Bindu Madhavi Boddupalli1 , Ramalingam Ramani2*
, Ramalingam Ramani2* , Elizabeth Owiti1
, Elizabeth Owiti1 , Elias Nelson3
, Elias Nelson3 and Michael Mungoma4
and Michael Mungoma4
1Department of Pharmaceutics and Pharmacy Practice, School of Pharmacy, Mount Kenya University, Thika, Kenya.
2Department of Pharmaceutical Chemistry, School of Pharmacy, Mount Kenya University, Thika, Kenya.
3Department of Natural Sciences, School of Pure and Applied Sciences, Mount Kenya University, Thika, Kenya.
4Department of Pharmacology and Clinical Practice, School of Pharmacy, Mount Kenya University, Thika, Kenya.
Corresponding Author E-mail:rramalingam@mku.ac.ke
DOI : https://dx.doi.org/10.13005/bpj/2961
Abstract
Bacterial resistance is at its peak challenging humankind and researchers dealing with microorganisms. To prevent further risk due to microbial infections, there is an emergency to solve this problem. The practice of using herbs in treating diseases is from ancient years, especially in African countries with rich natural remedies resources. Urtica Massaica from the Urticaceae family is abundantly found in Africa. Methanolic extract of leaf powder was tested against resistant microorganisms (Acinetobacter baumannii and MRSA (Methicillin-resistant Staphylococcus aureus). Ciprofloxacin was used as the standard antibacterial drug. Molecular docking studies were performed for the selected flavonoids of the plant from literature against penicillin-binding protein 2a and DNA gyrase subunit B. The results from the antimicrobial studies indicated the zone of inhibitions ranging from 6±0.00 mm to 11.67±0.33 mm. Molecular docking studies revealed the ability of Urtica flavonoids to bind with the selected target receptors. Kaempferol was found to have a higher docking score with a less binding energy of -11.25kCal/Mol and 9.83Kcal/Mol against PBP2a and DNA gyrase respectively. The results of the current study strongly indicate the need for further studies involving the isolation of pure compounds and their use against resistant bacteria.
Keywords
Acinetobacter; Antimicrobial Study; Bacterial Resistance; Molecular Docking; MRSA; Urtica Massaica
Download this article as:| Copy the following to cite this article: Boddupalli B. M, Ramani R, Owiti E, Nelson E, Mungoma M. Molecular Docking Studies and Antibacterial Evaluation of Urtica massaica Leaves. Biomed Pharmacol J 2024;17(3). |
| Copy the following to cite this URL: Boddupalli B. M, Ramani R, Owiti E, Nelson E, Mungoma M. Molecular Docking Studies and Antibacterial Evaluation of Urtica massaica Leaves. Biomed Pharmacol J 2024;17(3). Available from: https://bit.ly/3znIfFO |
Introduction
Infections as a result of microbes are still a major threat to many lives all over the world. To date, the rate of antibiotic resistance is very high with many micro-organisms developing resistance to the available antibiotics1. At this rate, the urge to have new antibiotic formulations as well as investigation of alternative antimicrobial agents derived from plants is on course. Herbal medicines are either the mainstay of healthcare delivery or serve as a complement to it across the globe2.
Herbs have been replacing synthetic compounds for the last few decades. As per the documentation of the World Health Organization, more than 80 % of the population in developing countries utilize herbal medicines3. This may be due to their easy availability, reduced or no toxic effect, and being able to cover a wide range of conditions4. The literature confirms the anti-oxidant and antibacterial properties of the phytoconstituents including flavonoids, tannins, catechins, vitamins, etc5. A mixture of these compounds in herbs can produce better protection than a single synthetic compound due to synergism. Urtica massaica is a perennial herb that grows to a height of 200 cm. It’s taxonomically placed in the kingdom Plantae family Urticaceae and genus Urtica. It’s well distributed in African countries including Kenya and Uganda where it grows in the forest near the mountains and the cattle areas in homesteads6. In Kenya, this plant has multiple uses including treating rashes and reducing sugar levels. The present study aimed at the antibacterial activities of acetone extract of Urtica Massaica leaves and docking studies.
Materials and Methods
Plant extract preparation
The plant was collected from rural parts of Kenya and authenticated by the East African Herbarium with a reference number of NMK/BOT/CTX/2/ID/2023. The extract was prepared by cold maceration. Briefly, leaf powder 200 g was soaked in 400 ml volume of methanol for 3 days. The flask containing the material was shaken daily, filtered after 72 hours, and concentrated by a rotary evaporator. The resultant extract was packed in clean and pre-weighed glass sample vials and stored for future analysis.
