Esra’a Jebreel Ibrahim Abu-Shoura1 , Shuaibu Abdullahi Hudu1*
, Shuaibu Abdullahi Hudu1* and Tasneem Farouq AL- Quadan2
and Tasneem Farouq AL- Quadan2
1Department of Basic Medical and Dental Sciences, Faculty of Dentistry, Zarqa University, Zarqa, Jordan.
2Department of Microbiology and Immunology, University of Louisville, School of Medicine, 500 S Preston St, Louisville, KY, United States.
Corresponding Author E-mail: shudu@zu.edu.jo
DOI : https://dx.doi.org/10.13005/bpj/2928
Abstract
Background The biofilm phenomenon represents a prevalent mode of microbial life in nature which is characterized by cells irreversibly attaching to surfaces or each other and getting embedded in a matrix of extracellular polymeric substances. Object This study aimed to identify and characterize the genes associated with the common bacterial species responsible for biofilm formation in the catheters of hospitalized patients. Method Different bacterial strains were collected from catheterised patients at three local Jordanian hospitals for biofilm formation. The isolates were identified using Gram stain and Remel Rapid test. Biofilm formation was detected using the Tube method and tissue culture plate method. Result The presence of fimA and csgD genes was detected by polymerase chain reaction (PCR). Gram-negative bacteria species were isolated on the urethral catheters and the result shows the majority of the isolates were E. coli (40%), followed by K. pneumonia (27%). In comparison, the least was Citrobacter sp (2.6%). Similarly, some Gram-positive bacteria were also identified such as Staphylococcus aureus (4%) and Staphylococcus epidermidis (2.6%). K. pneumonia is strongly associated with biofilm formation (45%) followed by E. coli (33%). Conclusion Biofilm-associated genes, fimA and csgD were detected in all biofilm-positive samples containing the F plasmid, while csgD was detected in all biofilm-negative samples. Biofilm formation tends to be a prevalent process in patients included in this study and may cause dangerous complications in the patients especially in the elderly due to prolonged catheterization periods.
Keywords
Biofilm; Catheter; Genes; Jordan; Urinary infection
Download this article as:| Copy the following to cite this article: Abu-Shoura E. J. I, Hudu S. A, AL- Quadan T. F. Exploring Genetic and Phenotypic Factors Contributing to Urethral Catheter Biofilm Formation in Hospitalised Patients in Jordan. Biomed Pharmacol J 2024;17(2). |
| Copy the following to cite this URL: Abu-Shoura E. J. I, Hudu S. A, AL- Quadan T. F. Exploring Genetic and Phenotypic Factors Contributing to Urethral Catheter Biofilm Formation in Hospitalised Patients in Jordan. Biomed Pharmacol J 2024;17(2). Available from: https://bit.ly/3VRTnTE |
Introduction
Biofilm is a complex microbial system consisting of microorganisms attaching to surfaces; these microorganisms are embedded in a protective matrix characterized as a polysaccharide matrix produced by alginate polymerases 1. Bacteria form microcolonies and surround themself with an extracellular polymeric substance (EPS), between these colonies water-filled channels control the influx and efflux of nutrients and waste 2. In nature, bacteria live in polymicrobial ecosystems either as free planktonic floating cells suspended in a liquid medium or as a sessile community attached to surfaces3. Most bacteria tend to live in the attachment sessile mode attached to either biotic or abiotic surfaces such as aquatic systems living tissues, indwelling medical devices and industrial water system piping under certain circumstances such as pH, nutrient level, ionic strength and temperature4. Biofilm diseases are mainly associated with implant medical devices like catheters, valves, and joint prostheses5. However sometimes bacteria don’t need a device to form a biofilm layer; for example, lung infections, oral cavity infections, fibrosis and contact lenses could be associated with biofilm formation6. Usually, biofilm is associated with nosocomial infection which originates from the skin of the patients, health care workers and tap water. More than 35 % of nosocomial infections in the USA are urinary tract infections 7. Urinary Tract Infection (UTI) is the most common acquired infection by microbial invasion of the genitourinary tract 8. The infection extends from the urethra to the bladder until it arrives in the renal cortex of the kidneys. The risk of this type of infection increases with catheter usage; since catheters get intensively colonized with bacteria, this allows the bacteria to reach the bladder causing the infection more than the ordinary UTI 9. The bacteria can cause severe damage to the bladder mucosal tissue the enhanced invasion of the bacteria over time causes kidney failure 10. Catheter-acquired infection is classified according to the duration of catheterization in situ from long-term which is assumed to be more than 30 days, while the short-term is less than 30 days 11. UTI Patients with short-term indwelling catheters usually acquire asymptomatic infection, however, the infection develops dramatically once catheterization lasts more than a month. Many diseases are associated with long-term catheterization term such as kidney renal inflammation, acute prostatitis and renal stones 11,12. Whenever the catheter could be removed the physician should make this decision, the catheter must be the last choice.
