Samira Daw Ameigaal1 , Almuthanna K. Alkaraki2*
, Almuthanna K. Alkaraki2* and May Fouad Sadiq2
and May Fouad Sadiq2
¹Department of Medical Laboratories, Higher Institute of Medical Sciences and Technology, Bani Waleed, Libya.
²Department of Biological Sciences, Faculty of Science, Yarmouk University, Irbid 21163, Jordan.
Corresponding Author E-mail: Alkaraki@yu.edu.jo
DOI : https://dx.doi.org/10.13005/bpj/2875
Abstract
MTHFD1 and CBS genes have key roles in folate and homocysteine metabolism. Many studies reported an association between cancer pathogenesis and different functional SNPs of genes involved in the main folate metabolism and the transsulfuration pathway. The current population-based, case-control study examined the association between MTHFD1 G1958A, MTHFD1 T401C, and the CBS 844ins68 insertion with breast cancer (BC) risk in Jordanian women. The studied population included 200 female BC subjects and age-matched female controls. The targeted genotypes MTHFD1 G1958A and MTHFD1 T401C were amplified via PCR followed by subsequent digestion with the proper restriction enzyme (PCR-RFLP), while the insertion/deletion of CBS844ins68bp was visualized and scored directly after gel electrophoresis. Results showed that the examined individual alleles and genotypes of MTHFD 1958A, MTHFD1 401C, and CBS844ins68bp per se were not associated with risk of BC compared with their wild-type genotypes and alleles.
Keywords
Breast cancer; CBS 844ins68; Folic acid; Jordan; MTHFD1 G1958A; MTHFD1 T401C
Download this article as:| Copy the following to cite this article: Ameigaal S. D, Alkaraki A. K, Sadiq M. F. Association of MTHFD1 G1958A, MTHFD1 T401C and CBS 844ins68bp with Breast Cancer in Jordan. Biomed Pharmacol J 2024;17(1). |
| Copy the following to cite this URL: Ameigaal S. D, Alkaraki A. K, Sadiq M. F. Association of MTHFD1 G1958A, MTHFD1 T401C and CBS 844ins68bp with Breast Cancer in Jordan. Biomed Pharmacol J 2024;17(1). Available from: https://bit.ly/49V7GuW |
Introduction
Female breast cancer (BC) is the most commonly worldwide diagnosed cancer, with a recorded 2.3 (11.7%) million new cases in 2020 1. In Jordan, BC is the most common cancer, accounting for 38.4% of all detected cancers in females (National Cancer Registry (JNCR., 2021). Despite the high prevalence of BC and the identification of various risk factors, the causes of BC are not completely clear. A critical step for interventions and good management of BC is to identify risk factors for its development. There are several known risk factors for breast cancer including genetic factors 2’3 which are highly heterogeneous 3’4. While only a minor fraction of these genetic factors arise from mutations in established high-penetrance susceptibility genes, the predominant share is believed to stem from common genetic variants, including single nucleotide polymorphisms (SNPs). Identification of the genetic risk factors for any type of cancer in any population is essential for these factors can be changed and adjusted to reduce the risk of cancers. Among those that can allow protective effect against cancers in particular are different common genetic variants, for example, single nucleotide polymorphisms (SNPs) in the genes encoding functional enzymes and coenzymes in the folate/ one-carbon metabolism and lifestyle risk factors 5’6’7’8. SNPs in genes related to the folate/ one-carbon metabolism alter gene function or regulation by changing the structure and the catalytic activities of the affected enzymes, and increasing DNA methylation in promoters of many genes and methylation reduction in promoters of other genes. These changes lead to activation of proto-oncogenes and inactivation of tumor suppressor genes 9’10 and alter cell division leading to aberrant chromosomal segregation and tumorigenesis 11’12’13’14. Different studies reported the characteristics and risk factors for BC in Jordanian women; these included lifestyle risk factors such as alcohol consumption, cigarette smoking 6’15 high-fat nutrient intake pattern, and insufficient exercise 7. Other risk factors reported for BC in Jordan were age, obesity, body mass index (BMI), high level of estrogen, age at menarche and menopause, reproduction history, exposure to ionizing radiation, and hereditary background 16’17. Unlike the epidemiological factors, few and limited reports were published on the genetic factors and different gene polymorphisms (SNPs) associated with the risk of BC in Jordan. These included the known germline, high penetrance susceptibility genes, which include the germline BRCA1 and BRCA2 mutations 18’19 and few studies examined the association of SNPs in genes related to the folate/one-carbon metabolism with BC 20’21
MTHFD1 and CBS are genes encoding the enzymes methylenetetrahydrofolate dehydrogenase (MTHFD1) and cystathionine 𝛽-Synthase (CBS) respectively have central roles in folate and homocysteine metabolism through the folate /one-carbon metabolism and are candidate genes for cancer susceptibility 22’23’24’25’26. MTHFD1 enzyme catalyzes the irreversible conversion of 5,10-methylenetetrahydrofolate to the primary form of circulatory folate 5-methyltetrahydrofolate 27, and is essential for the synthesis of purine and pyrimidine bases 28 and regeneration of SAM 29. CBS is a major enzyme in the transsulfuration pathway, at the homocysteine (Hcy) junction of preserving methionine or converting it to cysteine. CBS eliminates Hcy by catalyzing the condensation of serine and Hcy to form cystathionine 30, which is then hydrolyzed to cysteine, a precursor of the potent antioxidant glutathione 28’31.
Different studies showed an association of MTHFD1 G1958A with different diseases including cardiovascular diseases 27‘32 maternal risk for fetal loss 33 and neural tube defects 34‘24‘35 and psychiatric disorders 36. Several clinical studies showed over-expression of CBS and, increased production of H2S in many cancer types including colon, ovarian, gastric, colorectal, prostate, and gastroesophageal cancer 35‘37‘38‘39‘40. The main CBS-derived metabolites are the anti-inflammatory (H2S) and Hcy 31. H2S is associated with signaling and protective effects on antioxidant defenses inhibits the production of hydrogen peroxide (H2O2) and other reactive oxygen species (ROS) and preserves the activity of key antioxidant enzymes including catalase and superoxide dismutase, glutathione peroxidase, and glutathione-S-transferase 41‘42‘43. Functional SNPs in CBS gene can promote carcinogenesis 11‘40‘44‘45‘46.
