Antibacterial Potential of Pseudomonas aeruginosa ISP1RL4 Isolated from Seaweed Eucheuma cottonii Against Multidrug-resistant Bacteria. Biomed Pharmacol J 2024;17(4). Available from: https://bit.ly/3ZoQABO |
Introduction
Bacteria that are resistant to multiple drugs (MDR) have become a significant public health issue due to their increasing prevalence1. The excessive and improper use of antibiotics has driven the development of resistance in various bacteria, leading to the accumulation of multiple resistance gene2s. A number of Gram-positive and negative bacterial species have transformed into resistance strains against different types of antibiotics such as β-lactamase classes and vancomycin3. Furthermore, the emergence of broad-spectrum beta-lactamase (ESBL) enzymes in Enterobacteriaceae has worsened the global antibiotic resistance crisis4. Although raising public awareness to use antibiotic rationally remains essential, the urgent need for new and potent antibiotic producers to combat MDR bacteria cannot be overstated.
For decades, terrestrial microorganisms particularly bacteria and fungi have been the primary target discovery for novel antibacterial compounds5. Nevertheless, many of these antibiotic compounds have been previously isolated and reported, leading to a reduction in the novelty rate of these compounds6. Conversely, marine habitats offer variety of bioactive molecules, including antibiotics that hold significant pharmaceutical importance7. Various marine species have been proven to produce arrays of bioactive molecules, with pharmaceutical potential including that of antibiotic compounds8. However, to perform clinical test and to synthesize specific bioactive molecules require a large number of biomasses to acquire sufficient extracts9. Furthermore, ecological concerns prevent the direct cultivation of such substantial biomass in nature. As a result, the focus has shifted towards bioprospecting marine bacteria, particularly those associated with marine organisms, as they offer faster and relatively easier cultivability under laboratory conditions, making them suitable for synthesizing compounds of interest10,11.
Marine microorganisms, particularly bacteria, have emerged as significant sources of novel antibacterial compounds. This is largely due to their unique evolutionary adaptations to the marine environment, which have equipped them with diverse biochemical pathways for producing bioactive metabolites12–14. Among these microorganisms, members of the genus Bacillus are particularly noteworthy. They are known to synthesize a variety of antimicrobial substances, including polyketides, lipopeptides, and bacteriocins, which exhibit broad-spectrum antimicrobial activity against various pathogens15–17. Other marine bacterial species such as actinobacteria are among the prominent producers of antibacterial compounds, showcasing their potential in drug discovery and development18. Overall, marine bacteria represent a rich reservoir of antibacterial compounds with diverse mechanisms of action.
Eucheuma cottonii, a seaweed species commonly found in Indonesia, is rich in nutrients and valuable compounds like carrageenan, flavonoids, and tannins19,20. Like other seaweeds, it forms a beneficial partnership with bacteria, which help it grows, develops, and defends against threats by producing antibacterial substances21. However, research on E. cottonii-associated bacteria is rather scarce22,23. In a previous study, Aeromonas bacteria isolated from E. cottonii, actively inhibited Staphylococcus aureus and Escherichia coli22. Additionally, twenty-three bacterial isolates were discovered on E. cottonii in a recent study conducted in the coastal waters of Buleleng, Bali, with six of these isolates exhibiting antibacterial activity against S. aureus, Streptococcus mutans, E. coli, and Klebsiella pneumoniae23.
Among six reported potential isolates, a bacterial isolate encoded as ISP1RL4 specifically displayed the strongest antibacterial activity23. However, it is remained unclear to what species this isolate is assigned. Therefore, it requires further molecular identification and characterization. In addition, it is intriguing to evaluate antibacterial activity of the isolate crude extract against multidrug-resistant bacteria to confirm its previous reported antibacterial potential based on agar block method23. Furthermore, chemical profiling on the bacterial extract was performed to provide a clear insight on the possible promising antibacterial compounds.
Materials and Methods
Materials
The ISP1RL4 bacterium was isolated from the seaweed E. cottonii from Patas village, Buleleng Regency, Bali, Indonesia23. The pure culture was grown in slant agar and stored at 4oC until further used. Gram staining and Ziehls-Neelsen staining kits were purchased from local suppliers. The test pathogenic bacteria used were Methicilin-resistant Staphylococcus aureus (MRSA), Escherichia coli Extended-spectrum beta-lactamases (ESBL), Klebsiella pneumoniae ESBLand ESBL Acinetobacter baumanii were used as previously described24. All other analytical grade chemicals such as aqua distillation, ethyl acetate was purchased from local suppliers.
DNA isolation
Cell mass of ISP1RL4 pure culture was grown in 1.5 mL sterile International Streptomyces Project-2 (ISP-2) broth medium and was incubated at 28oC for 7 days. Bacterial DNA was extracted using a bacteria DNA preparation kit following the protocol (Jena Bioscience, Germany). DNA concentration was determined by a Nanodrop with a 260/280 nm ratio25.
16SrDNA gene amplification and phylogenetic analysis
Molecular identification was performed by PCR amplifying 16S rRNA gene using two primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and reverse primer 1492R (5′-CGGTTACCTTGTTACGACTT-3′) based on the previously described protocol24. PCR product was sent for Sanger sequencing to PT Genetika Science, Tangerang, Indonesia (https://ptgenetika.com/). The quality of raw sequence was checked using sequence scanner V.2.0 and low-quality sequences were trimmed using ChromasPro 2.1.10. Finally, sequences were assembled using software DNAMAN Ver.9.0. The nucleotide sequence was compared against a database of known sequences using the n-BLAST method on the NCBI BLAST platform (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Following the BLAST search, the ten most relevant sequences were chosen and their base information was used to construct a phylogenetic tree. The Neighbor-joining method with the Kimura-2 parameter model was implemented in MEGA X software (https://www.megasoftware.net)24. The reliability of the tree was assessed using bootstrap analysis with 1000 repetitions.