Studying biofilm formation is very important, especially in the medical field; the importance stems from the necessity to avoid the complications of biofilm formation. In the Middle East, more than 8% of infections are nosocomial infections according to the World Health Organization which are related to the decline in health workers’ hygiene or other contamination sources13. More than 40% of these infections are acquired urinary tract infections associated with catheterization13,14. Many diseases are related to biofilm formation such as cystic fibrosis, otitis media and urinary tract infection. Biofilm is formed by several types of microorganisms such as Escherichia coli, Klebsiella pneumoniae, Candida albicans and Staphylococcus aureus 15. This study aimed to identify the common bacterial species responsible for biofilm formation in the catheters of hospitalized patients and determine which genes in these organisms are partially responsible for biofilm formation.
Methodology
To investigate the biofilm formation, urine catheters were collected from three governmental hospitals in Jordan; AL-Basher hospital, Prince Faysal Hospital and Zarka Governmental Hospital, the catheterisation period was at least four days.
Sample Identification
The urine catheters were collected directly from patients; the tip of the catheter was cut with a sterile sharp blade and scissors and placed directly in a sterile urine cup. The interior of the collected catheters was scraped with cotton swabs and cultured in enrichment media such as Luria-Bertani (LB) broth and was incubated at 37ºC for 24 hr. The suspensions were routinely cultured on Mac Conkey agar and blood incubated at 37ºC for 24h. Bacteria were classified into Gram-positive or Gram-negative using the Gram stain method as described previously16. Gram-negative bacteria were identified using a special kit (RemelRapID one) which consists of 18 biochemical reaction wells; the bacterial diluted sample reacts with each reagent. All bacteria were preserved in 20% glycerol at – 20 ºC until further use.
Biofilm Formation Assay.
The biofilm assay was performed in two methods. Tube Method and Tissue Culture Plate and the following steps explain both procedures as described previously 17-19.
Growing a Film
Bacteria were growing in enrichment media (LB) overnight.The overnight suspension was diluted 1:100 into fresh media, 100 µl of the suspension was added to 96 well plate and 1ml of the suspension was added to the test tube; both the tubes and plate were incubated for 24 hrs. at 37 ºC.
Staining the Biofilm
After incubation, the plate and the tubes were washed with water two times to discard the planktonic bacteria. 125 µl of 0.1% of crystal violet (CV) was added to the microtiter plate for staining, and 1ml+50µl 1% of CV was added to test tubes for tube staining both were incubated for 15 min. After that, the microtiter plate and the tubes were rinsed 3 times with water to rid the plate of excess cells and dye.The plate and tubes were dried for a few hours or overnight.
Quantifying the biofilm
To quantify the biofilm adhesion rate, 30% acetic acid was added to each tube for 10 min to solubilize the CV staining. The optical density of the adherent stained biofilm was measured at 570 nm.
Genomic DNA Extraction
Bacteria were grown in 5 ml of LB media at 37°C for one night. After centrifuging the culture at 14,000 rpm for one minute, the supernatant was thrown away and the pellet was kept. It was mixed with 500 µl of processing buffer (10 µM Tris-HCL, 10 µM EDTA, 50M NaCl, and 2% SDS) and 4 µL of proteinase K. After that, this blend was kept in a water bath at 56°C for two hours. The samples were spun at 14000 rpm for 3 minutes after being left to sit for a while. Another mix of 500 µl of processing buffer, 4 µl of proteinase K, and 7 µl of dithiothenitol (DTT) was added to the pellet. The mixture was then left to sit overnight at 56°C.
After being left to sit overnight, 500 µl of phenol was added to each tube. The tubes were then left at room temperature for 10 minutes before being centrifuged at 300 rpm for 5 minutes. After the supernatant was made, it was put in a new tube and 50 µl of 3M sodium acetate and 1 ml of cold isopropanol were added. After being kept at -20°C for 20 minutes, this mixture was spun at 3000 rpm for 10 minutes. Carefully, the supernatant was taken away, and 500 µl of 70% ethanol was used to wash the pellet. The ethanol was thrown away after another 3 minutes of spinning at 3000 rpm, and the pellet was left to dry in the air. Finally, the pellet was mixed again in 50 µl of Tris/EDTA buffer (TE buffer), and 0.8% agarose gel electrophoresis was used to look at the DNA that had been recovered.
Plasmid DNA Extraction.
