Ghofran Khald khlf¹* and Sameer Abdulameer Alash
and Sameer Abdulameer Alash
Department of Biology, collage of science, University of Baghdad, Baghdad, Iraq
Corresponding Author E-mail: ghofran.atiya2102@sc.uobaghdad. edu.iq
DOI : https://dx.doi.org/10.13005/bpj/2705
Abstract
Colorectal cancer (CRC) is the most common disease and cause of death globally. The aim of the study is investigation and detection of some bacterial interfering with CRC occurrence and progression. The study conducted between September 2022 till February 2023, a total of 50 specimens were collected from confirmed CRC patients. In addition, 50 stool specimens were collected from Healthy volunteers, considers as control group. Isolation and identification of bacteria in all collected specimens were done by using cultural and differential media (blood agar, macconkey agar and Pfizer agar), as well as the VITEK- 2 compact system. The bacterial species, in the specimens of control were ( Escherichia coli 50 (86.20%), Klebsiella Pneumonia 3(5.17%), Salmonella typhi 2(3.44%), Staphylococcus aureus 1(1.72%), Proteus mirabilis 1(1.72%) and Pseudomonas aeruginosa 1(1.72%), while in the specimens of CRC and polyp were (Escherichia coli 30(38.69%), Streptococcus uberis 6(7.79%), Enterobacter cloacae 4(5.19%), Proteus mirabilis 11(14.28), Streptococcus constellatus pharyneis 1(1.29%), Micrococcus luteus 1(1.29%), Staphlococcus pseudintermedius 1(1.29%), Streptococcus thoraltensis 1(1.29%), Citrobacter freundii 1(1.29%), Streptoccus mutans 1(1.29%), Enterococcus faecium 5(6.49%), Enterococcus faecalis 4(5.19%), Granulicatella elegans 1(1.29%), Enterococcus gallinarum 2(2.59%), Serratia marcescens 1(1.29%), Streptococcus sangunis 1(1.29%), Staphylococcus lentus 1(1.29%), Comamons testosteroni 1(1.29%), Morganella morganii 1(1.29%), Pseudomonas aeruginosa 1(1.29%), Klebsiella pneumonia 2(2.59%). The bacteria which has been shown to be associated and more abundance in the specimens of CRC tissues are Escherichia.coli 30(38.96%), Streptococcus uberis 6(7.79%), Enterobacter cloacae 4(5.19%), Enterococcus faecium 5(6.49%), Enterococcus faecalis 4(5.19%). Cell-line culture techniques for the five species showed a cellular viability, sequentially Streptocccus uberis (16.12%), Enterococcus faecium (16.39%), Entreococcus faecalis (9.48%), Enterobacter cloacae (15.11%) and Escherichia coli (17.61%). The results statistically studied by using SPSS, which showed excellent or (highly) significant (p-value is in the range of 0.001).
Keywords
Colorectal cancer; Cell line culture
Download this article as:| Copy the following to cite this article: khlf G. K, Alash S. A. Study the Bacterial Activity Isolated from Colon and Rectal Cancer Biopsy in Cell Lines Culture. Biomed Pharmacol J 2023;16(2). |
| Copy the following to cite this URL: khlf G. K, Alash S. A. Study the Bacterial Activity Isolated from Colon and Rectal Cancer Biopsy in Cell Lines Culture. Biomed Pharmacol J 2023;16(2). Available from: https://bit.ly/3JKAFXH |
Introduction
The third most common and second-leading cause of death common type of cancer is colorectal cancer (CRC) prevalent cancer,¹ its occurrence is expanding at such a rapid rate that it is becoming by far common and important health problem on a global level. Around 4% to 5% of people will acquire colorectal cancer, and various risk factors, such as diet (little fiber and excessive red meat), smoking, obesity, drinking, diabetes, and lifestyle, increase the likelihood of getting the disease and have been connected to the start and creation of CRC. ²⁻ ³ The human digestive system, particularly one’s colon, is home to a diverse bacteria population that counts between – bacteria.⁴ Because dysbiosis situations can result in colonic carcinogenesis by means of a chronic inflammatory process, the gut microbiota is crucial in this situation. A multitude of factors, including nutrition, antibiotics, and stress, can contribute to dysbiosis.