The association between each of MTHFD1 and CBS genes with BC are still controversial and indefinite. To our knowledge, this is the first study to examine associations of MTHFD1 and CBS genes with breast cancer in Jordanians. Thus in this case-control study, we aimed to examine the possible association of MTHFD1 G1958A (R653Q), MTHFD1 T401C (R134K), and the insertion mutation CBS 844ins68bp with BC among Jordanian women.
Materials and methods
Subjects
A group of two hundred women with confirmed diagnoses of breast cancer (cases) as well as a group of 200 age-matched unaffected women (controls) were recruited from two major referral hospitals for cancer, King Abdullah Hospital in Irbid, (northern part of Jordan) and Al-Basheer Hospital in Amman (central part of Jordan). Each of the participants in the study provided informed consent to donate samples of their blood and use clinical data for research. All procedures used were in strict compliance with the principles of the Helsinki II Declaration. All procedures used were in strict compliance with the principles of the Helsinki II Declaration. The study was ethically approved by the Yarmouk University (Irbid, Jordan) IRB committee (YU IRB DSR 2023/190).
Blood sampling, DNA isolation and Genotyping
Blood samples (3ml) were withdrawn from subjects into EDTA vacationers and stored at 4°C until DNA extraction. Genomic DNA was extracted from the collected blood samples according to the manufacturer’s instructions using a commercial kit (OMEGA Biokit). Genotyping of both MTHFD1 G1958A (rs2236225) and MTHFD1 T401C (rs1950902) was achieved by specific PCR amplification of the genomic DNA, followed by subsequent digestion with the proper restriction enzyme according to 33 and 47 respectively. At the same time, the detection of CBS844ins68bp was accomplished according to 48 by direct PCR.
Table 1 summarizes the genotyping conditions including the sequences of the primers, amplification conditions, restriction endonucleases and sizes of the DNA fragments produced. All PCR reactions were carried out in a total volume of 25μl, containing 1μl of (5pmole/μl) of the proper forward and reverse primers specific for each SNP, 12.5μl of 2X master mix (Promega, USA), and 1μl of DNA sample in nuclease-free water up to 25 μl. Amplification products were visualized by electrophoresis on 2% agarose gel (Agarose A; Biobasic) following staining with 0.5μg/ml ethidium bromide (Sigma, USA).
Statistical analyses
Comparisons between groups and the different allele frequencies were evaluated using Pearson chi-square and the goodness of fit test (P>0.05). The allele and genotype frequencies of the variants in the case and control groups were calculated. Logistic regression analyses were recruited to estimate the associations between the risk of breast cancer and each examined variant, determining odds ratios (ORs) and their corresponding 95% confidence intervals (CIs). The statistical analyses were conducted using SPSS version 22.0 (SPSS, Chicago, IL).
Table 1: Primers sequences and PCR amplification conditions for the genotyping of the targeted polymorphisms MTHFD1 G1958A, MTHFD1 T401C, and CBS 844ins 68
|
Polymorphism |
Primer sequence (5’→ 3’) |
PCR conditions: denaturation, annealing, and extension |
Restriction enzyme and incubation conditions |
Fragment length produced in base pairs (bp) |
Primers reference |
|
MTHFD1 G1958A (Arg653Gln) |
F:5’CACTCCAGTGTTTGTCCATG-3’ R:5’GCATCTTGAGAGCCCTGAC-3’ |
Total of 35 cycles: denaturation at 94˚C for 30 sec, annealing at 58˚C for 1 min., and extension at 72˚C for 1 min. |
*MspI incubated at 37°C, 3h |
G allele: 196bp, 71bp and 63bp
A allele: 267 and 63bp |
33 |
|
MTHFD1 T401C |
F: 5’-GGCGTACAAGGAATGAAAC3’ R: 5’-GGATGTGGATGGGTAAGTG3’ |
35 cycles: 95˚C for 30 sec, 48˚C for 40 sec, 72˚C for 40 sec |
*BsmAI Incubated at 37 °C, 16 h |
T allele: 180 and 45 bp C allele: 131, 49, and 45 bp |
47 |
|
CBS 844ins 68
|
F: 5’-CGCCCTCTGCAGATCATTGG3’ R: 5’-CCTTCCACCTCGTAGGTTGTC3’ |
32 cycles: 95˚C for 30 sec, 61˚C for 40 sec, 72˚C for 40 sec |
–– |
Wild type: 100bp, Homozygous mutant:168bp Heterozygous: 100bp and 168bp. |
48 |
*Source: (New England Biolabs, Ipswich, MA, USA).
Results
The mean ages of the studied BC patients (50.22 ± 10.8 years) and the unaffected controls (49.03± 10.4 years) were statistically not different (P= 0.919). Figures 1 and 2 show representative results of the PCR – RFLP genotyping of MTHFD1 G1958A and MTHFD1 T401C respectively, while Figure 3 shows representative results of CBS 844ins68bp by direct PCR. Table 2 shows the observed frequencies of both alleles and genotypes in BC patients and the unaffected controls. The alleles and genotypes of each of the three examined SNP were in Hardy Weinberg equilibrium (P> 0.05). Table 2 shows that the distribution of the wild type and mutant alleles of the individual polymorphisms MTHFD1 G1958A, MTHFD1 T401C and CBS 844ins68bp as well as the frequencies of the different genotypes of each examined polymorphism in the BC group were not significantly different from their frequencies in the controls. In addition, the sums of the mutant genotypes of each of these three polymorphisms were not significantly different between the patients and the control groups. The odd ratios of the sums of mutant genotypes MTHFD1 G1958A (GA+AA), MTHFD1T401C (TC+CC) and CBS 844ins68 (w/Ins + Ins/Ins) were 0.918 (95% CI=0.601-1.401; p = 0.67), 1.042 (95% CI=0.328-3.306; P=1.00) and 1.197 (95%CI = 0.701-2.044; p=0.498) respectively.
However, the frequencies of the double compound mutant genotypes GA/CC and AA/CC of the two non-synonymous MTHFD1 polymorphisms G1958A and T401C were higher in BC patients group compared to the unaffected controls. The double compound genotypes MTHFD GA/CC and AA/CC seemed to increase the risk for BC by 3.4 and 5.1 respectively (Table 3) but their confidence intervals were wide possibly due to the small and limited numbers of the observed individuals carrying double or triple compound genotypes.