A commercial kit from OMEGA bio-tek was used to get plasmid DNA from E. coli. To begin, E. coli samples were grown in 5 ml of LB broth in 10 ml tubes to make sure they had enough air. The tubes were then heated to 37oC and shaken for 12 to 16 hours. After the incubation time, the cultured broth was centrifuged at 10,000 rpm for 10 minutes to separate the bacterial cells in the pellet from the supernatant. The pellet was kept and the liquid was thrown away. After that, 250 µl of solution I (RNase A solution) was added to the pellets. Then, 250 µl of solution II was added. 350 µl of solution III was added to this mixture, and it was mixed well until a white residue formed. After that, the mixture was spun at 13,000 rpm for 10 minutes. After getting the clear residue, it was put on a DNA mini-column and spun at 13,000 rpm for 10 minutes. The column was then turned on and 700 µl of alcohol was added. It was then spun at 13,000 rpm for 2 minutes. The last step was to add 50 µl of elution solution to the column. It was then spun at 13,000 rpm for 2 minutes. After the plasmid DNA was removed, it was tested using 1% agarose gel electrophoresis.
Polymerase Chain Reaction (PCR) for Gene Detection
Polymerase Chain Reaction (PCR) was performed to determine the presence or absence of certain genes known to be responsible for biofilm formation. The bacterial genera selected for PCR were E. coli and K. pneumonia. Two genes were selected for E. coli (fimA and csgD), and one (mrkD) for K. pneumonia based on previous reports 20,21. E. coli PCR condition and protocol; 5µL of 1.9 µg/µl DNA plasmid extract or 5.08 µg/µl DNA genomic extract of E. coli isolated were added to 12.5 µL of 2× master mix, 1 µL of forward primer (100µM) and 1 µL of reverse primer (100 µM ) (Table 1), 5.5 µL H2O in a total volume of 25 µL. DNA amplification was performed according to the following conditions in Bio-Rad thermocycler; 94 ºC for 2 min, followed by 35 cycles of 20s at 94 ºC, 30s at 56ºC, 45s at 72 ºC, and final extension for 5 min at 72 ºC. The PCR amplicons as shown in Table 1 were examined on 2% agarose gel electrophoresis. DNA amplification was performed according to the following conditions in the (Bio-Rad) thermocycler. 94 ºC for 2 min, 10 initial cycles at 94 ºC for 10s, 30s at 63 ºC and 15 min at 68 ºC, followed by 20 cycles of 10s at 94 ºC, 30s at 63 ºC and 15 min plus 20s for each new cycle at 72 ºC, the final elongation step was 7 min at 72 ºC. PCR were amplicons examined on 2% agarose gel electrophoresis.
Table 1: Primes nucleotides sequences used in PCR amplification of E. coli fim A and csgD gene22,23
|
Organism |
Primers |
Amplicon Size |
Reference |
|
E. coli |
fim A F: 5′-GTTAGGACAGGTTCGTACCGCAT-‘3 fimA R: 5′-AAATAACGCGCCTGGAACGAATG-‘3 |
315bp |
22 |
|
csgD F: 5′-CGCGAATTCTCGCTGGCAATTACAGG-‘3 csgD R: 5-‘CGCGGATCCGCTGATGAACAACGAAC-‘3 |
480bp |
23 |
Ethical Approval
The study was approved by the Hashemite University Institutional Review Board (IRB) on 20/05/2014, session number (2014/2013/7/36) with reference No. 1404123/440/1
Results
Urine Catheter culture
One hundred and thirty-five (135) samples were collected from patients’ urinary catheters. Bacterial growth was observed in 99 (73%) out of the 135 isolates. The 99 isolates were classified into Gram-positive and Gram-negative based on Gram stain and biochemical tests, 24 samples out of 99 showed mixed growth as such they were excluded from the study. The (RemelRapID ONE) kit was used to identify the different genera. Nine different Gram-negative and Gram-positive bacterial genera were isolated from the tips of the catheters. E. coli was the major pathogen in the three Jordanian hospitals 30/75 (40%), followed by K. pneumonia 20/75 (27%), Pseudomonas 7/75 (9%), Enterobactersp 6/75 (8%), Proteus sp 3/75 (4%), S. aureus 3/75 (4%), Shigellasp 2/75 (2.6%), Citrobactersp 2/75 (2.6%), and S. epidermidis 2/75 (2.6%), 43/75 (55%) of isolated samples were isolated from female patients.
