Prisho Mariam Paul1, 2 , Krupakar Parthasarathy1*
, Krupakar Parthasarathy1* , Sudhanarayani S Rao1
, Sudhanarayani S Rao1 and Vignesh Sounderrajan1
and Vignesh Sounderrajan1
1Centre for Drug Discovery and Development, Sathyabama Institute of Science and Technology, Chennai, Tamil Nadu, India.
2Biotechnology Department, CMS College Kottayam, Kerala India.
Corresponding Author E-mail: pkrupakar.cddd@sathyabama.ac.in
DOI : https://dx.doi.org/10.13005//bpj/2775
Abstract
The current pandemic situation is created by the highly evolving SARS coronavirus 2 which is having several mutations in its structural proteins. The structural proteins of SARS CoV-2 include spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N) which are primarily responsible for the infection, transmission, and pathogenesis of the virus. Envelope protein is the smallest of the four proteins containing 75 amino acids with a molecular weight of about 8 kDa. The major functions of the hydrophobic envelope protein include envelope formation, budding, replication, and release of the virion. The presence of mutation on the envelope protein results in improper formation of the pentameric structure and also hinders other functional properties. Our computational analysis majorly focuses on several FDA-approved inhibitory compounds that bind to SARS CoV-2 envelope protein that help in the inhibition of virion formation. The percentage of similarity of the envelope protein between SARS CoV and SARS CoV-2 is approximately 96 percent. The homology-modeled structure of the SARS CoV E protein was downloaded from Protein Model Database (PMDB) and the mutation which was found to be consistent among most of the SARS CoV-2 variants was selected as T9I which is present in the N-terminal region. This mutation was introduced into the SARS CoV Envelope protein and was remodeled. The ligands which were approved by FDA were selected for docking analysis to understand their binding capabilities with the envelope protein. Ligands such as Beta-D-Fucose, Mycophenolic Acid, Castanospermine, 1-Deoxynojirimycin, Nafcillin, Guaifenesin, Nabumetone, Cinametic Acid, Lauric acid were used in our study. The docking simulations revealed that Lauric acid, Nafcillin, Nabumetone, and Mycophenolic acid have high binding energy with the SARS CoV-2 wild type (Wuhan) and mutant E protein of the SARS CoV-2 (Omicron) variant. This Insilico data gives insights to test these high binding compounds in invitro studies to prove their efficacy and the protein-protein interactions of envelope protein with its other partnering proteins. These pharmaceutical compounds are a potential alternative in the future for a novel drug development to treat several emerging variants of SARS CoV-2.
Keywords
Docking; Drug Development; Envelope protein; Omicron; SARS CoV-2
Download this article as:| Copy the following to cite this article: Paul P. M, Parthasarathy K, Rao S. S, Sounderrajan V. In silico Docking Analysis of the FDA-Approved Drugs on Envelope Protein of SARS CoV-2 Omicron Variant. Biomed Pharmacol J 2023;16(4). |
| Copy the following to cite this URL: Paul P. M, Parthasarathy K, Rao S. S, Sounderrajan V. In silico Docking Analysis of the FDA-Approved Drugs on Envelope Protein of SARS CoV-2 Omicron Variant. Biomed Pharmacol J 2023;16(4). Available from: https://bit.ly/3Qtzfoe |
Introduction
The coronavirus is one of the most frequent drivers of recent pandemics, and the RNA virus is one of the most virulent and dangerous viruses found in nature due to its propensity for creating an unexpected pandemic 1. High-lethality coronaviruses called MERS (Middle East Respiratory Syndrome) and SARS (Severe Acute Respiratory Syndrome) produce infections ranging from the common cold to lethal pneumonia. SARS CoV-2 makes an appearance in the sequel, as does the 2019 outbreak produced by the novel coronavirus (2019-nCoV) and sickness known as COVID-19. Due to the outbreak, which has infected more than 111 million people and resulted in 6 million deaths as of March 2022, COVID-19 is the most well-known word in 2020 2. On translation, SARS-genomic CoV-2’s architecture transforms into four structural proteins, sixteen non-structural proteins, and ten accessory proteins. Spike protein (S), an Envelope protein (E), Membrane protein (M), and Nucleocapsid protein (N) are structural proteins that constitute the virus protein interface to the external environment 3. The receptor binding domain (RBD) of the S protein combines with ACE2 (Angiotensin-Converting Enzyme-2) in the first step of the virus-host recognition mechanism.