⁵ Dysbiosis can alter immune responses, damage the intestinal barrier, and change metabolism, which can result in disease. By host DNA damage, the creation and maintenance of an inflammatory milieu, and interference with cellular immunological reaction, the bacteria in gut primarily influences the occurrence and progression of cancer. The development of cell culture had a profound impact on the field of life sciences and greatly advanced medical research. Utilizing cell lines in research is a crucial step in the modeling of diseases, the study of cancer, and the development of treatments,⁶ Primary cancer cell lines are tissue sample derived ex vivo cell populations that have been surgically removed; these tissue samples are typically taken from autopsy tissues, resections, pleural effusions, fine needle aspirates, core biopsies,⁷⁻⁸And since these samples capture the organic milieu of the tumor and maintain the distinctive interplay between cancerous and healthy cells. These interpersonal behaviors play a role in how cancer develops, spreads, and metastasizes as well as how the body reacts to treatments.⁸⁻⁹ In order to support the growth of rat and human pluripotent stem cells, rat embryonic fibroblasts (REF) are used. REF not only provide a substrate for the growth of pluripotent stem cells, but they also secrete vital growth factors that help to preserve stem cell pluripotency. Early passages use REF that have been isolated from rat embryos.¹⁰ REF must be irradiated or given mitomycin C to stop cell growth to fulfill the role of feeders cells. Moreover, the modified conditioned media can be created by cell for the development of feeder free pluripotent stem cells.
Materials and Methods
Bacterial Isolation and Identification
The period from September 2022 till February 2023, a total of 7 specimens of stool and biopsy without formalin were collected from confirmed CRC patients and 43 specimens of polyp patients admitted to hospitals in Baghdad (Medical city Hospitals, Gastrointestinal Hospital). Also, 50 stool specimens were collected from healthy volunteers with no personal or familial history or diagnosis of colorectal disease as control group. All patients underwent Fecal Occult Blood Test and laparoscopic (colonoscopy) and surgery. All specimens were transported to the laboratory without delay. Histopathologically for 7 specimens of colorectal cancer were differentiated as adenocarcinoma, and 43 specimens were diagnosis as polyp, as shown in (Figures 1, 2). Bacteria isolation were detected by cultural characterization on the blood agar media, macconkey agar, mannitol salt agar and Pfizer selective enterococcus agar at 37℃ under aerobic condition and in anaerobic gas jar condition. All the necessary examinations for preliminary diagnosis of the isolated bacteria were carried out, and the confirmatory diagnosis of these isolates was done by VITEK- 2 compact system.
 |
Figure 1: A, B. Colonrectal cancer polyp |
 |
Figure 2: After excision of polyp |
Supernatant extracts preparations
Preparation of bacterial crude supernatant¹¹:
5 mL of BHI were used to make a bacterial suspension, which was then incubated at 37℃ for 24 hours. 50mL of LB broth (Luria Bertani) was inoculated with a 200µL rate of the suspension, and it was then incubated for 48 hours at 31℃ ±1 with 200 rpm of shaker agitation. For bacterial cell sedimentation, the resulting broth was centrifuged at 10,000g for 4 minutes. The supernatant was then separated from the precipitated and filtered at 0.22µm. In order to employ the supernatant in the cell-line cultures later, it was placed in freezer at -20℃.
Cell line culture assay
The procedure was done according to Cell line culture method described by ¹². The test was achieved in Al- Nahrain university\ Biotechnology research center, under a supervision of the laboratory staff.
Cell line culture of (REF) were prepared to the experiment by thawing separately in a water-path at 37℃.
The cells were placed in a 25 cm² diameter animal cell culture vessel.
Incubation of the container containing REF cell lines in an incubator of 5% CO2 at a temperature of 37℃ for 24 hours, after a day of incubation, and when ensuring that there is growth in cell cultures and that they are clean from contamination, secondary cultures were conducted.
An inverted microscope was used to inspect the cells to make sure they were healthy, clean, and had multiplied to the necessary amount (10,000-10,000,000) cells/µL.