Scanning Electron Microscope (SEM) preparation
The SEM observation method was adapted from a previous study26.Cell mass of ISP1RL4 isolate were obtained from overnight culture on ISP-2 medium. Cell biomass was washed by Phosphate Buffer Saline (Merck, Germany) 1x. The cell biomass was soaked with 2% glutaraldehyde in PBS for 2 h at room temperature. The cell biomass was separated from solution was centrifugation method (13,000 rpm for 5 min). The cell biomass was rinsed with PBS twice. The sodium tetraoxide (1% in PBS) was added in cell biomass and was soaked for 2 h in room temperature. Sodium tetraoxide was removed by centrifugation method. Before coating process, the cell biomass was dehydrated with serial ethanolic solution (70% for 10 min, 96% for 10 min, absolute ethanol for 10 min) and followed by centrifugation method to obtain ethanolic free cell biomass. The cell biomass was placed on carbon tape and subsequently applying a gold coating through a sputtering process. The scanning electron microscope was set to a high vacuum, 5 kV accelerating voltage, 30% spot intensity, and magnifications of 2,000x, 5,000x, and 10,000x (Hitachi SU3500, Japan)26.
Fermentation and Extraction Condition
Ten mL of ISP1RL4 pre-cultured in ISP-2 media was added aseptically into 90 mL sterile ISP-2 broth and the mixture was fermented in 14 days at 150 rpm27. After reaching the fermentation period, the supernatant was separated from cell mass using Whatman paper no 1. Subsequently, it was extracted using 100 mL of ethyl acetate pro Analisa (Merck, Germany). A separatory funnel was used to separate the organic layer from liquid layer. The extraction and separation process were repeated two times. Finally, the pool of organic layers was evaporated in a vacuum evaporator to achieve the final extract for further antimicrobial screening28.
Antibacterial Assay of Against Selected Multidrug-Resistant Bacteria
Screening of the antibacterial activity of ISP1RL4 isolate crude extract was performed following disc diffusion assays. Thirty microliters of ISP1RL4 crude extract were applied to sterile 6-millimeter diameter papers disc. These discs were then placed in triplicates on LB agar plates containing various multidrug-resistant bacteria, including MRSA, extended-spectrum beta-lactamase (ESBL) Escherichia coli and ESBL Klebsiella pneumoniae, and ESBL Acinetobacter baumannii. These plates were stored at a 37oC incubator for one day. The diameter of inhibition zone formed against each bacterial target was quantify using a digital caliper.
Thin Layer Chromatography and Antibacterial Assay
The extract was separated and purified using thin layer chromatography (TLC). Briefly, TLC of the substance was applied on the GF-254 silica gel and developed in a solvent system containing n-hexane and ethyl acetate in a ratio of 4:6. Distinct components were identified by viewing them under ultraviolet light (254 nm), subsequently their retention factors (Rf values) were determined. Each visible spot on the TLC plate was then collected, dissolved in the same solvent mixture, and tested for antibacterial activity against the MDR bacterial test using the Kirby-Bauer method as previously described.
Gas Chromatography/Mass Spectrometry preparation and analysis
Volatile compounds that constitute ethyl acetate crude extract of ISP1RL4. Briefly, 0.1 gram of the crude extract was prepared and sent to the Forensic Laboratory Polda Bali, Indonesia for further analysis. The extract was injected to the GC/MS instrument (Agilent Technologies 7890B/Agilent Technologies 5977B) based on the following setting: HP-5ms ultra inert column 30 m x250 µm x 0.25 µm, oven temperature (-60oC to 325oC), mode (splitless), pressure (25.523 psi), total flow (20.9 ml/min), average velocity (62.662 cm/sec), purge flow to split vent (15 mL/min at 0.75 min), and gas (He). Chromatograms were analyzed by matching the compound fragments from each chromatogram peak with literature to determine the type of content and to understand the biological properties of the discovered compounds.
Liquid Chromatography High Resolution Mass Spectrometry (LC-HRMS) preparation and analysis
Metabolomic analysis of ISP1RL4 ethyl acetate was performed in Ultra-High-Performance Liquid Chromatography coupled to untargeted High-Resolution Mass Spectrometry (Thermo Scientific Dionex Ultimate 3000 RSLC Nano UHPLC paired with Thermo Scientific Q Extractive (Thermo Fisher Scientific, Massachusetts, USA). The machine was run by following previously described procedure26.
Results and Discussion
Morphological Observation
The cell morphology of the ISP1RL4 bacterial isolate, as observed under a microscope after Gram staining, categorizes the isolate as a Gram-negative bacterium with bacilli-shaped cells. (Figure 1A). This observation was confirmed by examining bacterial cells of the ISP1RL4 isolate under a scanning electron microscope, which provided a clearer appearance with smooth surface, approximately 5 µm in length, and exhibiting attachment to each other (Figure 1B). On agar plate, the ISP1RL4 pure isolate had colonies with irregular surface shapes, firmly attached to the media, with a powdery consistency, and rough textured and dull surfaces, and had a grayish-yellow pigmentation with the reverse of colony color with light yellow green pigmentation on ISP-2 agar at 11 days of age (Figure 1C). Based on the results of Ziehl-Neelsen’s acid-resistant staining, ISP1RL4 isolate was a non-acid fast bacterium because it could not retain the red dye from carbolic fuchsin after being dripped with alcoholic acid.