Antibacterial Activity
The disk diffusion method was used to test the antibacterial potential of Urtica massaica extract. The plant extract was used at 250, 200, and 150g/ml concentrations. Acinetobacter baumannii and methicillin-resistant Staphylococcus aureus (MRSA) were the targets of the activity. The bacteria were uniformly inoculated onto the Muller Hilton agar media and sterile paper disks laid by sterile forceps using sterile cotton wool swabs. Using a micropipette, exactly 15µl of the plant extract at various concentrations was loaded onto the disks. After being allowed one hour to dry, each plate was incubated for 18 hours. A ruler was used to measure the inhibition zones surrounding the disks, and the measurements were recorded in millimeters7.
Molecular Docking Studies
In this study, molecular docking was performed for flavonoid compounds with penicillin-binding protein 2a (PBP 2a) PDB ID: 4DKI8,9 and DNA gyrase subunit B PDB ID: 7PQI10. These proteins were retrieved from the RCSB Protein Data Bank (https://www.rcsb.org). Molecular docking studies were performed using Auto Dock Tools (Version 1.5.7). To investigate docking capacity, PBP 2a co-crystallized with Ceftobiprole and DNA gyrase B co-crystallized with Novobiocin were selected. Binding cavities of the reference ligand (Ceftobiprole and Novobiocin) bind in the crystal structure of proteins were selected as docking regions. The docking process was validated by redocking with the co-crystallized ligand.
In pre-docking, proteins were prepared by removing all water molecules and ligands and adding hydrogen atoms as well as gasteiger charges. On the other hand, all the flavonoid ligands were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov) in the SMILES format and then converted into pdbqt format using online OpenBabel software (https://www.cheminfo.org). The calculation of free binding energies and prediction of interacting amino acids as well as the number of hydrogen bonds involved with ligands were performed by using a genetic algorithm as a search parameter with 10 runs.
Data Analysis
Data analysis was conducted using GraphPad prism statistical data analysis software. Descriptive statistics followed to obtain the means and their respective standard error of the means. A significant difference between the concentrations of the extract and that of Ciproflaxacin was obtained through one-way ANOVA followed by fishers pairwise as the post hoc test.
Results and Discussion
Antibacterial Activity of Urtica massaica
The antibacterial activity of Urtica massaica against resistant strains of A. baumanni and MRSA was investigated in this study. The mean zones of inhibition ranged between 9.67±0.33 mm to 6±0.00 mm against MRSA and between 11.67±0.33 mm to 8.33±0.33 mm against A. baumanni. The standard antibiotic; ciprofloxacin recorded a mean zone of inhibition of 26±0.00 mm and 30.67±0.67 mm against MRSA and A. baumanni respectively. The comparison revealed a significant difference in the mean zones of inhibitions at all the studied concentration levels in against both MRSA and A. baumanni (p<0.05). In this analysis for both MRSA and A. baumanni, the standard antibiotic; ciprofloxacin recorded a significantly larger mean zone of inhibition while the extract at the lowest concentration of 150 mg/ml recorded a significantly smaller mean zone of inhibition (p<0.05).
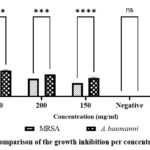 |
Graph 1: Comparison of the growth inhibition per concentration level |
Molecular Docking Studies
Reported flavonoids from Urtica species11, 12 were selected for molecular docking studies. Molecular docking studies were conducted to investigate the inhibition potential of phytochemicals specifically flavonoids present in the extracts against penicillin binding protein 2a isolated from methicillin resistance Staphylococcus aureus and DNA gyrase B isolated from Acinetobacter baumannii. The lowest binding energies score, inhibition constant, number of hydrogen bonding and amino acid involved in hydrogen bonding with ligands were captured and listed in Table 1 & 2 and its corresponding docking conformation shown in Figure 1 & 2.