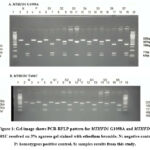 |
Figure 1: Gel image shows PCR-RFLP pattern for MTHFD1 G1958A and MTHFD1 T401C resolved on 3% agarose gel stained with ethedium bromide. |
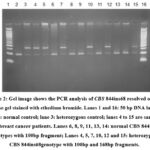 |
Figure 2: Gel image shows the PCR analysis of CBS 844ins68 resolved on 3% agarose gel stained with ethedium bromide. |
Table 2: Allele and genotype frequencies of MTHFD1 G1958A, MTHFD1 T401C, and CBS 844ins68polymorphisms in BC and the control groups.
|
Polymorphisms |
% Controls ( n) |
% Cases (n) |
OR (95% CI) |
P |
|
|
MTHFD1 G1958A |
|||||
|
G (ref) |
0.59% (235)
|
0.58% (231) |
– |
– |
|
|
A |
0.41% (165) |
0.42% (169) |
0.960 (0.725-1.271) |
0.774 |
|
|
GG (ref) |
0.30 (61) |
0.32 (65) |
– |
– |
|
|
GA |
0.57 (113) |
0.51 (101) |
0.847 (0.544-1.317) |
0.229 |
|
|
AA |
0.13 (26) |
0.17 (34) |
1.270 (0.677-2.375) |
0.263 |
|
|
GA and AA |
0.70 (139) |
0.68 (135) |
0.918 (0.601-1.401) |
0.667 |
|
|
MTHFD1 T401C |
|||||
|
T (ref) |
0.14% (56) |
0.13% (52) |
0.918 (0. 612-1.377) |
– |
|
|
C |
0.86% (344) |
0.87% (348) |
– |
0.679 |
|
|
TT (ref) |
0.03 (6) |
0.03 (6) |
– |
– |
|
|
TC |
0.22 (44) |
0.2 (40) |
0.955 (0.282-3.233) |
0.623 |
|
|
CC |
0.75 (150) |
0.77 (154) |
1.054 (0.329-3.375) |
0.623 |
|
|
TC and CC |
0.97 (194) |
0.97 (194) |
1.042 (0.328-3.306) |
1.000 |
|
|
CBS 844ins68 |
|||||
|
w (ref.) |
0.92% (369) |
0.91% (365) |
– |
– |
|
|
Ins |
0.078% (31) |
0.086% (35) |
0.876 (0.529-1.451) |
0.607 |
|
|
w/w (ref.) |
0.85 (170) |
0.83 (165) |
– |
– |
|
|
w/Ins |
0.15 (29) |
0.18 (35) |
1.222 (0.711-2.100) |
0.413 |
|
|
Ins /Ins |
0.005 (1) |
0.0 (0) |
0.0 |
0.0 |
|
|
w/ Ins + Ins/ Ins |
0.15 (30) |
0.18 (35) |
1.197 (0.701-2.044) |
0.498 |
|
Abbreviations: Ref – Reference category (wild-type allele/genotype); n – number of subjects; OR – Odds Ratio; CI – confidence interval of OR based on multinomial logistic regression; P > 0.05; w – wild-type allele of CBS 844 (100bp); Ins – CBS 844ins68bp.”
Table 3: Frequencies and numbers of the observed double compound genotypes among BC patients and the control groups.
|
Compound polymorphisms |
Controls (n) |
Cases (n) |
OR (95% CI) |
P |
|
MTHFD1 G1958A/MTHFD1 T401C |
||||
|
GG/TT (Ref.) |
0.02 (4) |
0.01 (1) |
– |
– |
|
GG/TC |
0.08 (15) |
0.07 (13) |
1.467 (0.343 – 35.056) |
– |
|
GG/CC |
0.21 (42) |
0.26 (51) |
1.857 (0.523 – 45.127) |
0.695 |
|
GA/TT |
0.01 (2) |
0.02 (4) |
1.000 (0.500 – 127.900) |
0.287 |
|
GA/TC |
0.13 (26) |
0.12 (24) |
1.69 (0.385 – 35.400) |
0.411 |
|
GA/CC |
0.43 (85) |
0.37 (73) |
3.435 (0.376 – 31.425) |
0.762 |
|
AA/TT |
0.00 (0) |
0.01 (1) |
– |
0.220 |
|
AA/TC |
0.02 (4) |
0.03 (5) |
1.000 (0.388 – 64.387) |
– |
|
AA/CC |
0.11 (22) |
0.14 (28) |
5.091 (0.531- 48.852) |
0.736 |
|
MTHFD1 G1958A/CBS ins68 |
||||
|
GG/ ww (Ref.) |
0.26 (51) |
0.26 (51) |
– |
– |
|
GG/ wIns |
0.05 (10) |
0.07 (14) |
1.400 (0.569 -3.442) |
0.400 |
|
GA/ ww |
0.48 (96) |
0.44 (87) |
0.906 (0.906 -1.471) |
0.366 |
|
GA/ wIns |
0.09 (17) |
0.07 (13) |
0.765 (0.337–1.736) |
0.448 |
|
AA/ ww |
0.12 (23) |
0.14 (27) |
1.174 (0.596-2.313 |
0.545 |
|
MTHFD1 T401C/ CBS ins68 |
||||
|
TT/ w/w (Ref.) |
0.02 (4) |
0.02 (4) |
– |
– |
|
TT/ wIns |
0.01 (2) |
0.01 (2) |
1.000 (0.091-11.028) |
1.000 |
|
TC/ ww |
0.18 (35) |
0.19 (37) |
1.057 (0.245– 4.556) |
0.795 |
|
TC/ wIns |
0.04 (8) |
0.02 (3) |
0.375 (0.055– 2.555) |
0.126 |
|
CC/ ww |
0.66 (131) |
0.62 (124) |
0.947 (0.232-3.867) |
0.467 |
|
CC/ wIns |
0.10 (19) |
0.15 (30) |
1.579 (0.352-7.079) |
0.093 |
|
CC/ InsIns |
0.01 (1) |
0 (0) |
– |
– |
OR: odd ratios; CI: confidence interval of OR according to multinomial logistic regression; P > 0.05; Ref: reference ; w: wild type (100 bp); Ins: mutant CBS 844ins68bp (168 bp).