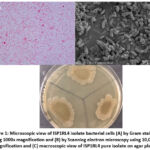 |
Figure 1: Microscopic view of ISP1RL4 isolate bacterial cells (A) by Gram staining using 1000x magnification and (B) by Scanning electron microscopy using 10,000x magnification and (C) macroscopic view of ISP1RL4 pure isolate on agar plate. |
Molecular Identification
DNA isolation of ISP1RL4 exhibited concentration of 296 ng/µL with a DNA purity level of 1.78 nm at a ratio of A260/280 nm. Based on the result of the alignment of ISP1RL4 isolate, it was revealed that ISP1RL4 isolate had DNA sequences with 100% homology (percentage identity) with Pseudomonas aeruginosa strain M4. The PCR results of the 16S rRNA product of ISP1RL4 has a DNA sequence of less than 1500 bp, which is 1411 bp. Percentage identity or what is known as the homology value is a percentage that indicates how well the input DNA sequence matches the target DNA sequence. The results of the phylogenetic tree construction as shown in Figure 2 showed that the ISP1RL4 isolate was located in the same clade and branch, and shared the same node with Pseudomonas aeruginosa strain M4 on the phylogenetic tree. According to Figure 3, the phylogenetic tree of ISP1RL4 isolate formed was not a paraphyletic group of phylogenetic trees, but a monophyletic. Based on molecular identification, ISP1RL4 isolate can be identified as Pseudomonas aeruginosa sp. and phylogenetically related to Pseudomonas aeruginosa strain M4.
 |
Figure 2: ISP1RL4 isolate phylogenetic tree which describes the phylogenetic position of ISPIRL4 isolate with other bacteria in one clade or another clade (Staphylococcus hominis strain DM 122 as the outer group). |
Evaluation of Antibacterial Activity of ISP1RL4 Crude Extract Against Multidrug-resistant Bacteria
The results showed that ISP1RL4 isolate could inhibit the growth of three multidrug-resistant bacteria, except for A. baumanii (Table 1). ISP1RL4 extract had moderate category of antibacterial activity with an average inhibition zone of >9 mm. The highest antibacterial activity of ISP1RL4 extract was shown in multidrug resistant E. coli ESBL bacteria with an inhibition zone diameter of 10.3 ± 3.0 mm. ISP1RL4 extract had the lowest ability to inhibit the activity of MRSA bacteria with diameter zone of inhibition of 9.0 ± 1.0 mm.
Ethyl acetate extract of ISP1RL4 isolate was able to inhibit the growth of the Gram-positive and Gram-negative multidrug resistant bacteria as shown in Figure 3. Ethyl acetate crude extract of ISP1RL4 was more effective to inhibit the growth of Gram-negative multidrug resistant bacteria especially E. coli ESBL as shown in Table 1 compared to MRSA.
Table 1: Antibacterial activity of the crude extract of ISP1RL4 isolate against multi-drug resistant bacteria.
|
Multidrug resistant bacteria |
Zone of Inhibition (mm) |
|
MRSA |
9,0 ± 1,0 |
|
E. coli ESBL |
10,3 ± 2,0 |
|
K. pneumoniae ESBL |
9,4 ± 0,1 |
|
A. baumanii |
0± 0,1 |
Notes: Average diameter of the inhibition zone for each isolate was measured from three replications.
 |
Figure 3: Antibacterial activity of ISP1RL4 crude extract against multidrug-resistant bacteria (A) Methicillin-resistant Staphylococcus aureus (MRSA), (B) Klebsiella pneumoniae ESBL |
Fraction Analysis of Active Compound Extract Filtrate in ISP1RL4 Isolate
The results of TLC analysis (thin layer chromatography) revealed that there was only one spot (Figure 4) that appeared on the GF254 TLC plate (Table 2). Fraction F1 had an Rf value of 0,06. The fraction was tested for antibacterial activity on the test bacteria. The highest inhibition of the fraction was observed against MRSA (Figure 5).
Table 2: Antibacterial activity of the active compound fraction of ISP1RL4 isolate on the test bacteria
|
Fraction |
Rf value |
||||
|
MRSA |
E. coli ESBL |
K. pneumoniae ESBL |
A. baumanii ESBL |
||
|
1 |
0,06 |
18±4 |
0 |
7,4±0,2 |
0 |
|
K+ (Nalidixic acid 30 µg) |
9±0,3 |
9,8±0,2 |
9,2±0,2 |
9,5±0,3 |
|
|
K- (Ethyl acetate) |
0 |
0 |
0 |
0 |
|
Notes: K+: Positive control; K-: Negative control. The average diameter of the inhibition zone for each isolate was measured from three replications
 |
Figure 4: Spot visualization of ISP1RL4 isolate extract fraction on GF254 silica gel plate |
Notes: Yellow circle: spot on the fraction that had successfully appeared.
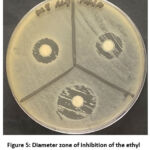 |
Figure 5: Diameter zone of inhibition of the ethyl acetate fraction against MRSA |
LC-HRMS Analysis of Active Compound in ISP1RL4 Isolate
The results of LC-HRMS analysis of the crude ethyl acetate extract isolate ISP1RL4 detected 381 compounds with different peak percentages and retention times which were successfully identified in the LC-HRMS chromatogram shown in Figure 6. Eleven antibacterial compounds detected in the ethyl acetate crude extract of ISP1RL4 isolate based on LC-HRMS could be seen in Table 3. The highest detected compound was 2-Amino-1,3,4-octadecanetriol (11,21 % relative abundance). The GC-MS analysis of the crude ethyl acetate isolate ISP1RL4 detected 11 antibacterial compounds as presented in Table 4.
 |
Figure 6: LC-HRMS chromatogram of the ethyl acetate extract of ISP1LR4 isolate |
Table 3: Antibacterial compounds detected in the ethyl acetate crude extract of ISP1RL4 isolate based on LC-HRMS results.