Cells were transferred to the container and the used cultural medium was discarded.
The cells were given enough trypsin enzyme (2–2.5 µL), which they then incubated with for 30–60 seconds at room temperature 22-26℃ until they had detached from the flask, and then monitored until it changed from a single cell layer to single cells. Cells were further dispensed by pipetting in the growth medium.
Cells were distributed from (10.000-10.000.000) in each one wall of sterile 96-well microtitration plate.
A volume of 200 µL of RPmi media (Roswell park memorial institute), were added to serial flat-bottom 96-wall polystyrene microtiter- plate.
A volume of 20µL of a prepared bacterial supernatant, was added to three wells of sterile flat bottomed 96-well polystyrene microtiter- plate. A total of six wells with Rpmi media were considered as control (without bacterial supernatant).
The plates were covered with their parafilm and incubated of 5% Co2 at temperature of 37℃ for 24 hours, after a day of incubation all plates were gently washed thrice with PBS and dried.
The MTT assay.¹³
The assay measures the percentage of cellular viability depending on the colorimetric variation occured in the cell-line culture. The procedure was done as follow.
Avolume of 10µL of MTT indicator was added to the whole tested microtiter- plate of cell-line culture, and then the plate was supernatant (Escherichia coli, Streptococcus uberis, Enterobacter cloacae, Enterococcus faecium, Enterococcus faecalis).
The optical absorbance valves were carried out at a wavelength of 620nm. Using the ELISA reader device (Bio-rad Germany).
All resultants data of the optical density valves were analyzed statistically by using SPSS (Statistical Package for the Social Science) (version 29, IBM, USA.2023).
The cellular viability percentage of the test calculated according to the following equation.¹⁴

Results
Bacterial identification
The identification of bacteria isolated from total specimens of CRC patients as well as healthy volunteers (control group), showed numerous different bacterial species with different percentage. The bacterial diversity in CRC patients distributed as shown in Table 1, while in control group as shown in Table 2.
Table 1: Types of bacterial isolates in colorectal cancer and polyp specimens.
|
N |
Type of bacterial isolate |
Number (%) |
|
1 |
Enterobacter cloacae |
4(5.19%) |
|
2 |
Streptococcus uberis |
6(7.79%) |
|
3 |
Escherichia coli |
30(38.96%) |
|
4 |
Proteus mirabilis |
11(14.28%) |
|
5 |
Streptococcus constellatus pharyneis |
1(1.29%) |
|
6 |
Micrococcus luteus |
1(1.29%) |
|
7 |
Staphylococcus pseudintermedius |
1(1.29%) |
|
8 |
Streptococcus thoraltensis |
1(1.29%) |
|
9 |
Citrobacter freundii |
1(1.29%) |
|
10 |
Streptococcus mutans |
1(1.29%) |
|
11 |
Enterococcus faecium |
5(6.49%) |
|
12 |
Enterococcus faecalis |
4(5.19%) |
|
13 |
Granulicatella elegans |
1(1.29%) |
|
14 |
Enterococcus gallinarum |
2(2.59%) |
|
15 |
Serratia marcescens |
1(1.29%) |
|
16 |
Streptococcus sangunis |
1(1.29%) |
|
17 |
Staphylococcus lentus |
1(1.29%) |
|
18 |
Comamons testosteroni |
1(1.29%) |
|
19 |
Morganella morganii |
1(1.29%) |
|
20 |
Pseudomonas aeruginosa |
1(1.29%) |
|
21 |
Klebsiella pneumonia |
2(2.59%) |
|
|
Total |
77 |
Table 2: Types of bacterial isolates in control specimens
|
N |
Type of bacterial isolate |
Number (%) |
|
1 |
Escherichia coli |
50(86.20%) |
|
2 |
Klebsiella pneumonia |
3(5.17%) |
|
3 |
Salmonella Typhi |
2(3.44%) |
|
4 |
Staphylococcus aureus |
1(1.72%) |
|
5 |
Proteus mirabilis |
1(1.72%) |
|
6 |
Pseudomonas aeruginosa |
1(1.72%) |
|
|
Total |
58 |
The notice cultural characteristics isolated from colorectal cancer and polyp on blood agar media, macconkey agar, mannitol salt agar and Pfizer agar, has been shown fermenter lactose on macconkey agar 36(46,75%), non-fermenter lactose on macconkey agar 4(5.19%), β hemolytic on blood agar 5(6.49%), α hemolytic on blood agar 4(5.19%), γ hemolytic on blood agar 1(0.77%), colony color from dark brown to black on Pfizer agar 2(2.59%), as shown in Table 3.