The nucleocapsid proteins that give the viral envelope its shape are created by two additional structural proteins, N and M. M interactions with N are necessary for capsid stability/assembly, M interactions with E are necessary for viral envelope formation as well as the release of new virions, and M interactions with S are necessary for the retention of S in the ERGIC/Golgi complex as well as the integration of new virions 4 .All of these structural and non-structural proteins are active from viral entry to viral egress inside the human host cell 5. Among structural proteins, the E protein has a short length of 76–109 amino acids and is an essential membrane ion channel protein. The hydrophilic amino acids from 07 to 12 are followed by a lengthy hydrophobic transmembrane domain (TMD) and a long hydrophilic carboxy terminus. In humans, the formation of the virion’s shape, pathogenesis, immune evasion, and departure all depend on the E protein. This makes the E protein an attractive target for the creation of new medicines. There are many ways to locate good medication candidates for identifying the inhibitors, and today, strong computational screening is accessible 6.
Emerging SARS CoV-2 variants, particularly those that are concerning, may have an impact on the pathogenicity and transmissibility of the virus, as well as the efficiency of diagnostic tools and vaccines. Although the SARS CoV-2 Delta variation (B.1.617.2) originally surfaced during the second wave of illnesses in India, Delta variants have grown in popularity globally and are continuously changing. Based on evidence that Omicron has several mutations that may alter its behavior, WHO designated the variant B.1.1.529 as a variant of concern on November 26, 2021, and named it Omicron. Researchers found that the E protein is conserved between the Delta strain and the Wuhan strain. The Omicron variant’s E protein, however, had been altered. The aim of this research is to identify FDA-approved drugs that have strong affinity to E protein of Omicron variant for therapeutic purpose.
Methodology
Sequence Retrieval and Mutational Analysis
A genome sequence of Indian SARS CoV-2 omicron variant and SARS CoV-2 Wuhan strain were retrieved from GISAID. A high-speed multiple sequence alignment programs called as Multiple Alignment using Fast Fourier Transform (MAFT) method was used to compare the genome sequences of Indian Omicron variants to those of the Wuhan strain. After MAFT’s multiple sequence alignment, MEGA was used to extract the envelope protein region, which was then translated into a protein sequence.
Target Preparation
The structure of SARS CoV-2 E protein sequence, was accessed from the PMDB from the Protein Model Database (PMDB) (http://srv00.recas.ba.infn.it/PMDB/main.php) using Accession No. PM0082981. Then, using Spdbv, calculate its energy and depict its Ramchandran distribution. The potential mutation on the envelope protein was found in the N-terminal region as T9I by mutational analysis, and the same mutation was introduced into the SARS CoV-2 Envelope protein and reconstructed the structure. The energy and Ramachandran plot distribution were then verified once more.
Ligand Preparation
The National Centre for Biotechnology Information (NCBI) PubChem database (https://pubchem.ncbi.nlm.nih.gov/) was used to obtain E-protein ligand compounds. All of the ligands used in this paper were gathered from the literature 7,8. The ligands are obtained in sdf format, and then transformed to PDB using the Open Babel Tool. (https://sourceforge.net/ projects/openbabel/).
Molecular Docking
The docking analysis of the Omicron SARS CoV-2 E protein with ligands was done using Argus lab docking software, which is widely available. By default, all of the parameters used in Argus lab docking were selected. The calculation type was set to “dock” mode, and the ligand was set to “flexible mode.” 0.40 was chosen as the grid resolution. The lowest energy meant that the ligand and receptor were easy to bind9.