Table 1: Docking studies with A. baumanni DNA gyrase B ATPase
|
S. No |
Ligand |
Binding Energy Kcal/Mol |
Inhibition Constant T 298.15 K Ki |
No of H- Bonding |
Amino Acids involved |
|
1 |
Ciprofloxacin |
-11.05 |
7.89nM |
2 |
GLY91 THR179 |
|
2 |
Gossypetin |
-11.99 |
1.62nM |
1 |
GLY91 |
|
3 |
Isovitexin |
-13.90 |
65.21pM |
1 |
UNL1 |
|
4 |
Kaempferol |
-11.25 |
5.70nM |
1 |
GLY91 |
|
5 |
Myricetin |
-12.55 |
633.15pM |
2 |
GLY91 THR179 |
|
6 |
Quercetin |
-12.00 |
1.60nM |
2 |
GLY91 THR179 |
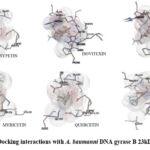 |
Figure 1: Docking interactions with A. baumanni DNA gyrase B 23kDa ATPase |
Molecular docking studies against DNA gyrase B isolated from A. baumanni were found to have binding energies of -13.90 Kcal/mol to 11.05 Kcal/mol. Ciprofloxacin exhibited binding at the active sites of GLY91 and THR179 with a binding energy of -11.05 Kcal/Mol. Among the tested phyto constituents, Isovitexin was found to have a more binding energy of -13.90Kcal/Mol at the active site of UNL1 on A. baumanni DNA gyrase B ATPase.
Table 2: Docking studies with MRSA PBP2a
|
S. No |
Ligand |
Binding Energy Kcal/Mol |
Inhibition Constant T 298.15 K Ki (nM) |
No of Hydrogen Bonding |
Amino Acids involved in HB |
|
1 |
Ciprofloxacin |
-9.95 |
50.97 |
2 |
SER403 ASN464 |
|
2 |
Gossypetin |
-11.23 |
5.88 |
2 |
ASN464 THR600 |
|
3 |
Isovitexin |
-12.63 |
552.07pM |
4 |
SER598 THR444 TYR446 HIS583 |
|
4 |
Kaempferol |
-9.83 |
62.66 |
2 |
ASN464 THR600 |
|
5 |
Myricetin |
-11.04 |
8.14 |
3 |
SER403 SER462 TYR446 |
|
6 |
Quercetin |
-10.58 |
17.57 |
4 |
ASN464 TYR446 HIS583 GLU447 |
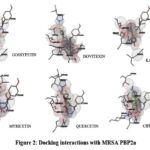 |
Figure 2: Docking interactions with MRSA PBP2a |
Molecular docking studies against PBP 2a isolated from MRSA were found to have binding energies from -9.83 Kcal/mol to 12.63 Kcal/mol. Ciprofloxacin exhibited binding at the active sites of SER403 and ASN464 with a binding energy of -9.95 Kcal/Mol. Among the tested phytoconstituents, Isovitexin was found to have more binding energy of -12.63Kcal/mol at the active sites of SER598, THR444, TYR446, and HIS583 on MRSA PBP2a.
Amino acids involved in the interactions with phytoconstituents match with the amino acids involved in binding with co-crystalized ligand (Ceftobiprole and Novobiocin) which is retrieved from PDBsum (https://ebi.ac.uk) and shown in Figure 3. The ligand Novobiocin has formed 5 hydrogen bonds with Asn 60, Asp 87, Asp 95, His 97, and Arg 150 of DNA gyrase B amino acids of DNA gyrase B. In our molecular docking study, phytoconstituents formed hydrogen bonding with Unl1, Gly91, and Thr179 which are different from amino acids involved with cocrystalized ligand Novobiocin. It was found that ligand Ceftobiprole has formed 5 hydrogen bonds with Ser 403, Asn 464, Ser 598, Thr 600, and Glu 602 amino acids of penicillin binding protein 2a. In our molecular docking study, all the phytoconstituents form hydrogen bonding with penicillin binding protein 2a at Ser 403, Asn 464, Ser 598, and Thr 600 like co-crystalized ligand Ceftobiprole.
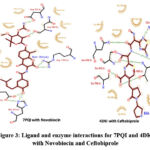 |
Figure 3: Ligand and enzyme interactions for 7PQI and 4DKI with Novobiocin and Ceftobiprole |
The spreading of resistant pathogenic bacteria is now an alarming threat to public health both in developed countries and developing countries. Recent literature according to the China Antimicrobial Resistance Surveillance System from a period of 2019- 2020 confirms the occurrence of various gram-positive and gram-negative resistant micro-organisms. In case of no effective measures, the deaths due to antibiotic resistance will reach 10 million by 2025 13. This situation demands the development of compounds against resistant organisms. Research is focusing on plants to tackle antibiotic resistance. Plants contain various phytochemical constituents, such as alkaloids, flavonoids, tannins, saponins, phenols and terpenoids are proven for therapeutic activities including antibacterial capability. Worldwide, the usage of herbal medicine is increasing and is projected to increase by 5.5% by 202714. Dependency on herbal medicine in African countries is more than 60% for health benefits. Africa is rich in many plant species with diverse health benefits including combating drug resistance and further research focusing on new identifications against bacterial resistance is critical for combating bacterial resistance in Africa and globally 15.