In order to find out the effect of the observed triple compounds of the examined polymorphisms, MTHFD1G1958A/ MTHFD1T401C/ CBS 844ins68 genotypes were examined in the cases and the controls. The results are shown in Table 4, which shows that there are no significant differences between the observed frequencies of the triple compound genotypes between the cases and the controls expect for the triple compound genotype MTHFD1 1958AA/MTHFD1 40CC/CBS 844ins68 (w/ins68) which have increased odd ratio to 5. This suggested that the presence of these three polymorphisms together in the same individual have a synergistic effect on increasing the risk for BC.
Table 4: Numbers of the observed triple compound genotypesof MTHFD1 G1958A, MTHFD1 T401C and CBS 844ins68 among the cases and the control groups.
|
Variants of MHFD1 G1958A/ MTHFD1 T401C/ CBS 844ins68 |
Number of Controls
|
Number of Patients
|
OR
|
95% C.I
|
P value |
|
GG/TT/ ++ (Ref.) |
2 |
1 |
|
|
|
|
GG/ TT/+ins |
2 |
0 |
– |
– |
– |
|
GG/ TC/ ++ |
13 |
11 |
1.692 |
0.135- 21.270 |
0.674 |
|
GG/ TC/ +ins |
2 |
2 |
1.000 |
0.090- 44.350 |
1.000 |
|
GG/CC/++ |
36 |
39 |
2.167 |
0.188- 24.929 |
0.701 |
|
GG/CC/+ ins |
6 |
12 |
2.000 |
0.299- 3.468 |
0.148 |
|
GA/TT/++ |
2 |
3 |
2.000 |
0.150 – 59.890 |
0.653 |
|
GA/TC/++ |
20 |
22 |
2.000 |
0.185- 26.157 |
0.744 |
|
GA/TC/+ins |
6 |
1 |
0.333 |
0.014- 8.182 |
0.057 |
|
GA/CC/++ |
74 |
63 |
1.703 |
0.151- 19.222 |
0.246 |
|
GA/CC/+ins |
11 |
11 |
1.000 |
0.157- 25.404 |
1.000 |
|
AA/TT/ ins,ins |
0 |
1 |
– |
– |
– |
|
AA/TC/++ |
3 |
5 |
1.333 |
0.204- 54.532 |
0.475 |
|
AA/CC/++ |
20 |
21 |
1.100 |
0.176- 25.010 |
0.869 |
|
AA/CC/+ins |
2 |
7 |
5.000 |
0.397- 25.37 |
0.092 |
OR: odd ratios; CI: confidence interval of OR according to multinomial logistic regression; P > 0.05; +: wild type CBS (100 bp); ins: mutant CBS 844ins68 (168 bp); Ref: reference.
Discussion
In this current population-based case-control study, we investigated the association between MTHFD 1958A, MTHFD1 401C variants and the insertion CBS844ins68bp with the risk of BC. MTHFD1 gene, mapped to chromosome 14 (14q24), codes for the enzyme MTHFD1, which is a core enzyme in the folate/ one-carbon metabolism 49. MTHFD1 is a trifunctional enzyme consists of two major domains, an N-terminal containing the dehydrogenase and cyclohydrolase activities and a synthetase domain in the C terminus 50, and is associated with epigenetic events and DNA methylation that can influence susceptibility to cancers 13. The SNP MTHFD1 G1958A is a G to A transition at position 1958 located within the 10-formyl-THF synthetase domain 28, resulting in the substitution of glutamine with arginine residue at position 653 (R653Q). MTHFD1 G1958A causes reduction in the activity and stability of the enzyme 40‘51 and facilitates different developmental diseases and different types of cancers, including breast cancer 14, colorectal 52, gastric 47, methotrexate sensitivity in acute lymphoblastic leukemia 53‘54. However, other different studies including a meta-analysis of Asians showed lack of association between MTHFD1 G1958A and different types of cancers including lung cancer, and head and neck cancer 55‘56. Furthermore, MTHFD1 G1958A decreased the risk for acute lymphoblastic leukemia 57. The less common SNP MTHFD1 T401C in which the arginine at position 134 is substituted by lysine (R134K), lies within the dehydrogenase/cyclohydrolase domain of the enzyme 50, which may result in disturbance of the folate-mediated homocysteine pathway that is associated with cancer 34‘14‘40. The activity of MTHFD1 G1958A was correlated to reduction in the synthase activity, which is essential for the remethylation of homocysteine (Hcy) to methionine, which is a precursor for the synthesis of SAM 29‘40. Reduced levels of SAM lead to low methyl supply, which can result in global DNA hypomethylation, and very little conversion of dUMP to dTMP, leading to uracil misincorporation into DNA 58, DNA strand breaks, chromosomal instability, alteration of gene expression and consequently promoting carcinogenesis 58‘59. However, current results showed that neither the individual polymorphism MTHFD1 G1958A nor MTHFD1 T401C per se were associated with BC, which supported the reported lack of association between MTHFD1 401C and human cancers 34 as well as the lack of association between MTHFD 1958A and BC in west Siberian region of Russia 60. However, our results were contrary to those obtained from studying a mix of White, African American, Hispanic, Asian, and unknown postmenopausal females, which reported association of MTHFD1 T401C with risk of BC 14. In addition, results of our current study were also different from those reported association of MTHFD1 gene with other different types of cancer including gastric, colon and head and neck cancers 47‘47. Different studies also showed that the homozygous mutant genotype MTHFD1 1958AA induced significant reduction in the overall cancer risk, and the risk of primary liver and colon cancers 61. In addition, homozygous patients with MTHFD1 1958AA genotype had significantly higher frequency of tumor CpG island hypermethylation compared to wild-type MTHFD1 1958GG homozygotes, which were significantly associated with DNA hypomethylation 13, suggesting that the G allele may exert a protective effect for cancer risk by protecting from DNA hypomethylation 61.
Cystathionine 𝛽-synthase (CBS) is another core enzyme in the folate/ one-carbon related metabolism, specifically at the reverse transsulfuration pathway, which transfers sulfur from the cytotoxic metabolite homocysteine to cysteine. The CBS gene located in the subtelomeric region q.22.3, of chromosome 21 62 encodes for the 63-kDa CBS subunits of the tetramer active enzyme 63. Each subunit of CBS consists of N- and C- terminal domains. The N-terminal domain binds to the cofactor heme and is essential for proper folding and assembly of the protein, but is not essential for its catalytic activity 64. The C-terminal regulatory domain contains the binding sites for the allosteric activator SAM and is responsible for CBS subunit tetramerization 63. The variant CBS ins68, which initially detected in a heterozygous patient with homocystinuria due to CBS deficiency 65 had been associated with lowered plasma homocysteine levels, consists of an identical insertion of 68bp DNA repeat within exon 8 of the CBS gene 66.