Results and Discussion
Sequence Retrieval and Mutational Analysis
Wuhan and Omicron genome sequences were obtained from GISAID (https://www.gisaid.org/).The MAFT tool was used to align genome sequences, and the MEGA tool was used to extract the E protein gene, which was subsequently translated into E protein.
 |
Figure 1: Multiple Sequence Analysis of Omicron E protein from India and wild type Wuhan E protein. |
Target Preparation
The modelled SARS CoV-2 Envelope protein can be retrieved from Protein Model Database (PMDB) under Accession No. PM0082981(Figure 2A). Checked the amino acid distribution in Swiss pdb viewer’s by Ramachandran plot for structure significance (Figure 2B). The majority of amino acids are found in the permitted zone. Also, compute the energy, which comes out to 1902.888 KJ/Mol.
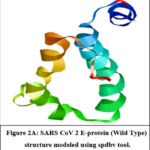 |
Figure 2A: Ramachandran Plot CoV 2 E Protein (Wild Type) structure modeled using spdbv tool.
|
 |
Figure 2B: Ramachandran Plot CoV 2 E Protein (Wild Type). Majority of amino acids in protein are distributed in permitted region.
|
The induced mutation T9I is the most common mutation in the omicron variant. So, by using the Spdbv tool, inserted T9I mutation into the wild type structure (Figure 3A). Energy minimization was done and examined the Ramachandran plot and energy value (Figure 3B). The computed energy obtained after mutation is -1880.569 KJ/mol. Mutations raises the energy value, implying that structure lowers stability. With mutation, however, the Ramachandran plot distribution is satisfied. As a result, the altered structure is being considered for future analysis.
 |
Figure 3A: Induced mutation (T9I) in SARS CoV 2 Envelope protein modeled structure using Spdbv tool.
|
 |
Figure 3B: Ramachandran Plot for Induced mutation (T9I) in SARS CoV 2 Envelope protein. Majority of amino acids in protein are found in permitted region.
|
Ligand Preparation
Totally 9 ligands with binding affinity for the SARS CoV E protein were selected from the available literature. The 3D structures of the ligands were retrieved from the PubChem database and translated to pdb format using the Open Babel programme. Table 1 lists all ligands with a PubChem ID.
Table 1: List of Ligands used in the protein docking with SARS CoV-E protein and their PubChem IDs
|
Name |
Pubchem id |
Name |
Pubchem id |
|
Beta-D-Fucose |
439650 |
Nafcillin |
8982 |
|
Mycophenolic Acid |
446541 |
Guaifenesin |
3516 |
|
Castanospermine |
54445 |
Nabumetone |
4409 |
|
1-Deoxynojirimycin |
29435 |
Cinametic Acid |
6436159 |
|
Lauric acid |
3893 |
Guanifenesin, Nabumetone, Cinametic Acid, and Lauric Acid were chosen based on functions related to SARS CoV 2 symptoms; FDA authorized drugs, and the severity of documented pharmacological adverse effects was well studied7. Nafcillin was chosen as one of the ligands based on an aromatic ring pattern recognition screening. The E protein’s TMD (Transmembrane Domain) has three aromatic Phe residues (at positions 20, 23, and 26); drugs with an aromatic carbon ring have a better chance of binding to Phe amino acid. The drug Nafcillin was chosen based on FDA approval status, and reported side effect7. The other ligands, Beta-D-Fucose, Mycophenolic Acid, Castanospermine, and 1-Deoxynojirimycin, exhibit binding potential with viral Envelop protein and have also passed via a Lipinski filter8.