Table 3: Characteristics in cultures of colorectal cancer and polyps
|
Characteristics in cultures |
% |
|
Fermenter lactose on macconkey agar |
36(46,75%) |
|
Non Fermenter lactose on macconkey agar |
4(5.19%) |
|
β hemolytic on blood agar |
5(6.49%) |
|
α hemolytic on blood agar |
4(5.19) |
|
γ hemolytic on blood agar |
1(0.77%) |
|
colony color from dark brown to black on Pfizer agar |
2(2.59%) |
The notice cultural characteristics isolated from control specimens on blood agar media, macconkey agar, and Pfizer agar, has been shown fermenter lactose on macconkey agar 53(91.37%), non-fermenter lactose on macconkey agar 1(1.72%), β hemolytic on blood agar 1(1.72%), as shown Table 4.
Table 4: Characteristics in cultures of control specimens
|
Characteristics in cultures |
% |
|
Fermenter lactose on macconkey agar |
53(91.37%) |
|
Non Fermenter lactose on macconkey agar |
1(1.72%) |
|
β hemolytic on blood agar |
1(1.72%) |
Bacterial selection
The results showed that among different bacterial species diagnosed in this study, the bacteria (Streptococcus uberis, Escherichia coli, Enterococcus faecalis, Enterobacter cloacae, Enterococcus faecium), were the most highly frequents in the specimens group of CRC patients, their percentages were (Streptococcus uberis 6(7.79%), Escherichia coli 30(38.96%), Enterococcus faecalis 4(5.19%), Enterobacter cloacae 4(5.19%) and Enterococcus faecium 5(6.49%)) respectively. In addition the species (Streptococcus uberis, Escherichia coli, Enterococcus faecalis, Enterobacter cloacae, Enterococcus faecium) were not found among species diagnosed in the control group. These species were selected as a chosen bacteria to be study their effectiveness in the cell-line culture technique.
Cell-line cultures viability
The study clarified and revealed the actions and impacts of the bacterial supernatants extracts as an inhibition factors of cellular growth on REF cell-line. The results data of the cellular viability percentage showed, for Streptococcus uberis (16.12%), Enterobacter cloacae (15.11%), Enterococcus faecium (16.39 %), Entreococcus faecalis (9.48%) and Escherichia coli (17.61%), as shown in Table 5 and Figure 3 showed the cellular viability. Figures 5,6,7,8 and 9 showed the impacts of bacterial supernatant extracts on cell-line cultures comparable with Figure 4, which represents the control state.
Table 5: Cellular viability on REF cell line culture for five isolates (Streptococcus uberis, Enterococcus faecium ,Entreococcus faecalis, Enterobacter cloacae complex, Escherichia coli).
|
NO |
Bacterial species |
Cellular viability % |
|
1 |
Streptococcus uberis |
16.12% |
|
2 |
Enterobacter cloacae complex |
15.11% |
|
3 |
Enterococcus faecium |
16.39% |
|
4 |
Enterococcus faecalis |
9.48% |
|
5 |
Escherichia coli |
17.61% |
Figure 3. Cellular viability percentage on REF cell line culture for five isolates (Streptococcus uberis, Enterococcus faecium , Entreococcus faecalis, Enterobacter cloacae complex, Escherichia coli).