The observed results demonstrated that CBS 488ins68 polymorphism is not a risk factor for BC, which supported previous reports that considered CBS 844ins68 as a neutral variant 67, which generates an alternative splice site, which allows elimination of the entire insertion to form a normal CBS mRNA transcript 68. Current results were also in harmony with the lack of association between CBS 844ins68 polymorphism with different types of cancer including colorectal cancer 47 carcinomas of the upper gastrointestinal tract 69 and prostatic carcinoma 70. Furthermore, our results were in harmony with the unaltered expression of CBS enzyme in BC and the majority of cancer types where the role of CBS has been examined (with the exception of liver cancer and glioma) 42. However, our current results were contrary to the reported significant association of CBS 844ins68 with risk of BC in Mexicans. In Mexicans, both homozygous and heterozygous genotypes of CBS 844ins68 were associated with the risk of BC 45. Such controversial results could be due to ethnic differences as well as different lifestyles, diet and uptake levels of folate in the Jordanian and Mexican populations. Furthermore, current results are also different from the reported expression of CBS in breast cancer-affected tissue, compared to the normal control unaffected breast tissue in mastectomy samples of BC patients 42‘ 71.
The conflicting and controversial reports in the literature concerning the association of MTHFD1 G1958A, MTHFD1 T401C and CBS 844ins68bp with the risk of BC could be reflection to the complexity of the regulation of CBS and MTHFD1 genes, as well as the complexity of the carcinogenesis process itself.
Both CBS and MTHFD1 are involved in methyl group metabolism 70. CBS catalyzes different but interrelated cellular biochemical pathways, including availability of SAM and DNA methylation, while MTHFD1 G1958A is associated in breast cancer with hormone receptor content and DNA methylation frequency 13. BC patients homozygous for the MTHFD1 1958AA genotype had a significantly higher frequency of tumor CpG island hypermethylation compared to the wild-type homozygotes MTHFD1 1958 GG 13. CBS transcription is regulated by different mechanisms including the methylation status of the CpG islands in its two principal GC-rich promoters 72‘73 and by several hormones and transcription factors 57‘74‘75‘76. CBS also regulates the production of both ROS, which is triggered by glutathione abundance 65 and the intrinsic cellular regulator H2S 77‘78. In addition, the process of carcinogenesis involves altered methylation cycle accompanied with promoter hypermethylation, which leads to inactivation of genes in almost all pathways protective of carcinogenesis such as DNA repair, cell cycle control and apoptosis 31. All these factors including ethnicity, which is a possible cause for differences in genetic variants in MTHFD1, CBS and other genes in the folate/one-carbon metabolism in different ethnic groups, could contribute to differential risks of developing breast cancer between different populations 79. In addition, other factors specific for different populations such as environmental, lifestyles, nutrition and uptake levels of folate may act upon these SNPs to generate a gradient of intermediates in the folate/ one-carbon metabolism and associated transsulfuration pathway.
Various studies showed that different individual polymorphisms in different genes of the folate/ one-carbon metabolism were not per se associated with breast cancer. These include the polymorphisms MTHFR C677T and MTHFR A1298C in the methylenetetrahydrofolate reductase, TYMS 1494 ins/del 6 in the thymidylate synthase and MTRR A66G in 5-methytetrahydrofolate homocysteine methyltransferase reductase (MTRR). However, the presence of mutant alleles for two polymorphisms of these genes increased the risk of BC or were associated with increasing the risk of developing more BC aggressive phenotypes 80’81. We examined the effect of double compound genotypes of MTHFD1 T401C, MTHFD1 1958A and CBS 488ins68 CBS 488ins68 on the risk of BC. Results showed that the double compound states of both MTHFD1 T401C and MTHFD1 1958A (1958GA/ 401CC) and (1958AA/401CC) seemed to increase the risk of BC by 3.4 and 5.1 folds respectively (Table 3), but such increase was accompanied by large confidence intervals possibly due to the observed small numbers of these compound genotypes. Further investigations are required to explore the effect of double compound genotypes of MTHFD1 T401C, MTHFD1 1958A and CBS 488ins68 on the risk of BC. In conclusion, CBS 844ins68bp, MTHFD1 G1958A and MTHFD1 T401C polymorphisms per se are not risk factors of BC in Jordan females. For better BC management, further studies can be considered for understanding the influence of double compound genotypes of these polymorphisms on the risk of BC in Jordan.
Conclusion
This study revealed that each of MTHFD1 G1958A, T401C and CBS 844ins68 polymorphisms alone had no direct risk for BC in the Jordanian women, compared to the wild-type genotypes.
Acknowledgment
We would like to extend our gratitude to all women who participated in this study and to the Ministry of Health in Jordan. Yarmouk University completely supported this work.
Conflict of interest
The authors have no conflict of interest to declare.
Funding Source
The Deanship of Scientific Research and Graduate Studies at Yarmouk University, Irbid, Jordan (Grant # 20/2014).
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209-249. doi:10.3322/caac.21660
CrossRef - Hankinson SE, Colditz GA, Willett WC. The lifelong interplay of genes, lifestyle, and hormones. Breast Cancer Res. 2004;6(5):213-218. doi:10.1186/bcr921
CrossRef - Ponder BAJ. Cancer genetics. Nature. 2001;411(6835):336-341.
CrossRef - Verma M. Biomarkers for risk assessment in molecular epidemiology of cancer. Technol Cancer Res Treat. 2004;3(5):505-514. doi:10.1177/153303460400300512
CrossRef - Dieterich M, Stubert J, Reimer T, Erickson N, Berling A. Influence of lifestyle factors on breast cancer risk. Breast Care. 2014;9(6):407-414. doi:10.1159/000369571
CrossRef - Atoum MF, Al-Hourani HM. Lifestyle related risk factors for breast cancer in Jordanian females. Saudi Med J. 2004;25(9):1245-1248.
- Tayyem RF, Mahmoud RI, Shareef MH, Marei LS. Nutrient intake patterns and breast cancer risk among Jordanian women: a case-control study. Epidemiol Health. 2019;41:e2019010. doi:10.4178/epih.e2019010
CrossRef - American Cancer Society. Breast Cancer Risk and Prevention Breast Cancer Risk Factors You Cannot Change. CancerOrg. Published online 2020:1-45.