|
Name |
Formula |
Cal. Molecular Weight |
Compound characteristic |
Activity |
% relative abundance |
Reference |
|
2-Amino-1,3,4-octadecanetriol |
C18H39NO3 |
317.29191 |
Organic compound |
Antibacterial |
11.21 |
29 |
|
Bis(4-ethylbenzylidene) sorbitol |
C24H30O6 |
414.20368 |
Organic compound |
Antibacterial |
4.81 |
30 |
|
Dextromethorphan |
C18H25NO |
271.19315 |
Organic compound |
Antibacterial |
4.53 |
31 |
|
Dibutyl phthalate |
C16H22O4 |
278.15142 |
Organic compound |
Antibacterial |
4.41 |
32 |
|
C14-Dihydroceramide |
C32H65NO3 |
511.49607 |
Organic compound |
Antibacterial |
4.17 |
33 |
|
C16-Dihydroceramide |
C34H69NO3 |
539.52751 |
Organic compound |
Antibacterial |
2.76 |
33 |
|
6-Methoxyquinoline |
C10H9NO |
159.06818 |
Organic compound |
Antibacterial |
2.10 |
34 |
|
D-(+)-Maltose |
C12H22O11 |
342.11637 |
Organic compound |
Antibacterial |
1.79 |
35 |
|
Propranolol |
C16H21NO2 |
259.15684 |
Organic compound |
Antibacterial |
1.63 |
36 |
|
1-Stearoylglycerol |
C21H42O4 |
358.30770 |
Organic compound |
Antibacterial |
1.52 |
37 |
|
Armillaramide |
C34H69NO4 |
555.52202 |
Organic compound |
Antibacterial |
1.12 |
33 |
Table 4: Antibacterial compounds detected in the ethyl acetate crude extract of ISP1RL4 isolate based on GC-MS results
|
Name |
Formula |
Molecular Weight (g/mol) |
Compound characteristic |
Activity |
% relative abundance |
Reference |
|
2-Hexanol |
C6H14O |
102.17 |
Organic compound |
Antibacterial |
10.94 |
38 |
|
3-Hexanol |
C6H14O |
102.17 |
Organic compound |
Antibacterial |
9.86 |
39 |
|
3-Pentanol, 2-methyl- |
C6H14O |
102.17 |
Organic compound |
Antibacterial |
9.86 |
40 |
|
2-Hexanone |
C6H12O |
100.16 |
Organic compound |
Antibacterial |
8.60 |
41 |
|
Isobutyl acetate |
C6H12O2 |
116.16 |
Organic compound |
Antibacterial |
7.62 |
42 |
|
Cyclotrisiloxane, hexamethyl- |
C12H22O11 |
222.46 |
Organic compound |
Antibacterial |
7.12 |
43 |
|
3-Hexanone |
C18H30O3S |
100.16 |
Organic compound |
Antibacterial |
4.56 |
44 |
|
Toluene |
C6H5CH3 |
92.14 |
Organic compound |
Antibacterial |
3.81 |
45 |
|
Ethylbenzene |
C8H10 |
106.16 |
Organic compound |
Antibacterial |
2.40 |
46,47 |
|
o-xylene |
C8H10 |
106.16 |
Organic compound |
Antibacterial |
2.40 |
48–50 |
|
Actinobolin |
C13H20N2O6 |
135.21 |
Organic compound |
Antibacterial |
1.10 |
51 |
Discussion
The study investigated the antibacterial properties of a bacterial strain, ISP1RL4, isolated from E. cottonii seaweed. Microscopic analysis revealed that the isolate was rod-shaped and Gram-negative. Molecular identification confirmed that the isolate ISP1RL4 identified as Pseudomonas aeruginosa. The isolation of the bacterium from seaweed aligns with the known distribution of Pseudomonas species, which are frequently found in marine ecosystems52,53. Marine P. aeruginosa has been a subject of interest due to its diverse characteristics and potential applications. Studies have highlighted the adaptability of P. aeruginosa in various environments, including marine habitats, where it demonstrates unique patterns of cultivability and survival, indicating physiological adaptations to oceanic conditions54.
Our study has provided further evidence to support the antibacterial potential of P. aeruginosa ISP1RL4, as initially suggested by block agar experiments23. The ethyl acetate extract derived from P. aeruginosa ISP1RL4 effectively suppressed the growth of both Gram-positive and Gram-negative MDR bacteria. Additionally, P. aeruginosa has been identified as a source of antimicrobial activity against multidrug-resistant pathogens, with certain marine isolates demonstrating efficacy against bacteria like S. aureus55. The production of antimicrobial metabolites by marine Pseudomonas strains further underscores their potential as sources of novel antibacterial agents. The antibacterial properties exhibited by the ethyl acetate extract suggest that its components have a wide range of antibacterial activity56. This result aligns with prior research highlighting the broad-spectrum antibacterial properties of P. aeruginosa against various bacterial species. In terms of the degree of inhibition, the extract exhibited moderate to strong antibacterial activity, with inhibition zones ranging from 9.8 to 11 millimeters57. Notably, the purified extract effectively suppressing MRSA bacterial target compared to that of other test bacteria. This finding highlights the selective antibacterial activity of the purified extract, demonstrating a particular efficacy against MRSA compared to other bacterial strain. In addition, discrepancy diameter zone of inhibition could be due to the absence of an outer membrane with lipopolysaccharide in Gram-positive bacteria, making them more susceptible to antibacterial compounds compared to Gram-negative bacteria58,59. The produced diameter zone of inhibition may vary which can be attributed to the different types of secondary metabolites produced, different chemical composition, concentration, and polarity. Additionally, the morphological and physiological characteristics of each bacterial strain influence these results60.
Variations in the type of active compound content were found in the LC-HRMS and GC-MS results. Active compounds were found to be more diverse and numerous in LC-MS results than GC-MS. The ability of LC-HRMS to generate exact mass measurements and molecular formulas for unknown compounds in an extract could contribute to this observation. Additionally, this method excels at determining chemical structures with high sensitivity, even when working with small sample sizes and limited time61,62. Comparing to GC-MS, a commonly employed technique for characterizing chemotypes from a sample, the instrument was limited to analyzing non-polar and/or volatile compounds63. This had covered a broader spectrum of compounds found in LC-HRMS which had been shown by the identification of non-volatile compounds such as sorbitol and ceramides.