Docking Analysis
Molecular docking was done for 9 ligands against SARS CoV-2 E protein (Wuhan and Mutated Variant). Table 2 shows the results of the interaction of Envelope protein of wild type and mutant variant with the 9 selected ligands. When comparing the activation energies among the studied ligands, Mycophenolic Acid with energy (-9.11521 kcal/mol (Figure 4A), Nafcillin with energy -10.7206 kcal/mol (Figure 4B), Nabumetone with energy -9.88149 kcal/mol (Figure 4C), and Lauric acid with energy -9.11057 kcal/mol (Figure 4D), were determined to have the greatest activation energy. Thus, the docking findings were examined, and it was concluded that among the 9 ligands, these 4 ligands-Mycophenolic Ac, Nafcillin, Nabumetone, Lauric acid have the best binding relationship with the Omicron E protein and wild type protein, and that they could be useful in the design and development of new anti-SARS CoV-2 drugs.
Beta lactamase-resistant penicillin with a limited spectrum is called Nafcillin 10. Along with community-acquired pneumonia (CAP), Nafcillin is also used to treat a number of illnesses in the lower respiratory tract 11. Nafcillin has the strongest binding affinity for E-TMD protein’s domain. As a result, Nafcillin could be repurposed for the treatment of SARS CoV-2 infection, as it could also attack bacterial co-infection in a COVID patient, which causes symptoms similar to those found in SARS CoV-2 infection 7. COVID-19 causes an unregulated immunological response in which leukocytes are attracted all over the body during the early stages. This is followed by an immunosuppressive condition, which is exacerbated by the loss of T and B cells. The permeability of the blood capillaries that line the alveolar wall of the lungs increases, allowing fluid to flow inside and cause pulmonary edoema and ARDS (Acute Respiratory Distress Syndrome). According to evidence in the literature, the viral E-protein causes this condition 12. As a result, the anti-inflammatory medication Nabumetone can prevent the E-protein from forming a functional pentameric form, which could help to reduce the condition of uncontrolled immune response. The ability of Lauric acid/Dodecanoic acid to prevent viral E-protein oligomerization boosts its therapeutic potential against viral infection 7. Mycophenolic acid may be a potential druggable protein ligand of SARS CoV-2 for preventing viral assembly and hence inhibiting the development of COVID-19 8. To understand the impact of these ligands on the elimination of virus from the host cell by affecting viral budding, the effect on blocking envelope formation and inhibiting virus release from the host cell a complete in vitro and in vivo studies is needed.
 |
Figure 4: Images of the severe Acute Repiratory Syndrome-Coronavirus 2 Envelope protein (Omicron Variant) with the top four liqands with hig binding energy |
Table 2: Binding affinity of selected ligands with SARS COV-2-Envelope protein of Wuhan and Induced Mutant.
|
S.No |
Name |
Docking Energy |
Docking Energy |
|
1 |
Beta-D-Fucose |
-6.54 |
-6.11 |
|
2 |
Mycophenolic Acid |
-9.12 |
-9.39 |
|
3 |
Castanospermine |
-6.42 |
-6.17 |
|
4 |
1-Deoxynojirimycin |
-6.28 |
-6.13 |
|
5 |
Nafcillin |
-10.72 |
-10.07 |
|
6 |
Guaifenesin |
-7.52 |
-7.22 |
|
7 |
Nabumetone |
-9.88 |
-9.98 |
|
8 |
Cinametic Acid |
-7.77 |
-7.44 |
|
9 |
Lauric acid |
-9.11 |
-9.61 |
Table 3: Bonding Information of selected molecules.