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome-biological and translational implications. Nat Rev Cancer. 2011;11(10):726-734. doi:10.1038/nrc3130
CrossRef - Xu X, Chen J. One-carbon metabolism and breast cancer: an epidemiological perspective. J Genet Genomics. 2009;36(4):203-214.
CrossRef - Choi S-W, Mason JB. Folate and carcinogenesis: an integrated scheme. J Nutr. 2000;130(2):129-132.
CrossRef - Duthie SJ, Narayanan S, Brand GM, Pirie L, Grant G. Impact of folate deficiency on DNA stability. J Nutr. 2002;132(8):2444S-2449S.
CrossRef - Li SY, Rong M, Iacopetta B. Germ-line variants in methyl-group metabolism genes and susceptibility to DNA methylation in human breast cancer. Oncol Rep. 2006;15(1):221-225.
CrossRef - Stevens VL, McCullough ML, Pavluck AL, et al. Association of polymorphisms in one-carbon metabolism genes and postmenopausal breast cancer incidence. Cancer Epidemiol Biomarkers Prev. 2007;16(6):1140-1147. doi:10.1158/1055-9965.EPI-06-1037
CrossRef - Al Qadire M, Alkhalaileh M, Hedaya H. Risk factors for breast Cancer among Jordanian women: a case-control study. Iran J Public Health. 2018;47(1):49.
- Arkoob K, Al-Nsour M, Al-Nemry O, Al-Hajawi B. Epidemiology of breast cancer in women in jordan: Patient characteristics and survival analysis. East Mediterr Heal J. 2010;16(10):1032-1038. doi:10.26719/2010.16.10.1032
CrossRef - Abdel-Razeq H, Mansour A, Bater R. Trends, Patterns, and Treatment Outcomes of Cancer Among Older Patients in Jordan: A Retrospective Analysis of National Cancer Registry and Institutional Outcome Data. JCO Glob Oncol. 2020;(6):745-751. doi:10.1200/go.20.00044
CrossRef - Abdel-Razeq H, Al-Omari A, Zahran F, Arun B. Germline BRCA1/BRCA2 mutations among high risk breast cancer patients in Jordan. BMC Cancer. 2018;18(1):1-11. doi:10.1186/s12885-018-4079-1
CrossRef - Abu-Helalah M, Azab B, Mubaidin R, et al. BRCA1 and BRCA2 genes mutations among high risk breast cancer patients in Jordan. Sci Rep. 2020;10(1):1-9. doi:10.1038/s41598-020-74250-2
CrossRef - Awwad N, Yousef AM, Abuhaliema A, Abdalla I, Yousef M. Relationship between genetic polymorphisms in MTHFR (C677T, A1298C and their haplotypes) and the incidence of breast cancer among jordanian females – case-control study. Asian Pacific J Cancer Prev. 2015;16(12):5007-5011. doi:10.7314/APJCP.2015.16.12.5007
CrossRef - Sadiq MF, Issa NMA, Alhakim MO, et al. Association of Genetic Variants of Enzymes Involved in Folate / One-Carbon Metabolism with Female Breast Cancer in Jordan. 2019;12(1):89-97.
- Zarou MM, Vazquez A, Vignir Helgason G. Folate metabolism: a re-emerging therapeutic target in haematological cancers. Leukemia. 2021;35(6):1539-1551. doi:10.1038/s41375-021-01189-2
CrossRef - Sellers TA, Kushi LH, Cerhan JR, et al. Dietary folate intake, alcohol, and risk of breast cancer in a prospective study of postmenopausal women. Epidemiology. Published online 2001:420-428.
CrossRef - Carroll N, Pangilinan F, Molloy AM, et al. Analysis of the MTHFD1 promoter and risk of neural tube defects. Hum Genet. 2009;125(3):247-256. doi:10.1007/s00439-008-0616-3
CrossRef - Miranti EH, Stolzenberg-Solomon R, Weinstein SJ, et al. Low vitamin B12 increases risk of gastric cancer: A prospective study of one-carbon metabolism nutrients and risk of upper gastrointestinal tract cancer. Int J Cancer. 2017;141(6):1120-1129. doi:10.1002/ijc.30809
CrossRef - Kim SE. Enzymes involved in folate metabolism and its implication for cancer treatment. Nutr Res Pract. 2020;14(2):95-101. doi:10.4162/nrp.2020.14.2.95
CrossRef - Frosst P, Blom HJ, Milos R, et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10(1):111-113. doi:10.1038/ng0595-111
CrossRef - Hol FA, van der Put NMJ, Geurds MPA, et al. Molecular genetic analysis of the gene encoding the trifunctional enzyme MTHFD (methylenetetrahydrofolate‐dehydrogenase, methenyltetrahydrofolate‐cyclohydrolase, formyltetrahydrofolate synthetase) in patients with neural tube defects. Clin Genet. 1998;53(2):119-125.
CrossRef - Oldenburg RA, Meijers-Heijboer H, Cornelisse CJ, Devilee P. Genetic susceptibility for breast cancer: how many more genes to be found? Crit Rev Oncol Hematol. 2007;63(2):125-149.
CrossRef - Nazki FH, Sameer AS, Ganaie BA. Folate: metabolism, genes, polymorphisms and the associated diseases. Gene. 2014;533(1):11-20.
CrossRef - Zhu H, Blake S, Chan KT, Pearson RB, Kang J. Cystathionine β -Synthase in Physiology and Cancer. Biomed Res Int. 2018;2018. doi:10.1155/2018/3205125
CrossRef - Cheng J, Zhu W-L, Dao J-J, Li S-Q, Li Y. Relationship between polymorphism of methylenetetrahydrofolate dehydrogenase and congenital heart defect. Biomed Env Sci. 2005;18(1):58-64.
