Upik Rahmi1,2* , Hanna Goenawan2
, Hanna Goenawan2 , Nova Sylviana2
, Nova Sylviana2 , Setiawan2
, Setiawan2 and Farida Murtiani3
and Farida Murtiani3
1Nursing Program Study, Universitas Pendidikan Indonesia, Jl. Dr. Setiabudi No. 229 Bandung, Jawa Barat, Indonesia.
2Medical Faculty, Padjadjaran University, Jl. Raya Bandung Sumedang KM.21, Hegarmanah, Kec. Jatinangor, Kabupaten Sumedang, Jawa Barat Indoensia.
3Sulianti Saroso Infectious Disease Hospital, Jakarta, Indonesia
Corresponding Author E-mail: upikrahmi@upi.edu
DOI : https://dx.doi.org/10.13005/bpj/2894
Abstract
Introduction: EGR1 (Early Growth Response 1) gene expression is a molecular response that occurs in the brain as a result of synaptic activity and environmental stimuli. Early growth response 1 (EGR1) expression can be affected by several factors, including exercise or physical training. This review aims to determine the effect of EGR 1 expression on hippocampal synaptic plasticity function. Method: Literature search using data-based Pubmed, Science Direct, and Scopus online. The data used is from the year 1978 until the year 2022. Searched using English keywords such as EGR 1 and hippocampus. Results: Animal and human studies show that physical exercise can increase the expression of the EGR1 gene in the brain. This enhanced EGR1 expression is associated with increased synaptic plasticity, which includes changes in the strength and connectivity of synapses between neurons. Synaptic plasticity refers to the ability of the nervous system to change the strength and efficiency of communication between neurons. Physical exercise has been shown to increase synaptic plasticity by increasing dendritic growth and continuity, increasing neurogenesis (the formation of new neurons), and increasing synaptic connections between neurons. Physical exercise can increase EGR1 expression and synaptic plasticity. Increased EGR1 expression and synaptic plasticity induced by physical exercise are associated with improvements in cognitive functions, including memory, learning, and thinking ability. Conclusion: There is evidence that exercise can increase EGR1 expression and synaptic plasticity in the brain, especially in the hippocampus, to improve cognitive function.
Keywords
EGR 1; Hippocampus; Synaptic plasticity
Download this article as:| Copy the following to cite this article: Rahmi U, Goenawan H, Sylviana N, Setiawan S, Murtiani F. The Impact of Early Growth Response 1 (Egr1) on Hippocampal Synaptic Plasticity and Cognitive Function: Narrative Review. Biomed Pharmacol J 2024;17(2). |
| Copy the following to cite this URL: Rahmi U, Goenawan H, Sylviana N, Setiawan S, Murtiani F. The Impact of Early Growth Response 1 (Egr1) on Hippocampal Synaptic Plasticity and Cognitive Function: Narrative Review. Biomed Pharmacol J 2024;17(2). Available from: https://bit.ly/4a19HFA |
Introduction
Early Growth Response 1 (EGR1) is a gene that plays a role in cognitive processes. Different members of the Egr family of transcriptional regulators have distinct functions in cognitive processes, with Egr1 being required specifically for long-term memory, while Egr3 is primarily essential for short-term memory1. EGR 1 in the central nervous system is a mediator of the interaction of genes with the environment and how environmental stimuli trigger rapid responses and lasting neural adaptations to neuronal function and plasticity2,3. EGR1 is activated by several external stimuli, such as growth factors, and cytokinin stimuli4,5. When activated, the EGR1 protein binds to a specific DNA region called an early growth response element (ERE) located in the promoter of a target gene6.
After EGR1, mRNA levels are characterized by their rapid upregulation within minutes. It is not related to protein synthesis, which is activated by general intracellular signaling pathways such as the triphosphoinositide kinase (PI3K) pathway or the mitogen-activated kinase (MAPK) pathway and can be triggered by various stimuli 2,6,7. Despite their extensive and overlapping nature, each EGR 1 differs in its activator, downstream regulatory pathway, target, and expression pattern1,2,6,8. Early growth response 1 (EGR1) underlies brain activity, including neurotransmission, synaptic plasticity, and learning and memory processes. In this article, We emphasize the function of EGR1 in both physiological states of the CNS. Before analyzing the genes, pathways, and biological processes that are targets of EGR1 in the CNS, we provide a summary of the variables that regulate its expression.