Biofilm Detection Assay
All samples were applied to the biofilm colourimetric assay, and classified as non-adherent, weakly, moderately and strongly. The cut-off OD was the negative control (ODc = 0.129 nm), the OD of the non-adherent ≤ ODc. Weakly adherent sample OD should be ODc ˂ OD ≤ 2×ODc (0.258). The moderate adherent samples OD should be 2× ODc ˂ OD ≤ 4× ODc (0.516). The strongly adherent sample OD should be 4×ODc ˂ OD (0.516), and E. coli DH5α used as a negative control. The OD values using tissues culture plate (TCP) showed that E. coli 11/30 (33%) tend to form biofilm. In K. pneumonia 12/20 (60%) of samples have the strong to moderate ability to form the biofilm.
Table 2: Shows the different isolates’ biofilm formation ability using the tube method and Tissue Culture Plate method.
|
Biofilm formation |
Strongly adherent (+++) |
Moderate adherent (++) |
Weakly adherent (+) |
Non-adherent (0) |
Total |
||||
|
TM |
TCP |
TM |
TCP |
TM |
TCP |
TM |
TCP |
||
|
E. coli |
5 |
5 |
10 |
6 |
4 |
4 |
11 |
15 |
30 |
|
K. pneumonia |
9 |
10 |
7 |
2 |
1 |
4 |
3 |
4 |
20 |
|
Pseudomonas sp |
– |
4 |
– |
0 |
– |
0 |
– |
3 |
7 |
|
Proteus sp |
– |
2 |
– |
0 |
– |
0 |
– |
1 |
3 |
|
Shigella sp |
– |
0 |
– |
0 |
– |
0 |
– |
2 |
2 |
|
Cirtrobacter sp |
– |
0 |
– |
0 |
– |
0 |
– |
2 |
2 |
|
Enterobacter sp |
– |
2 |
– |
0 |
– |
0 |
– |
4 |
6 |
|
S. aureus |
– |
2 |
– |
0 |
– |
1 |
– |
0 |
3 |
|
S. epidermidis |
– |
1 |
– |
0 |
– |
0 |
– |
1 |
2 |
TM: tube method TCP: tissue culture plate
PCR for Biofilm Formation Genes
In this study, two genes are directly involved in biofilm formation (fimA and csgD). All E. coli samples that were phenotypically positive for biofilm formation contained (fimA, csgD) genes; fimA was detected in the plasmid, as shown in Figure 1, while csgD was detected in the genomic DNA, as shown in Figure 2. A positive control confirmed the gene amplification for fimA and csgD genes of E. coli ATCC 25922. The products were analyzed using electrophoresis, and the gel results are shown in Figure 3. The phenotypic and genotypic characteristics of E. coli and K. pneumonia are summarized in Table 3.
 |
Figure 1: PCR Amplification products of fimA gene. Amplified products were analyzed by electrophoresis on 2% ethidium bromide-stained agarose gel. Lane L: |
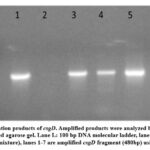 |
Figure 2: PCR Amplification products of csgD. Amplified products were analyzed by electrophoresis on 2% ethidium bromide-stained agarose gel. Lane L: |
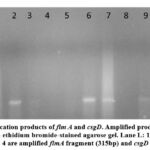 |
Figure 3: PCR Amplification products of fim A and csgD. Amplified products were analyzed by electrophoresis on 1% ethidium bromide-stained agarose gel. Lane L: |
Table 3: E. coli and K. pneumonia phenotypic and genotypic characteristics
|
Types of E. coli Serotype |
Biofilm (TM) |
Biofilm (TCP) |
F-Plasmid |
fimA |
csgD |
|
01 |
+++ |
+++ |
+ |
+ |
+ |
|
02 |
++ |
++ |
+ |
+ |
+ |
|
03 |
++ |
++ |
+ |
+ |
+ |
|
04 |
++ |
+ |
+ |
+ |
+ |
|
05 |
++ |
++ |
+ |
+ |
+ |
|
06 |
+++ |
+++ |
+ |
+ |
+ |
|
07 |
+++ |
+++ |
+ |
+ |
+ |
|
08 |
+++ |
+++ |
+ |
+ |
+ |
|
09 |
++ |
+ |
+ |
+ |
+ |
|
10 |
++ |
++ |
+ |
+ |
+ |
|
11 |
++ |
++ |
+ |
+ |
+ |
|
12 |
++ |
++ |
+ |
+ |
+ |
|
13 |
++ |
+ |
+ |
+ |
+ |
|
14 |
+++ |
+++ |
+ |
+ |
+ |
|
15 |
++ |
+ |
+ |
+ |
+ |
|
16 |
+ |
– |
+ |
+ |
+ |
|
17 |
+ |
– |
+ |
+ |
+ |
|
18 |
+ |
– |
+ |
+ |
+ |
|
19 |
+ |
– |
+ |
+ |
+ |
|
20 |
– |
– |
+ |
+ |
+ |
|
21 |
– |
– |
– |
– |
+ |
|
22 |
– |
– |
– |
– |
+ |
|
23 |
– |
– |
– |
– |
+ |
|
24 |
– |
– |
– |
– |
+ |
|
25 |
– |
– |
– |
– |
+ |
|
26 |
– |
– |
– |
– |
+ |
|
27 |
– |
– |
– |
– |
+ |
|
28 |
– |
– |
– |
– |
+ |
|
29 |
– |
– |
– |
– |
+ |
|
30 |
– |
– |
– |
– |
+ |
|
Types of K. pneumonia Strain |
Biofilm (TM) |
Biofilm (TCP) |
F-Plasmid |
fimA |
csgD |
|
Kp1 |
++ |
++ |
– |
– |
– |
|
Kp 2 |
++ |
+ |
– |
– |
– |
|
Kp 3 |
++ |
+ |
– |
– |
– |
|
Kp 4 |
+++ |
+++ |
– |
– |
– |
|
Kp 5 |
++ |
++ |
– |
– |
– |
|
Kp 6 |
++ |
+ |
– |
– |
– |
|
Kp 7 |
++ |
+++ |
– |
– |
– |
|
Kp 8 |
+++ |
+++ |
– |
– |
– |
|
Kp 9 |
+++ |
+++ |
– |
– |
– |
|
Kp 10 |
++ |
+ |
– |
– |
– |
|
Kp 11 |
+++ |