- Parle-McDermott A, Kirke PN, Mills JL, et al. Confirmation of the R653Q polymorphism of the trifunctional C1-synthase enzyme as a maternal risk for neural tube defects in the Irish population. Eur J Hum Genet. 2006;14(6):768-772. doi:10.1038/sj.ejhg.5201603
CrossRef - Brody LC, Conley M, Cox C, et al. A polymorphism, R653Q, in the trifunctional enzyme methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase/formyltetrahydrofolate synthetase is a maternal genetic risk factor for neural tube defects: Report of the birth defects res. Am J Hum Genet. 2002;71(5):1207-1215. doi:10.1086/344213
CrossRef - Jiang J, Zhang Y, Wei L, Sun Z, Liu Z. Association between MTHFD1 G1958A polymorphism and neural tube defects susceptibility: A meta-analysis. PLoS One. 2014;9(6). doi:10.1371/journal.pone.0101169
CrossRef - Kempisty B, Sikora J, Lianeri M, et al. MTHFD 1958G> A and MTR 2756A> G polymorphisms are associated with bipolar disorder and schizophrenia. Psychiatr Genet. 2007;17(3):177-181.
CrossRef - Bhattacharyya S, Saha S, Giri K, et al. Cystathionine Beta-Synthase (CBS) Contributes to Advanced Ovarian Cancer Progression and Drug Resistance. PLoS One. 2013;8(11). doi:10.1371/journal.pone.0079167
CrossRef - Katsouda A, Bibli S-I, Pyriochou A, Szabo C, Papapetropoulos A. Regulation and role of endogenously produced hydrogen sulfide in angiogenesis. Pharmacol Res. 2016;113:175-185.
CrossRef - Hellmich MR, Szabo C. Hydrogen sulfide and cancer. Handb Exp Pharmacol. 2015;230:233-241. doi:10.1007/978-3-319-18144-8_12
CrossRef - Hasan T, Arora R, Bansal AK, Bhattacharya R, Sharma GS, Singh LR. Disturbed homocysteine metabolism is associated with cancer. Exp Mol Med. 2019;51(2). doi:10.1038/s12276-019-0216-4
CrossRef - Wen YD, Wang H, Kho SH, et al. Hydrogen Sulfide Protects HUVECs against Hydrogen Peroxide Induced Mitochondrial Dysfunction and Oxidative Stress. PLoS One. 2013;8(2). doi:10.1371/journal. pone. 0053147
CrossRef - Ascenção K, Szabo C. Emerging roles of cystathionine β-synthase in various forms of cancer. Redox Biol. 2022;53(May). doi:10.1016/j.redox.2022.102331
CrossRef - Panagaki T, Lozano-Montes L, Janickova L, et al. Overproduction of hydrogen sulfide, generated by cystathionine β-synthase, disrupts brain wave patterns and contributes to neurobehavioral dysfunction in a rat model of down syndrome. Redox Biol. 2022;51(November 2021). doi:10.1016/j.redox.2022.102233
CrossRef - Kruszyna Ł, Lianeri M, Rydzanicz M, Gajęcka M, Szyfter K, Jagodziński PP. Polymorphic variants of folate metabolism genes and the risk of laryngeal cancer. Mol Biol Rep. 2010;37:241-247.
CrossRef - Gallegos-Arreola MP, Figuera-Villanueva LE, Ramos-Silva A, et al. The association between the 844ins68 polymorphism in the CBS gene and breast cancer. Arch Med Sci. 2014;10(6):1214-1224. doi:10.5114/aoms.2014.47830
CrossRef - Newman AC, Maddocks ODK. One-carbon metabolism in cancer. Br J Cancer. 2017;116(12):1499-1504. doi:10.1038/bjc.2017.118
CrossRef - Wang L, Ke Q, Chen W, et al. Polymorphisms of MTHFD, plasma homocysteine levels, and risk of gastric cancer in a high-risk Chinese population. Clin Cancer Res. 2007;13(8):2526-2532. doi:10.1158/1078-0432.CCR-06-2293
CrossRef - Shannon B, Gnanasampanthan S, Beilby J, Iacopetta B. A polymorphism in the methylenetetrahydrofolate reductase gene predisposes to colorectal cancers with microsatellite instability. Gut. 2002;50(4):520-524. doi:10.1136/gut.50.4.520
CrossRef - Rozen R, Barton D, Du J, Hum DW, MacKenzie RE, Francke U. Chromosomal localization of the gene for the human trifunctional enzyme, methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase- formyltetrahydrofolate synthetase. Am J Hum Genet. 1989;44(6):781-786.
- Hum DW, Bell AW, Rozen R, MacKenzie RE. Primary structure of a human trifunctional enzyme. Isolation of a cDNA encoding methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase-formyltetrahydrofolate synthetase. J Biol Chem. 1988;263(31):15946-15950. doi:10.1016/s0021-9258(18)37540-9
CrossRef - Christensen KE, Rohlicek C V, Andelfinger GU, et al. The MTHFD1 p. Arg653Gln variant alters enzyme function and increases risk for congenital heart defects. Hum Mutat. 2009;30(2):212-220.
CrossRef - Chen J, Kyte C, Valcin M, et al. Polymorphisms in the one-carbon metabolic pathway, plasma folate levels and colorectal cancer in a prospective study. Int J Cancer. 2004;110(4):617-620. doi:10.1002/ijc.20148
CrossRef - Krajinovic M, Lemieux-Blanchard É, Chiasson S, Primeau M, Costea I, Moghrabi A. Role of polymorphism in MTHFR and MTHFD1 genes in the outcome of childhood acute lymphoblastic leukemia. Pharmacogenomics J. 2004;4(1):66-72. doi:10.1038/sj.tpj.6500224
CrossRef - De Jonge R, Hooijberg JH, Van Zelst BD, et al. Effect of polymorphisms in folate-related genes on in vitro methotrexate sensitivity in pediatric acute lymphoblastic leukemia. Blood. 2005;106(2):717-720. doi:10.1182/blood-2004-12-4941
CrossRef - Matakidou A, El Galta R, Rudd MF, et al. Prognostic significance of folate metabolism polymorphisms for lung cancer. Br J Cancer. 2007;97(2):247-252. doi:10.1038/sj.bjc.6603830
CrossRef - da Silva LMRB, Galbiatti ALS, Ruiz MT, et al. MTHFD1 G1958A, BHMT G742A, TC2 C776G and TC2 A67G polymorphisms and head and neck squamous cell carcinoma risk. Mol Biol Rep. 2012;39:887-893.