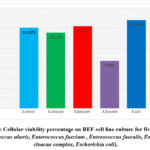 |
Figure 3: Cellular viability percentage on REF cell line culture for five isolates |
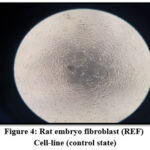 |
Figure 4: Rat embryo fibroblast (REF) Cell-line (control state) |
 |
Figure 5: Streptococcus uberis impact on REF cell- line |
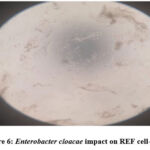 |
Figure 6: Enterobacter cloacae impact on REF cell-line |
 |
Figure 7: Enterococcus faecium impact on REF cell-line |
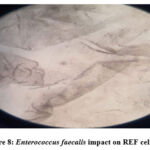 |
Figure 8: Enterococcus faecalis impact on REF cell-line |
 |
Figure 9: Escherichia coli impact on REF cell- line. |
Statistical analysis
Statistically the P-valves recorded in this study to the whole bacterial species in comparable with control were 0.001 which referred to a state of excellent or highly significant, as shown in Table 6.
Table 6. Statistical analysis for five isolates (Streptococcus uberis, Enterococcus faecium ,Entreococcus faecalis, Enterobacter cloacae complex, Escherichia coli) comparable with control.
|
Paired Sample Test |
|||||||||
|
Paired Difference Significance |
|||||||||
|
95% Confidence Interval Of the Difference |
|||||||||
|
|
Mean |
Std Deviation |
Std Error mean |
Lower |
Upper |
t |
dt |
One- Sided p |
Two-Sided p |
|
Pair1 Postive control-Streptococcus uberis |
412667 |
008622 |
004978 |
391249 |
434084 |
82.903 |
2 |
<.001 |
<.001 |
|
Pair2 Postive control- Enterobacter cloacae |
417667 |
005859 |
003383 |
403111 |
432222 |
123.462 |
2 |
<.001 |
<.001 |
|
Pair3 Postive control- Enterococcus faecium |
411333 |
010116 |
005840 |
386204 |
436463 |
70.428 |
2 |
<.001 |
<.001 |
|
Pair4 Postive control- Enterococcus faecalis |
445333 |
020207 |
011667 |
395136 |
495531 |
38.171 |
2 |
<.001 |
<.001 |
|
Pair5 Postive control- Escherichia coli |
405333 |
012014 |
006936 |
375489 |
435177 |
58.437 |
2 |
<.001 |
<.001 |
Discussion
Streptococcus uberis was showed the cellular viability percentageon REF cell line culture (16.12%), Phenotypic studies the Streptococcus uberis have made it possible to identify and characterize potential virulence factors in S. uberis strains, including the capsule, plasminogen activating factor, uberis factor, M- and R-like proteins, neutrophilic toxin, hyaluronidase, and extracellular matrix binding proteins. However, it is understood that environmental cues may influence how bacteria express their virulence factors.¹⁵ Briefly, virulence factors includes: plasminogen activator proteins such as PauA and PauB and SK, resistance to phagocytosis presented by a hyaluronic acid capsule, CAMP factor, a surface dehydrogenase protein GapC, sortases, Opp proteins implicated in dynamic transport of solutes, lactoferrin binding proteins, and adherence and invasion of epithelial cells mediated by S.uberis adhesion Molecule (SUAM).¹⁶ By acting on the proteins of the extracellular matrix, plasmin, which is created when plasminogen is activated, grants access to deep tissues. The first plasminogen activator identified in S. uberis strains that could activate bovine plasminogen was PauA activator, which had a molecular weight of about 30 KDa. Escherichia coli was showed the cellular viability percentageon REF cell line culture (17.61%). Cyclomodulins are toxins made by pathogenic E. coli strains, and they include cytotoxic necrotizing factor (CNF), cycle inhibiting factor (Cif), colibactin, and cytolethal distending toxins (CDTs). A toxin called colibactin is created by the enzyme polyketide synthetase (pks).¹⁷ Due to their capacity to affect cellular differentiation, apoptosis, and cell proliferation by interfering with the eukaryotic cell cycle and/or encouraging DNA damage, cyclomodulins are gaining more and more attention.¹⁸ Entreococcus faecalis was showed the cellular viability percentageon REF cell line culture (9.48%). It has been demonstrated that E. faecalis colonizes the digestive system by creating bacterial biofilms.¹⁹ Enhance the generation of ROS, which in turn induce colonic epithelial DNA double strand breaks aneuploidy and tetraploidy and extracellular superoxide , which stops the cell cycle,²⁰ and chromosomal instability , may be linked to a local inflammatory response, alterations in the mucosa’s nutrient compositions . Reactive oxygen species (ROS) produce by E. faecalis harm colon epithelial DNA and participate in the development of adenomatous polyp and.²⁰⁻²¹ Enterococcus faecium was showed the cellular viability percentageon REF cell line culture (16.39%). A study showed that hydrogen peroxide is produce by E. faecium at quantities that harm cells. Oxygen radical’s ability to disrupt membranes may make surrounding intestinal epithelial cells more susceptible to cellular harm, according to research on E. faecium transposon insertion mutants. Evidence suggests that the DNA of colonic epithelial cells is harmed by the formation of extracellular superoxide and hydrogen peroxide.²²Enterobacter cloacae was showed the cellular viability percentageon REF cell line culture (15.11%). Enterobacter cloacae its capacity the release of different cytotoxins including pore forming thiol- activated cytotoxins similar to the shiga like toxin Ⅱ, ²³hemolysins, and enterotoxins are crucial for its pathogenicity.²⁴The colorimetric technique called the MTT assay is used to measure the metabolic activity of cells and to determine how many viable cells are present, as shown in Figure 10, 11.