The active compound of P. aeruginosa ISP1RL4 ethyl acetate extract found in LC-HRMS was mainly dominated by 2-Amino-1,3,4-octadecanetriol (11.2%), which belong to the phytosphingosine compound class. Phytosphingosine is a long-chain sphingolipid base consisting an amino alcohol with 18 carbon atoms typical in plants, which has antibacterial properties. Phytosphingosine, at a concentration of 15.9 μg/mL, effectively killed 95% of the three bacterial species: P. syringae pv. tomato, A. tumefaciens, and R. radiobacter29. C14-Dihydroceramide, C16-Dihydroceramide, and Armillaramide are ceramides reported for the first time that had been isolated from Eucheuma cottonii-associated bacterial. A study discovered that short-chain ceramides and a ω-azido-C6-ceramide exhibited antibacterial activity towards Neisseria meningitidis and N. gonorrhoeae33.Ceramides found in Cissus incisa leaves had an antibacterial activity against nine multidrug-resistant bacteria, with the most significant inhibition observed against Gram-negative bacteria, particularly carbapenem-resistant Acinetobacter baumannii at a concentration of 50 μg/mL64. Based on LC-HRMS results, a sorbitol compound was found in the ethyl acetate extract of P. aeruginosa ISP1RL4, namely Bis(4-ethylbenzylidene) sorbitol, which had also been found in black cumin extract with the best antibacterial activity against MRSA via in silico methods30.
Despite the confirmed antibacterial potential of ethyl acetate extract of P. aeruginosa ISP1RL4, there are a number of limitations that need to be improved for future studies. Firstly, the observed activity resulted from crude extracts therefore an optimization is required such as performing NMR analysis to determine the exact active compounds65. Secondly, the fermentation volume at the current study was set at 100 mL which resulted in a low yield of an active extract, therefore a higher volume e.g. 1 liter is essential to obtain more extracts. Thirdly, the current screening was mainly focused on measuring the diameter zone of inhibition, however no information is available on the median lethal dose (LD50) of the extract which is crucial for a more comprehensive antibacterial analysis. Fourthly, more MDR bacterial strains that are among human pathogens such as Enterobacterium faecium, Enterobacterium faecalis, and Streptococcus pneumoniae1 need to be included to evaluate antibacterial spectrum of the ethyl acetate extract of P. aeruginosa ISP1RL4. Lastly, the current study has not evaluated toxicity of the compounds, so the future study should be focused to screen for in vitro toxicity test such as MTT assays and lactate dehydrogenase. Such screening will ensure safety and efficacy of the extract.
Conclusion
In conclusion, this study has confirmed antibacterial potential of the isolate Pseudomonas aeruginosa ISP1RL4 against selected multidrug-resistant bacteria. Analysis of chemical profiles of the crude extract has identified a number of promising antibacterial molecules that potentially can be synthesized by P. aeruginosa ISP1RL4. Future studies would be focused to improve some limitations of the current results such as identifying the exact antibacterial compounds, increasing fermentation volume to obtain higher yield, analyzing LD50 of the extract, adding more MDR bacteria tests and performing in vitro toxicity tests. Nevertheless, this study adds a valuable insight on the potential of seaweeds-associated bacteria especially from the species of P. aeruginosa as the producer of (novel) antibacterial compounds.
Acknowledgment
Authors also thanks The Directorate of Laboratory Management, Research Facilities and Science and Technology Park, National Research and Innovation Agency (BRIN) Yogyakarta, Indonesia, for SEM and LC-HRMS instrument.
Funding Source
This research was funded by the Fundamental Research Grants awarded by the Ministry of Education, Culture, Research, and Technology Indonesia (fiscal year 2024) to Anak Agung Gede Indraningrat (grant numbers: 110/E5/PG.02.00.PL/2024, 2927/LL8/AL.04/2024, 565/Unwar/DPPM/PD-13/2024).
Conflict of Interest
The author(s) do not have any conflict of interest.
Data availability Statement
Sequence data of ISP1RL4 bacterial isolate was deposited in GenBank under accession number PP783522. Data related to antibacterial screening, SEM and light microscopy observation, TLC, LC-HRMS and GC-MS can be accessed via the Figshare online repository http://surl.li/rpyonh
Ethics Statement
Experiment described in this manuscript has been ethically approved by the Ethics Commission of Faculty of Medicine and Health Sciences, Warmadewa University, Denpasar-Bali under ethics number: 345/Unwar/FKIK/EC-KEPK/I/2023 on 2 October 2023
Informed Consent Statement
This study did not involve human participants, and therefore, informed consent was not required
Clinical Trial Registration
This research does not involve any clinical trials
Author Contributions
AAGI designed and provided consumables for experiments, PPCPP performed lab works. AAGI wrote draft manuscript. EM performed LC-HRMS and SEM experiments. MDW performed data analysis on GC-MS and LC-HRMS. DAPSM cultured MDR bacteria and fermentation of ISP1RL4 isolate. NLPEKS performed TLC and data analysis. All authors read and reviewed the draft manuscript. All authors agreed with the manuscript before submission.
References
- Bharadwaj A., Rastogi A., Pandey S., Gupta S., Sohal J. S. Multidrug-Resistant Bacteria: Their Mechanism of Action and Prophylaxis. Biomed Res Int. 2022;2022:1-17
CrossRef - Urban-Chmiel R., Marek A., Stępień-Pyśniak D, Wieczorek K., Dec M., Nowaczek A., Osek J.. Antibiotic Resistance in Bacteria—A Review. Antibiot. 2022;11(8):1079.