Current research aimed at screening Urtica massaica against resistant bacteria. Urticaspecies are found mostly in Europe, North America, and North Africa along with some parts of Asia. In 2018, 46 species of Urtica were reported along with their habitat, phytochemical constituents, and antibacterial activity. Urtica Dioica dominated the reports mentioned about antibacterial studies. Information about Urtica massaica and studies from Africa are not reported in the review16. In 2019, the antibacterial activity of Urtica massaica leaf extract was published against Staphylococcus aureus and Escherichia coli. The zone of inhibition reported was between 6mm- 8.4mm at 100mg/mL17. Other studies from Africa (Rwanda) included about Phytochemical screening and antibacterial activity of stem and root bark extracts against Escherichia coli, Salmonella sp., and Staphylococcus aureus. The maximum zone of inhibition is 28mm at 600mg/mL18.
Molecular docking studies in recent research are used as a powerful tool to predict the activity of compounds against the target receptor. A very recent study confirmed the presence of phenolic and flavonoid contents along with anti-inflammatory and molecular docking studies against cyclooxygenase-211. Among the tested active constituents, Kaempferol was found to have the least binding energy indicating more chances of binding into the target site for both the selected proteins. Kaempferol-containing plants are gaining importance in clinical microbiology and literature supports the activity of extracts containing Kaempferol and its derivatives against resistant microorganisms like A. baumanni and MRSA19. Docking and antimicrobial studies confirm the potential of Urtica massaica against the selected resistant microorganisms.
Conclusion
The results from the in vitro antibacterial activity of Urtica massaica in the present study showed that Urtica massaica is a potential antimicrobial agent. The ability of extract could be attributed to various phytochemical compounds present in plants such as the phenolic and flavonoid compounds in the extract. It’s therefore evident that this plant could be of help in inhibiting the progression of the oxidative stress that aids in the pathogenesis of chronic life-threatening disorders. Molecular docking studies also confirmed the same giving a way forward to investigate further on Urtica massaica to isolate the individual compounds and prove them for the antimicrobial activity against resistant microorganisms.
Acknowledgments
The authors express their deep sense of gratitude to the management and research team of Mount Kenya University for their encouragement in the successful completion of this work.
Conflict of Interest
The authors do not have any conflict of interest.
Funding Sources
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Data Availability Statement
This statement does not apply to this article.
Ethics Statement
This research did not involve human participants, animal subjects, or any material that requires ethical approval.
Informed Consent Statement
This study did not involve human participants, and therefore, informed consent was not required
Authors’ Contribution
Bindu Madhavi Boddupalli: Conceptualization; Manuscript writing; Statistical design
Ramalingam Ramani: Molecular docking studies and analysis
Elizabeth Owiti: Execution of the work
Elias Nelson: Procurement of materials and part of execution
Michael Mungoma: Manuscript proof reading and analysis
References
- Aslam B., Wang W., Arshad M. I., Khurshid M., Muzammil S., Rasool M. H., Nisar M. A., Alvi R. F., Aslam M. A., Qamar M. U., Salamat M. K. F., Baloch Z. Antibiotic Resistance: A Rundown of a Global Crisis. Infect Drug Dis. 2018; 11: 1645-1658.
CrossRef - Murugaiyan J., Anand K. P., Rao G. S., Iskandar K., Hawser S., Hays J. P., Mohsen Y., Adukkadukkam S., Awuah W. A., Jose R. A. M., Sylvia N., Nansubuga E. P., Tilocca B., Roncada P., Calero N. R., Morales J. M., Amin R., Kumar B. K., Kumar A., Toufik A. R., Zaw T. N., Akinwotu O. O., Satyaseela M. P., Dongen M. B. M. V. Progress in Alternative Strategies to Combat Antimicrobial Resistance: Focus on Antibiotics. Antibiotics. 2022; 11: 1-39.
CrossRef - Sen S., Chakraborthy R. Revival, Modernization and Integration of Indian Traditional Herbal Medicine in Clinical Practice: Importance, Challenges and Future. J Tradit Complement Med. 2017; 7(2): 234-244.
CrossRef - Xu D. P., Li Y., Meng X., Zhou T., Zhou Y., Zheng J., Zhang J. J., Li H. B. Natural Antioxidants in Foods and Medicinal Plants: Extraction, Assessment and Resources. Int J Mol Sci. 2017; 18(1): 1-32.