|
Name |
Docking Energy(Mutant) |
Binding Information |
|
|
Bond Type |
Distance(A0) |
||
|
Mycophenolic Acid |
-9.11521 kcal/mol |
61 ARG(N…O) |
2.189983 |
|
Nabumetone |
-9.88149 kcal/mol |
61 ARG(N…O) |
2.998031 |
|
Lauric acid |
-9.11057 kcal/mol |
61 ARG(N…O) |
2.913834 |
Discussion
This study focuses at the pharmaceutically available drugs for the most potent inhibitors of Envelope protein of SARS CoV-2 wild type and its mutant (Omicron). The coronavirus envelope protein is essential for the virus’s life cycle. The coronavirus envelope protein is a tiny integral membrane protein with just about 100 residues and two to three clusters of juxta membrane cysteine, which aids in virus morphogenesis and tropism 13. According to investigations on the structural proteins of both SARS CoV and SARS CoV-2, the E protein may play a role in a variety of crucial processes, including membrane construction and membrane curvature induction, division and release, apoptosis, inflammation, and potentially autophagy. The protein is mostly found in the membranes of the ER and Golgi apparatus, where it actively contributes to viral morphogenesis, budding, and trafficking 14. The vast array of newly discovered viruses, including dengue, Zika, and the SARS CoV-2 pandemic, demonstrate the inadequate effectiveness of the available antiviral therapeutics 15. Additionally, the tremendous evolutionary potential of some viruses, such coronaviruses, hinders the development of effective antiviral therapies. Through viral assembly, viral budding, viral propagation, envelope construction by appropriating parts of the host cell membranes, and infectious virus release from the host cell, the virus’s envelope protein is crucial for the development of disease in the host 16. The SARS CoV- envelope (E) protein is a small polypeptide which has at least one α-helical transmembrane domain. If there is an absence, or inactivation, of E protein leads to virus attenuation because it will disturb the virion morphology or tropism 13. Therefore, in order to select the most efficient compounds, the docking studies are taken into account against SARS CoV-2 envelope protein.
The study revealed that Mycophenolic Acid with energy -9.11521 kcal/mol, Nafcillin with energy -10.7206 kcal/mol, Nabumetone with energy -9.88149 kcal/mol, and Lauric acid with energy -9.11057 kcal/mol. Theses protein ligands that are druggable might be able to prevent SARS CoV-2 production by preventing the viral assembly. Similarly some reports report that natural compounds such as amentaflavone, hypericin, and Torvoside H binds with the main protease of the SARS-CoV 2 and inhibits its activity 17. Doxycyclin, available antibiotics shows better binding efficiency with SARS CoV-2 E protein which has satisfied the Lipinski rule for the pharmaceutical application. The high molecular weight Simeprevir and grazoprevir have been extensively studied for their potential anti-viral activity against hepatitis C genotype 1 18.
Mycophenolic acid produced from Penicillium stoloniferum used as an immunosuppressive drug and reported for its antibacterial, antifungal and antiviral property 19. Nafcillin is a second generation penicillinase-resistant penicillin group of antibiotic used to treat Staphylococcus infection. Nabumetone is a potent a non-steroidal anti-inflammatory drug which helps to treat chronic arthritis. Lauric acid is a natural compound found in vegetable oil, animal fat and oils traditionally used as ailments which have proven antimicrobial efficacy (19). As a result, these ligands may be the best alternative for additional extensive studies in wet lab Experiments and proper clinical trials to produce a medication for treating SARS CoV-2 patients.
Conclusion
This study shows that the envelope protein’s TMD domain exhibits the highest binding affinity for Nafcillin. It can combat bacterial co-infection in COVID patients, which causes symptoms identical to those of SARS-CoV-2 infection. It also emphasizes the need to research on antiviral compounds from natural sources which could help to discover broad spectrum antiviral drugs. These should be able to combat both newly discovered viruses that are absolutely unknown as well as newly developing mutants or varieties of viruses that are already known.
Acknowledgment
The work is supported by Indian Council for Medical Research (ICMR) and Centre for Drug Discovery and Design, Sathyabama Institute of Science and Technology, Chennai.
Conflict of interest
The authors declare no conflict of interest.
Funding Sources
The article is funded by ICMR- VIR/COVID19/33/2021/ECD-I
References
- Paital B. Nurture to nature via COVID-19, a self-regenerating environmental strategy of environment in global context. Sci Total Environ. 2020 Aug;729:139088.
CrossRef - Cascella M, Rajnik M, Aleem A, Dulebohn SC, Di Napoli R. Features, Evaluation, and Treatment of Coronavirus (COVID-19). In Treasure Island (FL); 2022.
- Satarker S, Nampoothiri M. Structural Proteins in Severe Acute Respiratory Syndrome Coronavirus-2. Arch Med Res. 2020 Aug;51(6):482–91.