+++ |
– |
– |
– |
|
Kp 13 |
+++ |
+++ |
– |
– |
– |
|
Kp 14 |
+++ |
+++ |
– |
– |
– |
|
Kp 15 |
+++ |
+++ |
– |
– |
– |
|
Kp 16 |
+++ |
+++ |
– |
– |
– |
|
Kp 17 |
+ |
– |
– |
– |
– |
|
Kp 18 |
– |
– |
– |
– |
– |
|
Kp 19 |
– |
– |
– |
– |
– |
|
Kp 20 |
– |
– |
– |
– |
– |
Kp: Klebsiella pneumonia‘ TM: tube method TCP: tissue culture plate (+): strength of adherence, (-): No adherence
Discussion
Bacterial adhesion has been considered a virulence factor contributing to infections associated with indwelling medical devices, particularly catheters. This ability to form the biofilm helps to resist the host’s immune response and is considered the main factor responsible for chronic infection. In this study, we investigated the ability of isolates to form biofilm. It included 75 specimens, both Gram-positive and Gram-negative bacteria. This study found that E. coli is the most prevalent organism (40%), followed by K. pneumonia (26%) associated with catheter-associated urinary tract infection, which is in agreement with the findings of other researchers 24-26. More than 50% of the samples were isolated from the ICU, indicating that patients with serious diseases are more affected by nosocomial acquired urinary tract infections, as reported previously 27-29.
In this study, the samples were collected from both males and females, and we found that more than 57% of the isolates were from female patients. This might not be unrelated to the fact that UTI is more common in females than males due to their short urethra 30-32. Catheter insertion should be done under sterile conditions using iodine or specific insertion gel, and in the case of female patients positioning of the catheter needs to be checked to ensure that it is correctly positioned in the urethra. During the insertion procedure, the bacteria could get access to the urinary tract even with sterilization. Male patients are not subjected to this process; moreover, using condom catheters for males decreases the catheterization-associated infection33,34. Studies showed that E. coli is still heading the urinary tract infection causative list 35,36 which is in line with the findings of this study. On the other hand, the number of Gram-positive isolates was very low which indicates that the contamination sources were not related to the skin of health workers or patients, Staphylococcus epidermidis constituted only 2.5% the of isolates which concurs with previous studies 37,38. Fifty-seven per cent of the isolates were biofilm producers while 43% of the isolates were non-biofilm producers, 45% of K. pneumonia isolates were strong biofilm producers and 33 % of E. coli isolates were strong biofilm producers.
The ability of isolates to form biofilm indicates that patients in Jordanian hospitals are exposed to biofilm formation complications just like patients in other health settings globally 25,39,40. Since E. coli and K. pneumonia were the most frequently isolated species, we investigated the presence of certain genes which are known to be involved in biofilm formation. These genes fimA and csgD are regulatory genes for pili and curli production which are involved essentially with biofilm formation in E. coli 41-43. In this study, the fimA gene and csgD genes were detected in all E. coli biofilm-producing isolates, which confirmed that the catheter-isolated organisms can form biofilm; and more than 50% of the patients were under antibiotic treatment but were not responding to it. Thus, patients would suffer from catheter-acquired infection complications which include fever, urine burning, back pain, and pyelonephritis and it could ultimately lead to kidney failure septicemia or even death24,44,45. Antibiotics fight side by side with the immune system to overstep the bacterial infection and survive without any server damage, thus any paralysis of the antimicrobial agent job would weaken the body effort. Only 26% of the collected samples were free of bacterial growth which indicates that nosocomial infection shows a prevalent mode and this in turn increases the risk of indwelling urinary catheter-related disease45-48.