CrossRef - Zhang H, Ma H, Li L, Zhang Z, Xu Y. Association of Methylenetetrahydrofolate Dehydrogenase 1 Polymorphisms with Cancer: A Meta-Analysis. PLoS One. 2013;8(7). doi:10.1371/journal.pone.0069366
CrossRef - Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Abberant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10(7):687-692. doi:10.1093/hmg/10.7.687
CrossRef - Ulrich CM, Curtin K, Samowitz W, et al. MTHFR variants the risk of G:C→A:T transition mutations within the p53 tumor suppressor gene in colon tumors. J Nutr. 2005;135(10):2462-2467. doi:10.1093/jn/135.10.2462
CrossRef - Vaĭner AS, Boiarskikh UA, Voronina EN, et al. Polymorphic variants of folate metabolizing genes (C677T and A1298C MTHFR, C1420T SHMT1 and G1958A MTHFD) are not associated with the risk of breast cancer in West Siberian Region of Russia. Mol Biol (Mosk). 2010;44(5):816-823.
CrossRef - Moruzzi S, Guarini P, Udali S, et al. One-carbon genetic variants and the role of MTHFD1 1958G>A in liver and colon cancer risk according to global DNA methylation. PLoS One. 2017;12(10):1-14. doi:10.1371/journal.pone.0185792
CrossRef - Munke M, Kraus JP, Ohura T, Francke U. The gene for cystathionine β-synthase (CBS) maps to the subtelomeric region on human chromosome 21q and to proximal mouse chromosome 17. Am J Hum Genet. 1988;42(4):550-559.
- Ereño-Orbea J, Oyenarte I, Martínez-Cruz LA. CBS domains: Ligand binding sites and conformational variability. Arch Biochem Biophys. 2013;540(1-2):70-81. doi:10.1016/j.abb.2013.10.008
CrossRef - Majtan T, Singh LR, Wang L, Kruger WD, Kraus JP. Active cystathionine β-synthase can be expressed in heme-free systems in the presence of metal-substituted porphyrins or a chemical chaperone. J Biol Chem. 2008;283(50):34588-34595. doi:10.1074/jbc.M805928200
CrossRef - Sebastio G, Sperandeo MP, Panico M, De Franchis R, Kraus JP, Andria G. The molecular basis of homocystinuria due to cystathionine β-synthase deficiency in Italian families, and report of four novel mutations. Am J Hum Genet. 1995;56(6):1324-1333.
- Tsai MY, Bignell M, Yang F, Welge BG, Graham KJ, Hanson NQ. Polygenic influence on plasma homocysteine: association of two prevalent mutations, the 844ins68 of cystathionine β-synthase and A2756G of methionine synthase, with lowered plasma homocysteine levels. Atherosclerosis. 2000;149(1):131-137.
CrossRef - Kluijtmans LAJ, Boers GHJ, Trijbels FJM, van Lith-Zanders HMA, van den Heuvel LPWJ, Blom HJ. A common 844INS68 insertion variant in the cystathionine β-synthase gene. Biochem Mol Med. 1997;62(1):23-25.
CrossRef - Tsai MY, Bignell M, Schwichtenberg K, Hanson NQ. High prevalence of a mutation in the cystathionine β-synthase gene. Am J Hum Genet. 1996;59(6):1262-1267.
- Ott N, Geddert H, Sarbia M. Polymorphisms in methionine synthase (A2756G) and cystathionine β-synthase (844ins68) and susceptibility to carcinomas of the upper gastrointestinal tract. J Cancer Res Clin Oncol. 2008;134:405-410.
CrossRef - Kimura F, Franke KH, Steinhoff C, et al. Methyl group metabolism gene polymorphisms and susceptibility to prostatic carcinoma. Prostate. 2000;45(3):225-231. doi:10.1002/1097-0045(20001101)45:3<225::AID-PROS4>3.0.CO;2-7
CrossRef - Xue G, Lu CJ, Pan SJ, et al. DNA hypomethylation of CBS promoter induced by folate deficiency is a potential noninvasive circulating biomarker for colorectal adenocarcinomas. Oncotarget. 2017;8(31):51387-51401. doi:10.18632/oncotarget.17988
CrossRef - Qi F, Zhou Y, Xiao Y, et al. Promoter demethylation of cystathionine-β-synthetase gene contributes to inflammatory pain in rats. Pain. 2013;154(1):34-45. doi:10.1016/j.pain.2012.07.031
CrossRef - Ratnam S, Maclean KN, Jacobs RL, Brosnan ME, Kraus JP, Brosnan JT. Hormonal regulation of cystathionine β-synthase expression in liver. J Biol Chem. 2002;277(45):42912-42918. doi:10.1074/jbc.M206588200
CrossRef - Lechuga TJ, Qi QR, Kim T, Magness RR, Chen DB. E2β stimulates ovine uterine artery endothelial cell H2S production in vitro by estrogen receptor-dependent upregulation of cystathionine β-synthase and cystathionine γ-lyase expression. Biol Reprod. 2019;100(2):514-522. doi:10.1093/biolre/ioy207
CrossRef - Zhao Q, Zhang C, Li D, et al. CBS gene polymorphism and promoter methylation-mediating effects on the efficacy of folate therapy in patients with hyperhomocysteinemia. J Gene Med. 2020;22(4):0-1. doi:10.1002/jgm.3156
CrossRef - Szabo C, Coletta C, Chao C, et al. Tumor-derived hydrogen sulfide, produced by cystathionine-β-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc Natl Acad Sci U S A. 2013;110(30):12474-12479. doi:10.1073/pnas.1306241110
CrossRef - Szabo C. A timeline of hydrogen sulfide (H2S) research: From environmental toxin to biological mediator. Biochem Pharmacol. 2018;149(September):5-19. doi:10.1016/j.bcp.2017.09.010
CrossRef - Lewis SJ, Harbord RM, Harris R, Smith GD. Meta-analyses of observational and genetic association studies of folate intakes or levels and breast cancer risk. J Natl Cancer Institutefile///C/Users/al-awsat/Downloads/scholar (34).ris. 2006;98(22):1607-1622. doi:10.1093/jnci/djj440
CrossRef - artin YN, Olson JE, Ingle JN, et al. Methylenetetrahydrofolate reductase haplotype tag single-nucleotide polymorphisms and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15(11):2322-2324. doi:10.1158/1055-9965.EPI-06-0318
CrossRef - Castiglia P, Sanna V, Azara A, et al. Methylenetetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphisms in breast cancer: A sardinian preliminary case-control study. Int J Med Sci. 2019;16(8):1089-1095. doi:10.7150/ijms.32162
CrossRef