CrossRef - Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020 Jun;178:104792.
CrossRef - Mukherjee S, Bhattacharyya D, Bhunia A. Host-membrane interacting interface of the SARS coronavirus envelope protein: Immense functional potential of C-terminal domain. Biophys Chem [Internet]. 2020;266(July):106452. Available from: https://doi.org/10.1016/j.bpc.2020.106452
CrossRef - Abdullah Alharbi R. Structure insights of SARS-CoV-2 open state envelope protein and inhibiting through active phytochemical of ayurvedic medicinal plants from Withania somnifera. Saudi J Biol Sci. 2021;28(6):3594–601.
CrossRef - Suganya J, Radha M, Naorem DL, Nishandhini M. In silico docking studies of selected flavonoids – Natural healing agents against breast cancer. Asian Pacific J Cancer Prev. 2014;15(19):8155–9.
CrossRef - Das G, Das T, Chowdhury N, Chatterjee D, Bagchi A, Ghosh Z. Repurposed drugs and nutraceuticals targeting envelope protein: A possible therapeutic strategy against COVID-19. Genomics. 2021;113(1):1129–40.
CrossRef - Azeez SA, Alhashim ZG, Al Otaibi WM, Alsuwat HS, Ibrahim AM, Almandil NB, et al. State-of-the-art tools to identify druggable protein ligand of SARS-CoV-2. Arch Med Sci. 2020;16(2):497–507.
CrossRef - Khataniar A, Pathak U, Rajkhowa S, Jha AN. A Comprehensive Review of Drug Repurposing Strategies against Known Drug Targets of COVID-19. Covid. 2022;2(2):148–67.
CrossRef - Wynn M, Dalovisio JR, Tice AD, Jiang X. Evaluation of the efficacy and safety of outpatient parenteral antimicrobial therapy for infections with methicillin-sensitive Staphylococcus aureus. South Med J. 2005;98(6):590–5.
CrossRef - Nieto-Torres JL, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Castaño-Rodriguez C, Fernandez-Delgado R, et al. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology [Internet]. 2015;485:330–9. Available from: http://dx.doi.org/10.1016/j.virol.2015.08.010
CrossRef - Parthasarathy K, Lu H, Surya W, Vararattanavech A, Pervushin K, Torres J. Expression and purification of coronavirus envelope proteins using a modified β-barrel construct. Protein Expr Purif [Internet]. 2012;85(1):133–41. Available from: http://dx.doi.org/10.1016/j.pep.2012.07.005
CrossRef - Cao Y, Yang R, Lee I, Zhang W, Sun J, Wang W, et al. Characterization of the SARS-CoV-2 E Protein: Sequence, Structure, Viroporin, and Inhibitors. Protein Sci. 2021;30(6):1114–30.
CrossRef - Dassanayake MK, Khoo TJ, Chong CH, Di Martino P. Molecular Docking and In-Silico Analysis of Natural Biomolecules against Dengue, Ebola, Zika, SARS-CoV-2 Variants of Concern and Monkeypox Virus. Int J Mol Sci. 2022;23(19).
CrossRef - Dewald Schoeman BCF. Coronavirus envelope protein: current knowledge. Int J Environ Res Public Health. 2020;17(8):1–22.
CrossRef - Pervushin K, Tan E, Parthasarathy K, Lin X, Jiang FL, Yu D, et al. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5(7).
CrossRef - Saravanan KM, Zhang H, Senthil R, Vijayakumar KK, Sounderrajan V, Wei Y, et al. Structural basis for the inhibition of SARS-CoV2 main protease by Indian medicinal plant-derived antiviral compounds. J Biomol Struct Dyn [Internet]. 2022;40(5):1970–8. Available from: https://doi.org/10.1080/07391102.2020.1834457
CrossRef - Bhowmik D, Nandi R, Jagadeesan R, Kumar N, Prakash A. Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID- 19 . The COVID-19 resource centre is hosted on Elsevier Connect , the company ’ s public news and information . 2020;(January).