 |
Figure 10: A microtiter- plate of REF cell line culture and five isolates after added MTT indicator before incubation. |
 |
Figure 11: Metabolism of MTT to a formazan salt by viable cells, MTT indicator after incubation. |
Cell line culture are helpful model systems for researching typical cellular biochemical and physiological processes, and they can even be employed for diagnosis. The uniformity and reproducibility attained while employing cell culture systems is the key benefit. Cell culture essentially involves the multiplication and survival of cells in a lab setting. Cell growth is used in a variety of frequently occurring contexts, and Cell culture is usually thought of as a method by which cells are grown under controlled circumstances outside of a living organism (e.g., temperature, pH, nutrient, and waste levels), ²⁵the cells being investigated in the cell culture can be easily controlled when the traits and variables of cellular differentiation are known. A typical circumstance that is offered by the internal cellular program and external factors is cell differentiation.²⁶ In order to support the development of rat and human pluripotent stem cells, rat embryonic fibroblasts (REF) are used. REF not only provide a substrate for the growth of pluripotent stem cells, but they also secrete vital growth factors that help to preserve stem cell pluripotency. Early stages use REF that have been isolated from rat embryos,²⁷ REF must be irradiated or given mitomycin C to stop cell growth in order to function as feeder cells. Additionally, to prepare conditioned medium for feeder free pluripotent stem cell development, the treated cells can be employed. A fibroblast is the main functional cell in fibrous tissue. These cells are renowned for producing a lot of extracellular matrix (ECM) proteins, including collagen, glycosaminoglycans, and proteoglycans. They are irregularly shaped and adherent. By themselves, this elevates them to crucial status in numerous cell culture procedures, reasons using fibroblasts in cell culture because fibroblasts are simple to cultivate, and fibroblasts foster an ideal environment for development. Fibroblasts produce circumstances that are favorable for other cell types to grow in addition to their own by changing the properties of the environment around them. Indeed, it has been demonstrated that growing fibroblasts releases substances into the medium for cell culture that encourage development,²⁸and fibroblasts offer a model system for many illnesses and ailments it is known that fibroblasts act as agents of inflammation, disease related inflammation, and the evolution of cancer.²⁹⁻³⁰ The MTT assay is used to determine the cell viability and measure cytotoxicity (loss of viable cells) or cytostatic activity( shift from proliferation to quiescence) of potential medicinal agents and toxic materials is done by counting viable cells after staining with a vital dye, yellow MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, a tetrazole).³¹⁻³²⁻³³ The MTT assay measures colorimetrically the metabolic activity of cells. Under specific circumstances, the activity of NAD (P) H-dependent cellular oxidoreductase enzymes may indicate the presence of live cells. These enzymes have the ability to decrease the tetrazolium dye. In the mitochondria of live cells, MTT, or 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide, is converted to purple formazan, when the amount of purple formazan produced by cells treated with an agent is compared to the amount of formazan produced by untreated control cells, conversion can be directly related to the number of viable (living) cells because this reduction only occurs when mitochondrial reductase enzymes are active. By creating a dose response curve, it is possible to determine how effectively the substance kills cells. MTT solutions that have been dissolved in balanced salt solutions or tissue culture media are yellowish in hue and devoid of phenol red. Viable cells mitochondrial dehydrogenases split the tetrazolium ring, releasing purple MTT formazan crystals that are insoluble in water. The physiological condition of cells and variations in mitochondrial dehydrogenase activity in various cell types can influence the limitations of the MTT technique. MTT tests are typically carried out in the dark because the MTT reagent is light sensitive. Nevertheless, the MTT method of cell determination is useful in the measurement of cell growth in response to mitogens, antigenic stimuli, growth factors, and other reagents that promote cell growth, cytotoxicity studies, and the derivation of cell growth curves.