Method
Literature search using data based Pubmed, Science direct and Scopus online. The data used is from year 1978 until year 2022. Searched using English keywords such as EGR 1 and hippocampus.
Result and Discussion
Nearly three decades ago EGR 1 was first discovered to be cloned to be regulated by nerve growth factor (NGF) in the presence of a protein synthesis inhibitor while screening the cycloheximide9 in mouse PC12 cells. The process of cloning and characterization of this protein is carried out simultaneously in different groups in different cell lines stimulated by growth factors, which explains its alternative name: EGR110, NGFI-A9, Krox -2411, TIS85, and Zif26812. The screening strategy identified EGR3, EGR4 and EGR2, which together with EGR1 form the EGR family6,8.
All EGRs among species in the region contain three cysteine-2-histidine-two-zinc-finger (C2H2) DNA-binding domains and are homologous, indicating similarities in the DNA sequences recognized by each EGR protein and thus can overlap. In the purpose and function of EGR1, EGR2, and EGR3, but not EGR4, exhibit interaction domains with the transcriptional co-repressors NGFI-A-1/2 (NAB1 and NAB2), in addition, exerting negative control on transcriptional activity. Upregulation of EGR13, EGR1, EGR2 and EGR3 proteins may lead to suppression of their transcriptional role, partly supported by in vivo experimental evidence 14,15.
EGR1 is common in humans and many other animal species. This gene is located on chromosome 5 at the 5q31 locus. The EGR1 protein acts as a transcription factor, a molecule that regulates gene expression by binding to DNA and regulating the activity of target genes. Thus, it is imperative to align the amino acid sequences of all human, rat, and mouse EGR protein due to different regulation, transcriptional regulation, reactivity, neural function between EGR protein and protein interactions8.
EGR1 expression is undetectable in the nervous system of both embryos16,17, during postnatal development and its expression continues to slowly increase until adulthood, namely on postnatal day 17 in the mouse brain18. The gradual increase in EGR1 expression, closely corresponds to the period of maximal N-methyl-D-aspartate (NMDA) response and coincides with the time of synaptic formation in the CA1 and hippocampal cortical regions, increasing long-term inducibility (LTP)18. Establishing a link between EGR1 expression and synaptic plasticity. In adulthood, EGR1 is widely expressed throughout the brain, which tends to control cognition, including the hippocampus6,19,20. Thus, EGR1 plays a crucial role in learning and memory, as its activity increases in brain regions involved in cognitive function.
EGR 1 And Synaptic Plasticity
The dentate hippocampus is an integral part of the brain involved in memory formation and storage. EGR 1 is critical in regulating tooth learning15. The signaling process of EGR1 expression and signal transmission to neuronal synapses involves several complex steps. The EGR1 signaling pathway is triggered by an external stimulus, such as new learning or interesting stimuli. This stimulus activates neurons in the brain and triggers a series of biochemical and electrochemical changes in the cells4. An external stimulus causes a change in the membrane potential of neurons, which triggers the release of neurotransmitters at presynaptic synapses. These neurotransmitters bind to receptors on the postsynaptic cell membrane and initiate signaling through signaling pathways. When a neurotransmitter binds to a receptor, it activates the receptor. Receptors are composed of protein subunits and have a central function in transmitting signals to the cell. Activation of the receptor triggers a series of biochemical and molecular changes in the cell. EGR1 expression is often accompanied by MAPK signaling pathway and the protein kinase A (PKA) pathway4.
Activation of the receptor triggers the activation of MAP kinase, which is responsible for the phosphorylation and activation of the Ras protein. Ras then activates a series of protein kinases, including MEK (MAPK/ERK kinase) and ERK (extracellular signal-regulated kinase). ERK then translocates to the cell nucleus and phosphorylates a transcription factor such as Elk-1, which interacts with the early growth response element (ERE) in the EGR1 gene promoter to initiate gene expression4. Activation of the receptor can also trigger activation of the PKA pathway. PKA phosphorylates transcription factors such as CREB (cAMP response element binding protein). Phosphorylation of CREB triggers interactions with transcriptional co-activators, including EGR1. This CREB-EGR1 complex binds to the ERE region of the EGR1 gene promoter to amplify the gene21.