CrossRef - Muedd A., Shibli S., Quwaie D. A. A., Ashkan M. F., Alharbi M., Alanazi H., Binothman N., Aljadani M., Majrashi K. A., Huwaikem M., Abourehab M. A. S., Korma S. A., Saadony M. T. E. Extraction and Characterization of Polyphenols from Certain Medicinal Plants and Evaluation of Their Antioxidant, Antitumor, Anti-Diabetic, Antimicrobial Properties and Potential Use in Human Nutrition. Front Nutr. 2023; 10: 1-24.
CrossRef - Wabai Y. W., Maina M. J. K., Mwaniki N. E. N. Teratogenic Potential of Urtica massaica (mildbr) and Croton megalocarpus (hutch) in Mice. J Phytopharmacol. 2018; 7(6): 460-463.
CrossRef - Raho G. B., Benali M. Antibacterial Activity of the Essential Oils from the Leaves of Eucalyptus globulus Against Escherichia coli and Staphylococcus aureus. Asian Pacific J Tropical Biomedicine. 2012; 2(9): 739-742.
CrossRef - Milan D., Gibson A. S., Gargi H. S., Jayvi K. P., Dweipayan G. In Silico Analysis of Bioactive Phytocompounds as Inhibitors of Penicillin-Binding Protein 2a (pbp2a) and Class a Betalactamase of Target Microorganisms. Adv Biores. 2023; 1: 415-434.
- Lavanya P., Ramaiah S., Anbarasu A. A. Molecular Docking and Dynamics Study to Screen Potent Anti-Staphylococcal Compounds against Ceftaroline Resistant MRSA. J Cell Biochem. 2016; 117(2): 542-548.
CrossRef - Andrej E. C., Martina D., Davide B. T., Federica F., Daniela S., Masa S. Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa. J Med Chem. 2023; 66(2): 1380- 1425.
CrossRef - Wambui J., Ikedi R. I. O., Macharia R. W., KamaKama F., Nyaboga E. N. Phytoconstituents of Kenyan Stinging Nettle (Urtica species) and Their Molecular Docking Interactions Revealed Antiinflammatory Potential as Cyclooxygenase-2 Inhibitors. Sci Afr. 2024; 23(9): e02088.
CrossRef - Taheri Y., Quispe C., Bravo J. H., Rad J. S., Ezzat S. M., Merghany R. M., Shaheen S., Azmi L., Mishra A. P., Sener B., Kilic M., Sem S., Acharya K., Nasiri A., Martins N. C., Fokou P. V. T., Ydyrys A., Bassygarayev Z., Dastan S. D., Alshehri M. M., Calina D., Cho W. C. Urtica dioica Derived Phytochemicals for Pharmacological and Therapeutic Applications. Evid Based Complement Alternat Med. 2022; 24: 4024331.
CrossRef - Song L., Hu X., Ren X., Liu J., Liu X. Antibacterial Modes of Herbal Flavonoids Combat Resistant Bacteria. Front Pharmacol. 2022; 13: 873374.
CrossRef - Hexa R. Herbal Medicine Market Size and Forecast, by Product (Tablets & Capsules, Powders, Extracts), by Indication (Digestive Disorders, Respiratory Disorders, Blood Disorders), and Trend Analysis, 2014 – 2024. 2017. Available from: https://www.hexaresearch. com/research-report/global-herbal-medicinemarket.
- Moiketsi B. N., Makale K. P. P., Rantong G., Rahube T. O., Makhzoum A. Potential of Selected African Medicinal Plants as Alternative Therapeutics against Multi Drug Resistant Bacteria. Biomedicines. 2023;11(10): 2605.
CrossRef - Dorota K., Ewelina P., Hubert A. Urtica Spp: Ordinary Plants with Extraordinary Properties. Molecules. 2018; 23: 1664.
CrossRef - Kipruto A., Mwamburu L., Christine B., Kipngetich B. The Antimicrobial Activity of the Leaves of Urtica massaica on Staphylococcus aureus, Escherichia coli. J Medcinal Plants studies. 2019; 7(2): 21-24.
- Maniriho O., Nkurunziza J. P., Ayodele A. E., Benimana F., Murhula H. P., Farhan H. F. A., Nimbeshaho F., Cyiza F. Chemical Screening and Antimicrobial Activities of Rwandan Traditional Medicinal Plant, Urtica massaica mildbr (urticaceae). EAS J Pharm Pharmcol. 2021; 3(2): 56-63.
CrossRef - Periferakis A., Periferakis K., Badarau I. A., Petran E. M., Popa D. C., Caruntu A., Costache R. S., Scheau C., Caruntu C., Costache D. O. Kaempferol: Antimicrobial Properties, Sources, Clinical and Traditional Applications. Int J Mol Sci. 2022; 23(23): 15054.
CrossRef