CrossRef - Jubeh B., Breijyeh Z., Karaman R. Resistance of gram-positive bacteria to current antibacterial agents and overcoming approaches. Mol. 2020;25(12):1-22.
CrossRef - Amankwah F. K. D., Gbedema S. Y., Boakye Y. D., Bayor M. T., Boamah V. E. Antimicrobial Potential of Extract from a Pseudomonas aeruginosa Isolate. Scientifica. 2022;2022.
CrossRef - Schneider Y. K. Bacterial natural product drug discovery for new antibiotics: Strategies for tackling the problem of antibiotic resistance by efficient bioprospecting. Antibiot. 2021;10(7).
CrossRef - De La Hoz-Romo M. C., Díaz L., Villamil L. Marine Actinobacteria a New Source of Antibacterial Metabolites to Treat Acne Vulgaris Disease—A Systematic Literature Review. Antibiot. 2022;11(7).
CrossRef - Hai Y., Wei M. Y., Wang C. Y., Gu Y. C., Shao C. L. The intriguing chemistry and biology of sulfur-containing natural products from marine microorganisms (1987–2020). Mar Life Sci Technol. 2021;3(4):488-518.
CrossRef - Tan L. T. Impact of Marine Chemical Ecology Research on the Discovery and Development of New Pharmaceuticals. Mar Drugs. 2023;21(3).
CrossRef - Negara B. F. S. P., Riyanti ., Marhaeni B., Kusuma A. B. Antibacterial activity of Actinomycetes symbiont with seaweeds: a prosperous agent of animal antibacterial. Aceh J of Anim Sci. 2016;1(2):45-49.
CrossRef - Bengtsson-Palme J. Microbial model communities: To understand complexity, harness the power of simplicity. Comput Struct Biotechnol J. 2020;18:3987-4001.
CrossRef - Srinivasan R., Kannappan A., Shi C., Lin X. Marine bacterial secondary metabolites: A treasure house for structurally unique and effective antimicrobial compounds. Mar Drugs. 2021;19(10):1-36.
CrossRef - Ameen F., AlNadhari S., Al-Homaidan A. A. Marine microorganisms as an untapped source of bioactive compounds. Saudi J Biol Sci. 2021;28(1):224-231.
CrossRef - Barzkar N., Sukhikh S., Babich O. Study of marine microorganism metabolites: new resources for bioactive natural products. Front Microbiol. 2023;14.
CrossRef - Zha X., Ji R., Zhou S. Marine Bacteria: A Source of Novel Bioactive Natural Products. Curr Med Chem. 2024;31(41):6842-6854.
CrossRef - Alharbi N. K., Azeez Z. F., Alhussain H. M., Shalol A., Albureikan M., Elsehrahwy M., Aloraini G., El-Nablaway M., Khatrawi E., Ghareeb A. Tapping the biosynthetic potential of marine Bacillus licheniformis LHG166, a prolific sulphated exopolysaccharide producer: structural insights, bio-prospecting its antioxidant, antifungal, antibacterial and anti-biofilm potency as a novel anti-infective lead. Front Microbiol. 2024;15.
CrossRef - Hassan S. W. M. Antibacterial, anticoagulant and anti-inflammatory activities of marine bacillus cereus s1. J Pure Appl Microbiol. 2016;10(4):2593-2606.
CrossRef - Indraningrat A. A. G., Purnami P. P. C. P., Aryastuti A. A. S. A., Wijaya M. D., Horng J. T. Antibacterial, antifungal and antioxidant activities of Bacillus cereus SMPRL-2 isolated from seaweeds Eucheuma cottonii. In: IOP Conference Series: Earth and Environmental Science. Vol 1271. Institute of Physics; 2023.
CrossRef - Priyanka S., Jayashree M., Shivani R., Anwesha S., Bhaskara Rao K. V., I A. E. Characterisation and identification of antibacterial compound from marine actinobacteria: In vitro and in silico analysis. J Infect Public Health. 2019;12(1):83-89.
CrossRef - Andriani Z., Fasya A. G., Hanapi A. Antibacterial Activity of the Red Algae Eucheuma cottonii Extract from Tanjung Coast, Sumenep Madura. Alchemy. 2016;4(2):93.
CrossRef - Putri T., Arsianti A., Subroto P. A. M., Lesmana E. Phytochemical analysis and antioxidant activity of marine algae Eucheuma Sp. In: AIP Conference Proceedings. Vol 2092. ; 2019.
CrossRef - Singh R. P., Reddy C. R. K. Seaweed-microbial interactions: Key functions of seaweed-associated bacteria. FEMS Microbiol Ecol. 2014;88(2):213-230.
CrossRef - Hafsan H., Aziz I., Sukmawaty E., S. Aisyah S., Hasyimuddin H., Zulkarnain Z., Hajrah H. Antibiotic Activity of Endophytic Bacteria isolated from Euchema cottonii of North Galesong Sea, Takalar. 2019;(June).
CrossRef - Purnami P. P. C. P., Indraningrat A. A. G., Darmayasa I. B. G. Antibacterial Activity Screening Of Bacterial Isolates Associated With Seaweed Eucheuma cottonii From Coastal Area In Buleleng, Bali. Biotropika: Journal of Tropical Biology. 2022;10(2):132-140.
CrossRef - Indraningrat A. A. G., Purnami P. P. C. P., Aryastuti A. A. S. A., Wijaya M. D. Antibacterial Activity of Pseudomonas Aeruginosa ISP1RL3 Against Multidrug Resistance Bacteria. Jurnal Penelitian Pendidikan IPA. 2023;9(12):11126-11136.
CrossRef - Fisher Scientific T. NanoDrop Micro-UV/Vis Spectrophotometer NanoDrop One User Guide.; 2020. http://www.nanodrop.com/support
- Nirwati H., Damayanti E., Sholikhah E. N., Mutofa M., Widada J. Soil-derived Streptomyces sp. GMR22 producing antibiofilm activity against Candida albicans: bioassay, untargeted LC-HRMS, and gene cluster analysis. Heliyon. 2022;8(4):e09333.