After activation of signaling pathways, transcription factors such as Elk-1, CREB, and EGR1 move to the cell nucleus. In the cell nucleus, these factors interact with the ERE region of the EGR1 gene promoter. This interaction triggers the transcription and synthesis of EGR1 mRNA. The newly synthesized mRNA is then translated into the EGR1 protein in the cytoplasm of the cell. The EGR1 protein is then transported back to the cell nucleus where it functions as a transcription factor. After returning to the cell nucleus, the EGR1 protein binds to the ERE region of the target gene promoter. It modulates the transcriptional activity of target genes involved in memory formation and storage15.
EGR1 expression and regulation of its target genes influence synapse modification. This can include structural changes, such as the increase or elimination of synaptic spikes, as well as changes in the release of neurotransmitters and the sensitivity of the receptor15. Through these steps, the EGR1 signaling pathway influences gene expression and synaptic modifications that promote learning and memory processes in neurons. However, it is important to remember that these explanations are descriptive and the complexity of signaling mechanisms can vary depending on the context and type of stimulus.
EGR 1, Synaptic Plasticity and Exercise
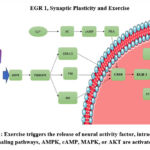 |
Figure 1: Exercise triggers the release of neural activity factor, intracellular signaling pathways, AMPK, cAMP, MAPK, or AKT are activated4 |
Physical exercise can influence gene expression through complex signaling pathways22. Intense and regular physical exercise can provide the body with external stimulation. This stimulation can occur during aerobic activity or other stressful activities that affect the nervous system 23. Physical exercise activates the nervous system, especially the autonomic and central nervous systems24. This activation releases neurotransmitters and peptides that play a role in nerve signal transmission25.
During physical exercise, muscle activity increases, which can activate the AMPK. Biochemical changes in AMPK cells can be triggered by phosphorylation of transcription factors such as CREB. Physical exercise can also trigger MAPK pathways, such as the ERK (extracellular signal-regulated kinase) pathway. Activation of the MAPK pathway can trigger the phosphorylation of transcription factors involved in EGR1 expression, such as Elk-1. Activation of transcription factors such as CREB and Elk-1 can interact with the early growth response elements (ERE) of the EGR1 gene promoter. This initiates the process of transcription and synthesis of EGR1 mRNA. The newly synthesized EGR1 mRNA is then translated into EGR1 protein in the cytoplasm, then functions as a transcription factor after the EGR1 protein is transported to the cell nucleus. When the EGR1 protein returns to the cell nucleus, it binds to the ERE region of the target gene promoter. This gene functions to regulate target genes involved in regulated physiological responses and adaptation to physical conditions exercise4 (Figure 1).
Physical activity can trigger the expression of the EGR 1 gene to increase blood flow and neurotrophic factors, such as brain-derived neurotrophic factor (BDNF) which plays a role in nerve growth and development26. This neurotrophic factor can stimulate EGR1 expression in the hippocampus, which regulates synaptic plasticity, an important mechanism for learning and memory15. Exercise can also have neuroprotective effects on the brain, including the hippocampus. Increased expression of EGR1 may play a role in protecting and restoring brain function disrupted by oxidative stress or cell damage27,28.
Exercise can increase EGR1 gene expression in the brain as a whole. This may occur because physical activity increases blood flow and neurotrophic factors, such as BDNF, which can stimulate EGR1 expression in neurons23,26. Regular exercise has been shown to protect nerves and improve nerve health. Several studies in which EGR1 expression can promote neuroprotection and recovery of impaired neuronal function23. Exercise can also increase neural plasticity, namely the ability of the nervous system to adapt and make new connections between neurons29. Although direct studies have not examined the effect of exercise on EGR1 expression in plastic neurons, higher EGR1 expression may play a role in the regulation of plastic neurons and the formation of new synaptic connections30,31.
EGR1 expression, and neurological health greatly requires further research to understand the precise relationship between exercise. Variables such as exercise type, duration, intensity, and individual characteristics can also affect cognitive function. A recent study and a more comprehensive review of the literature may provide additional information on the effect of exercise on EGR1 expression in neurons.