Conclusion
In Jordanian hospitals, E. coli was the most common infectious organism followed by K. pneumonia. The biofilm formation process represents a prevalent mode in Jordanian patients included in this study. The conjugative bacteria which have an F plasmid tend to form a multilayer biofilm while non-conjugative can’t form the biofilm even if the genes are present in another plasmid. Thetime and csgD adhesion genes were detected in all phenotypically positive for biofilm in the presence of optimum environmental conditions. The key to controlling the biofilm formation phenomenon is applying a prevention system in hospitals by controlling the environmental conditions to protect patients by providing them with probable nutrition, vaccination, and antimicrobial agents. Educating the health workers including doctors, nurses, therapists and the cleaning crew is important to controlling the nosocomial infection. This education must include: what is meant by biofilm, its complications, what the preventive actions should be done, what is the corrective action if biofilm was detected and what is the personal and economical benefit that we can gain in preventing such process. However, this study is limited by its sample size and details of antibiotic resistance profiles, which could be crucial for understanding treatment challenges.
Acknowledgement
The researchers appreciate Zarqa University for supporting this research publication
Conflict of interest
Authors declare no conflict of interest
Funding Source
No funding was received for this study
Authors’ contributions
Data curation: EIA; Formal analysis: SAH; Methodology: EIA, SAH; Project administration: TFA; Writing original draft: EIA, TFA; Writing review & editing: SAH, TFA, EIA; All authors approved the final submission.
References
- Parrilli E, Tutino ML, Marino G. Biofilm as an adaptation strategy to extreme conditions. Rendiconti Lincei Scienze Fisiche e Naturali. 2022;33(3):527-536.
CrossRef - Mahto KU, Kumari S, Das S. Unraveling the complex regulatory networks in biofilm formation in bacteria and relevance of biofilms in environmental remediation. Critical Reviews in Biochemistry and Molecular Biology. 2022;57(3):305-332.
CrossRef - Wu Y, Fu C, Peacock CL, et al. Cooperative microbial interactions drive spatial segregation in porous environments. Nature Communications. 2023;14(1):4226.
CrossRef - Xia A, Chen H, Huang Y, Zhu X, Liao Q. Mathematical modelling of intercellular interactions within the biofilm. Trends in Microbiology. 2022; 4:S0966-842X..
- Shay R, Wiegand AA, Trail F. Biofilm formation and structure in the filamentous fungus Fusarium graminearum, a plant pathogen. Microbiology spectrum. 2022;10(4):e00171-00122.
CrossRef - Ch’ng J-H, Chong KK, Lam LN, Wong JJ, Kline KA. Biofilm-associated infection by enterococci. Nature Reviews Microbiology. 2019;17(2):82-94.
CrossRef - Devanga Ragupathi NK, Veeraraghavan B, Karunakaran E, Monk PN. Biofilm-mediated nosocomial infections and its association with antimicrobial resistance: Detection, prevention, and management. Frontiers in Medicine. 2022;9:987011.
CrossRef - Mancuso G, Midiri A, Gerace E, Marra M, Zummo S, Biondo C. Urinary Tract Infections: The Current Scenario and Future Prospects. Pathogens. 2023;12(4):623-623.
CrossRef - Panjwani DM, Lakhani SJ, Mehta SJ, Kikani KM, Shah KS. A comprehensive study of microbiological profile, risk factors and antibiotic sensitivity pattern of catheter associated urinary tract infection in a teaching hospital of Gujarat. Journal of Applied Biology and Biotechnology. 2021;9(5):83-8.
- Panjwani DM, Lakhani SJ, Mehta SJ, Kikani KM, Shah KS. A comprehensive study of microbiological profile, risk factors and antibiotic sensitivity pattern of catheter associated urinary tract infection in a teaching hospital of Gujarat. Journal of Applied Biology and Biotechnology. 2021 Sep 1;9(5):83-8.
- Mert D, Iskender G, Kolgelier S, Ertek M. Evaluation of risk factors, causative pathogens, and treatment in recurrent percutaneous nephrostomy catheter-related urinary tract infections in cancer patients. Medicine. 2023;102(14):e33002-e33002.
CrossRef - Wojciuk B, Majewska K, Grygorcewicz B, et al. The role of uropathogenic Escherichia coli adhesive molecules in inflammatory response-comparative study on immunocompetent hosts and kidney recipients. Plos one. 2022;17(5):e0268243.