CrossRef - Asnani A., Luviriani E., Oedjijono O. Activity of Actinomycetes Isolated from Mangrove Segara Anakan Cilacap toward Methicillin-resistant Staphylococcus aureus (MRSA). J Kim Sains dan Apl. 2020;23(1):1-7.
CrossRef - Sulistyani N., Akbar A. N. Aktivitas Isolat Actinomycetes dari Rumput Laut ( Eucheuma cottonii ) sebagai Penghasil Antibiotik terhadap Staphylococcus aureus dan Escherichia coli ( Activity of Actinomycetes Isolate from Seeweed ( Eucheuma cottonii ) as Antibiotic Producer against Staphylococcus aureus and Escherichia coli. St. Jurnal Ilmu Kefarmasian Indonesia. 2014;12(1):1-9.
- Glenz R., Kaiping A., Göpfert D., Weber H., Lambour B., Sylvester M., Fröschel C., Mueller M., Osman M., Waller F. The major plant sphingolipid long chain base phytosphingosine inhibits growth of bacterial and fungal plant pathogens. Sci Rep. 2022;12(1):1-9.
CrossRef - Khalissa Anidya D., Purwono R. M., Andrianto D., Kusumawati N. T. Aktivitas Antibakteri Senyawa Aktif Ekstrak Jintan Hitam (Nigella sativa) Terhadap Bakteri MRSA secara In Silico (Antibacterial Activity of Black Cumin (Nigella sativa) Active Compounds Against MRSA in In Silico). 2023;1(2):92-101.
CrossRef - Jaybhaye D. L., Chandra S., Johar S., Nagre A. S. Comparative effect of mixture of ginger and honey with dextromethorphan in dry cough in children. Int J Basic Clin Pharmacol. 2021;10(5):545.
CrossRef - Shafeian E., Mostafavi G. P., Farimani M. M., Moradi M. A., Nazemi M. Extraction and investigation of biological activities of dioctyl phthalate and dibutyl phthalate from marine sponge Haliclona (Soestella) caerulea Larak Island, Persian Gulf. Iran J Fish Sci. 2022;21(5):1141-1155.
- Becam J., Walter T., Burgert A., Schlegel J., Sauer M., Seibel J., Schubert-Unkmeir A. Antibacterial activity of ceramide and ceramide analogs against pathogenic Neisseria. Sci Rep. 2017;7(1):17627.
CrossRef - Villa-Pérez C., Ortega I. C., Vélez-Macías A., Payan A., Echeverria G., Soria D., Valencia-Uribe G. Crystal structure, physicochemical properties, Hirshfeld surface analysis and antibacterial activity assays of transition metal complexes of 6-methoxyquinoline. New J Chem. 2018;42(9):7166-7176.
CrossRef - Tang Y., Yu P., Chen L. Identification of Antibacterial Components and Modes in the Methanol-Phase Extract from a Herbal Plant Potentilla kleiniana Wight et Arn. Foods. 2023;12(8).
CrossRef - Alotaibi H. F., Alotaibi H., Darwish K. M., Khafagy E., Abu Lila A., Ali M., Hegazy W., Aishawwa S. The Anti-Virulence Activities of the Antihypertensive Drug Propranolol in Light of Its Anti-Quorum Sensing Effects against Pseudomonas aeruginosa and Serratia marcescens. Biomedicines. 2023;11(12).
CrossRef - Orbán-Gyapai O., Liktor-Busa E., Kúsz N., Urbán E., Hohmann J., Vasas A. Antibacterial screening of Rumex species native to the Carpathian Basin and bioactivity-guided isolation of compounds from Rumex aquaticus. Fitoterapia. 2017;118:101-106.
CrossRef - Faisal Madhloom A., Bashir Hashim Al-Taweel F., Sha A. M., Raad Abdulbaqi H. Antimicrobial Effect of Moringa Oleifera L. and Red Pomegranate against Clinically Isolated Porphyromonas gingivalis: in vitro Study. Arch Razi Inst. 2022;77(4):1405-1419.
- Veerasophon J., Sripalakit P., Saraphanchotiwitthaya A. Formulation of anti-acne concealer containing cinnamon oil with antimicrobial activity against Propionibacterium acnes. J Adv Pharm Technol Res. 2020;11(2):53-58.
CrossRef - Gumgumjee N. M., Aly N. A. H., Malawi F. H. Synergistic antimicrobial effects and GC-MS analysis of phytocomponents of Commiphora quadricincta. J Food Process Technol. 2016;7(9).
CrossRef - Osama A., Awadelkarim S., Ali N., Khalid S., Mohammed S., Hashim N. Phytochemical Composition and Evaluation of Antimicrobial Activity of Blepharis linariifolia (Pers.) Seeds. 2017;2(2):1-6.
CrossRef - Asri N. A. A. M., Sani M. S. A., Othman R., Nordin N. F. H., Desa M. N. Antibacterial activities , chemical composition, and efficacy of green extract Carica papaya peel on food model systems. Published online 2022:1-18.
CrossRef - Balouch H., Demirbag Z., Durani M., Sarsekeeva F., Nygymetova A. Antibacterial activity of freshwater green microalgae from Almaty region. BIO Web Conf. 2024;100:02014.
CrossRef - Kawuri R., Darmayasa I. B. G. Bioactive compound from extract filtrat Streptomyces sp.Sp1. as biocontrol of vibriosis on larvae of Macrobrachium rosenbergii shrimps. Hayati. 2019;26(1):15-25.
CrossRef - Pacífico C., Fernandes P., de Carvalho C. C. C. R. Mycobacterial response to organic solvents and possible implications on cross-resistance with antimicrobial agents. Front Microbiol. 2018;9(MAY):1-12.