Conclussion
The expression of EGR1 can enhance synaptic plasticity in the brain, leading to improved cognitive function. Exercise is also a factor that can influence EGR1 expression.
Acknowledgement
This research was supported by the Indonesian University of Education and Padjadjaran University.
Conflict of Interest
The authors declare that they have no conflict of interest.
Funding Support
This study does not receive any funding from any type of institution.
References
- Herdegen T, Leah JD. Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res Brain Res Rev. 1998;28(3):370-490. doi:10.1016/s0165-0173(98)00018-6
CrossRef - Bahrami S, Drabløs F. Gene regulation in the immediate-early response process. Adv Biol Regul. 2016;62:37-49. doi:10.1016/j.jbior.2016.05.001
CrossRef - Guzowski JF, Setlow B, Wagner EK, McGaugh JL. Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos, and zif268. J Neurosci Off J Soc Neurosci. 2001;21(14):5089-5098. doi:10.1523/JNEUROSCI.21-14-05089.2001
CrossRef - Shan J, Dudenhausen E, Kilberg MS. Induction of early growth response gene 1 (EGR1) by endoplasmic reticulum stress is mediated by the extracellular regulated kinase (ERK) arm of the MAPK pathways. Biochim Biophys acta Mol cell Res. 2019;1866(3):371-381. doi:10.1016/j.bbamcr.2018.09.009
CrossRef - Lim CP, Jain N, Cao X. Stress-induced immediate-early gene, egr-1, involves activation of p38/JNK1. Oncogene. 1998;16(22):2915-2926. doi:10.1038/sj.onc.1201834
CrossRef - Beckmann AM, Matsumoto I, Wilce PA. AP-1 and Egr DNA-binding activities are increased in rat brain during ethanol withdrawal. J Neurochem. 1997;69(1):306-314. doi:10.1046/j.1471-4159.1997.69010306.x
CrossRef - Cepeda CCP, Lodovico A, Fowler N, Rodacki ALF. Effect of an Eight-Week Ballroom Dancing Program on Muscle Architecture in Older Adults Females. J Aging Phys Act. 2015;23(4):607-612. doi:10.1123/japa.2014-0101
CrossRef - O’Donovan KJ, Tourtellotte WG, Millbrandt J, Baraban JM. The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci. 1999;22(4):167-173. doi:10.1016/s0166-2236(98)01343-5
CrossRef - Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987;238(4828):797-799. doi:10.1126/science.3672127
CrossRef - Sukhatme VP, Cao XM, Chang LC, et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53(1):37-43. doi:10.1016/0092-8674(88)90485-0
CrossRef - Lemaire P, Revelant O, Bravo R, Charnay P. Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc Natl Acad Sci U S A. 1988;85(13):4691-4695. doi:10.1073/pnas.85.13.4691
CrossRef - Christy BA, Lau LF, Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with “zinc finger” sequences. Proc Natl Acad Sci U S A. 1988;85(21):7857-7861. doi:10.1073/pnas.85.21.7857
CrossRef - Gashler AL, Swaminathan S, Sukhatme VP. A novel repression module, an extensive activation domain, and a bipartite nuclear localization signal defined in the immediate-early transcription factor Egr-1. Mol Cell Biol. 1993;13(8):4556-4571. doi:10.1128/mcb.13.8.4556-4571.1993
CrossRef - Timmons JA, Jansson E, Fischer H, et al. Modulation of extracellular matrix genes reflects the magnitude of physiological adaptation to aerobic exercise training in humans. BMC Biol. 2005;3:19. doi:10.1186/1741-7007-3-19
CrossRef - Duclot F, Kabbaj M. The Role of Early Growth Response 1 (EGR1) in Brain Plasticity and Neuropsychiatric Disorders. Front Behav Neurosci. 2017;11:35. doi:10.3389/fnbeh.2017.00035
CrossRef - McMahon AP, Champion JE, McMahon JA, Sukhatme VP. Developmental expression of the putative transcription factor Egr-1 suggests that Egr-1 and c-fos are coregulated in some tissues. Development. 1990;108(2):281-287. doi:10.1242/dev.108.2.281