CrossRef - Fried ED. Hospital-acquired infections. Inpatient Safety: A Case-based Innovative Playbook for Safer Care 2023 Oct 17 (pp. 183-198). Cham: Springer International Publishing.
CrossRef - Mong I, Ramoo V, Ponnampalavanar S, Chong MC, Wan Nawawi WNF. Knowledge, attitude and practice about catheter‐associated urinary tract infection (CAUTI) prevention: A cross‐sectional study. Journal of Clinical Nursing. 2022;31(1-2):209-219.
CrossRef - Kar M, Dubey A, Patel SS, Siddiqui T, Ghoshal U, Sahu C. Characteristics of Bacterial Colonization and Urinary Tract Infection after Indwelling of Double-J ureteral Stent and Percutaneous Nephrostomy Tube. Journal of global infectious diseases. 2022;14(2):75-80.
CrossRef - Mahon CR, Lehman DC. Textbook of diagnostic microbiology-e-book. Elsevier Health Sciences; 2022 Nov 2.
- Zmantar T, Kouidhi B, Miladi H, Mahdouani K, Bakhrouf A. A microtiter plate assay for Staphylococcus aureus biofilm quantification at various pH levels and hydrogen peroxide supplementation. The new Microbiologica. 2010;33(2):137-145.
- Lakshmanan SB, Baskaran AM, Kumpati P, Karuppanan S. Comparison of distinct approaches for screening the extended-spectrum beta-lactamase in Klebsiella pneumoniae and its biofilm formation on various catheter surfaces, antibiotic resistance profile before and after biofilm formation. Journal of Applied Biology and Biotechnology. 2023 Dec 26;12(1):165-71.
CrossRef - Juliana A, Leela KV, Gopinathan A, Jayaprakash T. Biofilm formation and its association with gram negative sepsis pathogenicity. Biomedical and Pharmacology Journal. 2022 Dec 20;15(4):2099-106.
CrossRef - Bellifa S, Hassaine H, Balestrino D, et al. Evaluation of biofilm formation of Klebsiella pneumoniae isolated from medical devices at the University Hospital of Tlemcen, Algeria. African Journal of Microbiology Research. 2013;7(49):5558-64.
CrossRef - Reisner A, Höller BM, Molin S, Zechner EL. Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. Journal of bacteriology. 2006;188(10):3582-3588.
CrossRef - Hernandes RT, Velsko I, Sampaio SC, et al. Fimbrial adhesins are produced by atypical enteropathogenic Escherichia coli strains. Applied and Environmental Microbiology. 2011;77(23):8391-8399.
CrossRef - Saldaña Z, Xicohtencatl‐Cortes J, Avelino F, et al. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environmental microbiology. 2009;11(4):992-1006.
CrossRef - Werneburg GT. Catheter-associated urinary tract infections: current challenges and prospects. Research and Reports in Urology. 2022:109-133.
CrossRef - Zou Z, Potter R, WH MT, et al. Escherichia coli catheter-associated urinary tract infections are associated with distinctive virulence and biofilm gene determinants. JCI Insight. 2022:e161461-e161461.
CrossRef - Firouzjaei MD, Hendizadeh P, Halaji M, et al. Quinolone Resistance in Biofilm-Forming Klebsiella pneumoniae-Related Catheter-Associated Urinary Tract Infections: A Neglected Problem. Jundishapur Journal of Microbiology. 2023;16(8):1-8
CrossRef - Bizuayehu H, Bitew A, Abdeta A, Ebrahim S. Catheter-associated urinary tract infections in adult intensive care units at a selected tertiary hospital, Addis Ababa, Ethiopia. Plos one. 2022;17(3):e0265102.
CrossRef - Rosenthal VD, Myatra SN, Divatia JV, et al. The impact of COVID-19 on health care–associated infections in intensive care units in low-and middle-income countries: International Nosocomial Infection Control Consortium (INICC) findings. International journal of infectious diseases. 2022;118:83-88.
CrossRef - Neela VK, Moh’d Noordin S, Noordin SA, Hudu SA, Zainudin Z. Isolation of Janthinobacterium lividum from early onset neonatal sepsis patients in Malaysia. African Health Sciences. 2019;19(3):2378-2389.
CrossRef - Rubi H, Mudey G, Kunjalwar R. Catheter-Associated Urinary Tract Infection (CAUTI). Age. 2022; 14(10):e30385-e30385.
CrossRef - Al-Hussaniy HA, Altalebi RR, Albu-Rghaif AH, Abdul-Amir AG. The use of PCR for respiratory virus detection on the diagnosis and treatment decision of respiratory tract infections in Iraq. Journal of Pure & Applied Microbiology. 2022 Mar 1;16(1): 201-206.