Conclusions
The cell line culture study revealed a reduction in the cellular growth viability on REF cell line culture as a result of supernatants activity extracted from (Escherichia coli, Streptococcus uberis, Enterobacter cloacae complex, Entreococcus faecalis and Enterococcus faecium ), respectively. There are changes in morphology shape cells , Irregular shape disruptions and damage cells line REF and decrease in cells number because of the supernatant toxic effects for the isolated bacteria. The results showed the cellular viability percentage were for Streptocccus uberis (16.12%), Enterococcus faecium (16.39%), Entreococcus faecalis (9.48%), Enterobacter cloacae (15.11%) and Escherichia coli (17.61%), respectively. These results indicated the possible important role of these bacteria in developing the colon rectal cancer.
Acknowledgement
Praise is toʻʻAllah” who enables me with his blessing to achieve this modest scientific effort. I would like to express my deepest gratitude and thanks to my supervisor Prof. Dr. Sameer Abdulameer Alash for suggesting, planning the scheme of this study, inspiring guidance, advice, encouragement and support.
Conflict of interest
Conflict of interest was collage of science, University of Baghdad, Baghdad, Iraq
References
- Siegel RL, Miller KD, Goding S A, Fedewa SA, Butterly LF,Anderson JC Cercek A, Smith RA, Jemal A. Colorectalcancer statistics. CA Cancer J Clin. 2020; 70:145–164. https://doi.org/10.3322/caac.21601.
- Montalban-Arques A, Scharl M. Intestinal microbiota and colorectal carcinoma: Implications for pathogenesis, diagnosis,and therapy. EBioMedicine. 2019; 48, 648–655.
- Fong W, Li Q, Yu J . Gut microbiota modulation: A novel strategy for prevention and treatment of colorectal cancer. Oncogen. 2020; 39, 4925–494.
- Zhu Q, Gao R, Wu W, Qin H. The role of gut microbiota in the pathogenesis of colorectal cancer. 2013; 34-130-1285.
- Levy M, Kolodziejczyk AA, Thaiss. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17(4):219–232. doi:10.1038/nri..7.
- Jedrzejczak-Silicka M. “History of Cell Culture,” in New Insights into Cell Culture Technology, eds S. Joghi and T. Gowder (London: IntechOpen). 2017.
- Kodack D P, Farago A F, Dastur A, Held M A, Dardaei L, Friboulet L , et . al. Primary Patient-Derived Cancer Cells and Their Potential for Personalized Cancer Patient Care. Cell Rep. 2017; 21, 3298–3309. doi: 10.1016/j.celrep.2017.11.051.
- Miserocchi G , Mercatali L , Liverani C , De Vita A, Spadazzi C, Pieri F, et . al.. Management and potentialities of primary cancer cultures in preclinical and translational studies. J. Translat. Med. 2017; 15, 1–16. doi: 10.1186/s12967-017-1328-z
- Leithner K , Wohlkoenig C, Stacher E, Lindenmann J, Hofmann N. A, Gallé B, et. al. Hypoxia increases membrane metallo-endopeptidase expression in a novel lung cancer ex vivo model – role of tumor stroma cells. BMC Cancer. 2014; 14, 1–13. doi: 10.1186/1471-2407-14-40.