CrossRef - Crosby SD, Veile RA, Donis-Keller H, et al. Neural-specific expression, genomic structure, and chromosomal localization of the gene encoding the zinc-finger transcription factor NGFI-C. Proc Natl Acad Sci U S A. 1992;89(10):4739-4743. doi:10.1073/pnas.89.10.4739
CrossRef - Herms J, Zurmöhle U, Schlingensiepen R, Brysch W, Schlingensiepen KH. Developmental expression of the transcription factor zif268 in rat brain. Neurosci Lett. 1994;165(1-2):171-174. doi:10.1016/0304-3940(94)90737-4
CrossRef - Herdegen T, Kovary K, Buhl A, Bravo R, Zimmermann M, Gass P. Basal expression of the inducible transcription factors c-Jun, JunB, JunD, c-Fos, FosB, and Krox-24 in the adult rat brain. J Comp Neurol. 1995;354(1):39-56. doi:10.1002/cne.903540105
CrossRef - Knapska E, Kaczmarek L. A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog Neurobiol. 2004;74(4):183-211. doi:10.1016/j.pneurobio.2004.05.007
CrossRef - Benito E, Valor LM, Jimenez-Minchan M, Huber W, Barco A. cAMP response element-binding protein is a primary hub of activity-driven neuronal gene expression. J Neurosci Off J Soc Neurosci. 2011;31(50):18237-18250. doi:10.1523/JNEUROSCI.4554-11.2011
CrossRef - Pesce M, La Fratta I, Paolucci T, et al. From Exercise to Cognitive Performance: Role of Irisin. Appl Sci. 2021;11(15). doi:10.3390/app11157120
CrossRef - Clark PJ, Bhattacharya TK, Miller DS, Rhodes JS. Induction of c-Fos, Zif268, and Arc from acute bouts of voluntary wheel running in new and pre-existing adult mouse hippocampal granule neurons. Neuroscience. 2011;184:16-27. doi:10.1016/j.neuroscience.2011.03.072
CrossRef - Tsuchida R, Yamaguchi T, Funabashi D, Koumi Y, Kita I, Nishijima T. Exercise type influences the effect of an acute bout of exercise on hippocampal neuronal activation in mice. Neurosci Lett. 2022;783:136707. doi:10.1016/j.neulet.2022.136707
CrossRef - Kim S-H, Kim H, Kim S-S, et al. The influence of age on the treadmill exercise-induced c-Fos expression in the hippocampus of rats. Neurosci Res Commun. 2004;35(1):41-50. doi:https://doi.org/10.1002/nrc.20018
CrossRef - Cefis M, Prigent-Tessier A, Quirié A, Pernet N, Marie C, Garnier P. The effect of exercise on memory and BDNF signaling is dependent on intensity. Brain Struct Funct. 2019;224(6):1975-1985. doi:10.1007/s00429-019-01889-7
CrossRef - Worley PF, Christy BA, Nakabeppu Y, Bhat R V, Cole AJ, Baraban JM. Constitutive expression of zif268 in neocortex is regulated by synaptic activity. Proc Natl Acad Sci U S A. 1991;88(12):5106-5110. doi:10.1073/pnas.88.12.5106
CrossRef - Belviranlı M, Okudan N. Exercise Training Protects Against Aging-Induced Cognitive Dysfunction via Activation of the Hippocampal PGC-1α/FNDC5/BDNF Pathway. Neuromolecular Med. 2018;20(3):386-400. doi:10.1007/s12017-018-8500-3
CrossRef - Chatzi C, Zhang Y, Hendricks WD, et al. Exercise-induced enhancement of synaptic function triggered by the inverse BAR protein, Mtss1L. Elife. 2019;8. doi:10.7554/eLife.45920
CrossRef - Maddox SA, Monsey MS, Schafe GE. Early growth response gene 1 (Egr-1) is required for new and reactivated fear memories in the lateral amygdala. Learn Mem. 2011;18(1):24-38. doi:10.1101/lm.1980211
CrossRef - Williams JM, Beckmann AM, Mason-Parker SE, Abraham WC, Wilce PA, Tate WP. Sequential increase in Egr-1 and AP-1 DNA binding activity in the dentate gyrus following the induction of long-term potentiation. Brain Res Mol Brain Res. 2000;77(2):258-266. doi:10.1016/s0169-328x(00)00061-9
CrossRef