CrossRef - Bellahcen T. O., Cherki M., Sánchez J. A. C., Cherif A., EL Amrani A. Chemical Composition and Antibacterial Activity of the Essential Oil of Spirulina platensis from Morocco. Journal of Essential Oil-Bearing Plants. 2019;22(5):1265-1276.
CrossRef - Andila P. S., Nugroho L. H. Antibacterial and phytochemical constituent of Etlingera rubroloba A.D. Poulsen extract, an endemic ginger from Wallacea Region, Indonesia. Biodiversitas. 2022;23(7).
CrossRef - Rizwana H., Alwhibi M. S., Soliman D. A. Antimicrobial activity and chemical composition of flowers of Matricaria aurea a native herb of Saudi Arabia. International Journal of Pharmacology. 2016;12(6):576-586.
CrossRef - Tiwari S., Mishra S., Misra D. R., Upadhyay R. Identification of new bioactive compounds from fruit of Abutilon indicum through GCMS analysis. Biol Forum. 2016;8(1):548-554.
- Zayed M. Z., Samling B. Phytochemical constituents of the leaves of Leucaena leucocephala from Malaysia. Int J Pharm Pharm Sci. 2016;8(12):174-179.
CrossRef - Munk M. E., Sodano C. S., Mclean R. L., Haskellz T. H. Structure of Actinobolamine’. 1967;(7):4158-4165.
CrossRef - Bollinger A., Thies S., Katzke N., Jaeger K. E. The biotechnological potential of marine bacteria in the novel lineage of Pseudomonas pertucinogena. Microb Biotechnol. 2020;13(1):19-31.
CrossRef - Elabed H., González-Tortuero E., Ibacache-Quiroga C., Bakhrouf A., Johnston P., Gaddour K., Blázquez J., Rodríguez-Rojas A. Seawater salt-trapped Pseudomonas aeruginosa survives for years and gets primed for salinity tolerance. BMC Microbiol. 2019;19(1):1-13.
CrossRef - Khan A., Ahmad A., Akhtar F., Yousuf S., Xess I., Khan L., Manzoor N. Induction of oxidative stress as a possible mechanism of the antifungal action of three phenylpropanoids. FEMS Yeast Res. 2011;11(1):114-122.
CrossRef - Romero-González L. E., Rojas-Vargas J., Muriel-Millán L. F., Bustos-Martínez J., Bustamante V. H., Pardo-López L. Genomic and phenotypic characterization of Pseudomonas sp. GOM7, a novel marine bacterial species with antimicrobial activity against multidrug-resistant Staphylococcus aureus. PLoS One. 2023;18(7 July).
CrossRef - Thenmozhi S., Moorthy K., Sureshkumar B. T., Suresh M. Antibiotic Resistance Mechanism of ESBL Producing Enterobacteriaceae in Clinical Field: A Review. Int J Pure Appl Biosci. 2014;2(3):207-226.
- Indriani V., Chiuman L., Wijaya L. L., Lister G., Grandis L. Antibacterial Effect of Curcuma zedoaria Extract on Bacillus cereus and Staphylococcus epidermidis. Althea Medical Journal. 2020;7(1):6-10.
CrossRef - Epand R. M., Walker C., Epand R. F., Magarvey N. A. Molecular mechanisms of membrane targeting antibiotics. Biochim Biophys Acta Biomembr. 2016;1858(5):980-987.
CrossRef - Ouchari L., Boukeskasse A., Bouizgarne B., Ouhdouch Y. Antimicrobial potential of actinomycetes isolated from the unexplored hot Merzouga desert and their taxonomic diversity. Biol Open. 2019;8(2).
CrossRef - Sari S. A., Pujiyanto S., Suprihadi A. Antibacterial activity tests of isolate endophytic bacteria from the tea plant (Camellia sinensis) againts Staphylococcus aureus and Staphylococcus epidermidis. In: Journal of Physics: Conference Series. Vol 1524. Institute of Physics Publishing; 2020.
CrossRef - Aryal B., Adhikari B., Aryal N., Bhattarai B. R., Khadayat K., Parajuli N. LC-HRMS Profiling and Antidiabetic, Antioxidant, and Antibacterial Activities of Acacia catechu (L.f.) Willd. Formanowicz D, ed. Biomed Res Int. 2021;2021:1-16.
CrossRef - Campmajó G., Saurina J., Núñez O. Liquid chromatography coupled to high-resolution mass spectrometry for nut classification and marker identification. Food Control. 2023;152:109834.
CrossRef - Petrakis E. A., Mikropoulou E. V., Mitakou S., Halabalaki M., Kalpoutzakis E. A GC–MS and LC–HRMS perspective on the chemotaxonomic investigation of the natural hybrid Origanum × lirium and its parents, O. vulgare subsp. hirtum and O. scabrum. Phytochemical Analysis. 2023;34(3):289-300.
CrossRef - Nocedo-Mena D., Arrasate S., Garza-González E., Rivas-Galindo V., Romo-Mancillas A., Munteanu C., Sotomayor N., Lete N., Barbolla I., Martin C., del Rayo Camacho-Corona M. Molecular docking, SAR analysis and biophysical approaches in the study of the antibacterial activity of ceramides isolated from Cissus incisa. Bioorg Chem. 2021;109:104745.
CrossRef - Wang T., Li F., Lu Q., , Wu G., Jiang Z., Liu S., Habden X., Razumova E., Osterman I., Sergiev P., Dontsova O., Hu X., You X., Sun C. Diversity, novelty, antimicrobial activity, and new antibiotics of cultivable endophytic actinobacteria isolated from psammophytes collected from Taklamakan Desert. J Pharm Anal. 2021;11(2):241-250.
CrossRef