CrossRef - Awad MM, Elsahar MI. Asymptomatic Bacterial Infection in Pregnancy: A new update. Medical and Pharmaceutical Journal. 2022;1(2):41-51.
CrossRef - Tran C, Rodrigue D, Jones T, Bell N. Addressing CAUTIs with an External Female Catheter. American Journal of Nursing. 2023;123(1):50-55.
CrossRef - Steakin L. Male External Catheter Care and Maintenance. Rehabilitation Nursing Journal. 2023;48(6):186-189.
CrossRef - Mirzahosseini HK, Najmeddin F, Najafi A, et al. Correlation of biofilm formation, virulence factors, and phylogenetic groups among Escherichia coli strains causing urinary tract infection: A global systematic review and meta-analysis. Journal of Research in Medical Sciences. 2023;28(1):66.
CrossRef - Seid M, Markos M, Aklilu A, et al. Community-Acquired Urinary Tract Infection Among Sexually Active Women: Risk Factors, Bacterial Profile and Their Antimicrobial Susceptibility Patterns, Arba Minch, Southern Ethiopia. Infection and Drug Resistance. 2023:2297-2310.
CrossRef - Pouget C, Chatre C, Lavigne J-P, Pantel A, Reynes J, Dunyach-Remy C. Effect of Antibiotic Exposure on Staphylococcus epidermidis Responsible for Catheter-Related Bacteremia. International journal of molecular sciences. 2023;24(2):1547.
CrossRef - Theis TJ, Daubert TA, Kluthe KE, Brodd KL, Nuxoll AS. Staphylococcus aureus persisters are associated with reduced clearance in a catheter-associated biofilm infection. Frontiers in Cellular and Infection Microbiology. 2023;13:519.
CrossRef - Cangui-Panchi S, Ñacato-Toapanta A, Enríquez-Martínez L, Reyes J, Garzon-Chavez D, Machado A. Biofilm-forming microorganisms causing hospital-acquired infections from intravenous catheter: A systematic review. Current Research in Microbial Sciences. 2022;3:100175-100175.
CrossRef - Bhavsar RA, Gusani JK, Tank BY. A prospective study of biofilm production among catheterized and non-catheterized urinary bacterial isolates along with their antimicrobial susceptibility pattern in tertiary care hospital, Nadiad, Gujarat. National Journal of Physiology, Pharmacy and Pharmacology. 2023;14(4):0-0.
CrossRef - Prashanth K, Sawant AR, Panda L. Genetic basis of biofilm formation and their role in antibiotic resistance, adhesion, and persistent infections in ESKAPE pathogens. Understanding Microbial Biofilms. 2023:395-414.
CrossRef - Yan C-H, Chen F-H, Yang Y-L, et al. The Transcription Factor CsgD Contributes to Engineered Escherichia coli Resistance by Regulating Biofilm Formation and Stress Responses. International Journal of Molecular Sciences. 2023;24(18):13681.
CrossRef - Bhowmik A, Jana S, Bhattacharjee A, Dutta TK, Chauhan A. Escherichia coli biofilms. Application of Biofilms in Applied Microbiology: Elsevier; 2022:153-171.
CrossRef - Flores-Mireles A, Molina J, Kohler K, et al. Fibrinolytic deficiencies predispose hosts to septicemia from a catheter-associated UTI. Research Square. 2023:rs. 3. rs-3263501.
CrossRef - Al-hussaniy H, Kadhim Z. Methicillin-Resistant Staphylococcus aureus and New Delhi Metallo beta-lactamases-types of antibiotic resistance, methods of prevention. Medical and Pharmaceutical Journal. 2022 ;1(1):14-24.
CrossRef - Rosenthal VD, Yin R, Brown EC, et al. Incidence and risk factors for catheter-associated urinary tract infection in 623 intensive care units throughout 37 Asian, African, Eastern European, Latin American, and Middle Eastern nations: A multinational prospective research of INICC. Infection Control & Hospital Epidemiology. 2024;4:1-9.
CrossRef - Wang F, Wang X, Zhou S, Lou L, Wu Z. Development and validation of a risk prediction model for repeated indwelling urinary catheterization in patients with cervical cancer after surgery. Clinical Research Communications. 2023;6(1):5.
CrossRef - Alshrari AS, Hudu SA, Elmigdadi F, Imran M. The Urgent Threat of Clostridioides difficile Infection: A Glimpse of the Drugs of the Future, with Related Patents and Prospects. Biomedicines. 2023;11(2):426.
CrossRef