- Ping Li, Chang T, Ruty M, Li J, Nancy W, Youzhen Y, Robert E, Maxson E N, Schulze, Houyan S, Chih-Lin H, Martin F P, Qi-Long Y. Germline Competent Embryonic Stem Cells Derived from Rat Blastocysts. Cell. 2008; 135(7): 1299-1310.
- Mohammad SM, Mahmud A N, Zawawi N. Probiotic properties of bacteria isolated from bee bread of stingless bee Heterotrigona itama. J Apicultural Res. 2020; 60(1):172–87 (Taylor and Francis Ltd).
- Vasanthi P, Ganapathy M, Evanjelene VK, Ayyavuv N, Angamuthu J. Phytochemical screening and antioxidant activity of extracts of the leaf and bark of Albizia lebbeck (Benth) Acad J Med Plant. 2014; 2:26–31.
- Pillai T G, Raghu D, Karunagaran D. Cytotoxic effects of polysaccharides isolated from Emilia sonchifolia in cervical cancer cell line. Cancer Rep. Rev. 2017; 1(6):1–4.
- Amir M, Khan A, Ashraf K, Sharma D, Aqil M . Phytochemical analysis and in vitro antioxidant activity of Uncaria gambir. Int. J. Green Pharm . 2012; vol. 6, 67–72.
- Leigh J. Exploiting the genome in the control of Streptococcus uberis. Proceedings of the British Mastitis Conference, Garstang. 2003;15-22.
- Almeida RA, Luther DA, Park HM, Oliver SP. Identification, isolation, and partial characterization of a novel Streptococcus uberis adhesion molecule (SUAM). Vet Microbiol . 2006;115:183-19.
- Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B , Darfeuille-Michaud A. Colonization of the human gut by E. coli and colorectal cancer risk. Clinical Cancer Research. 2014; 20(4), 859-867.
- Veziant J, Gagnière J, Jouberton E, et. al. Association of colorectal cancer with pathogenic Escherichia coli: focus on mechanisms using optical imaging. World Journal of Clinical Oncology . 2016; 7(3):p. 293. doi: 10.5306/wjco.v7.i3.293.
- Barnes A M T, Dale J L, Chen Y, Manias D A, Greenwood Quaintance K E , Karau M K , et. al. Enterococcus faecalis readily colonizes the entire gastrointestinal tract and forms biofilms in a germ-free mouse model. Virulence. 2017; 8, 282–296. doi: 10.1080/21505594.2016.1208890
- Wang X, Allen T D, May R J, Lightfoot S, Houchen C W, Huycke M M. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer research. 2008; 68(23), 9909-9917.
- Wang X, Huycke M M. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology. 2007;132:551–561.
- Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis , 23:529–536. International journal of antimicrobial agents. 2002; 54(2): 245–248.
- Barnes A I, Paraje M G, Del C, Battan P, Albesa I. Molecular properties and metabolic effect on blood cells produced by a new toxin of Enterobacter cloacae. 2001;17: 409–418
- Mezzatesta M L, Gona F, Stefani S. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 2012; 7, 887–902. doi: 10.2217/fmb.12.61.
- Y H Lin, M H, Wu. In Comprehensive Biotechnology (Second Edition); 2011.
- Onur U, Ayla E . in Omics Technologies and Bio-Engineering; 2018.
- Ping Li, Chang T, Ruty M, Li J, Nancy W, Youzhen Y, Robert E, Maxson E N, Schulze, Houyan S, Chih-Lin H, Martin F P, Qi-Long Y . Germline Competent Embryonic Stem Cells Derived from Rat Blastocysts. Cell. 2008; 135(7): 1299-1310.
- Caneparo C,et. al. Conditioned medium produced by fibroblasts cultured in low oxygen pressure allows the formation of highly structured capillary-like networks in fibrin gels. Sci. Rep .10. 2020.
- Mizoguchi F, et. al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun. 2018; 91 9, 1–11 .
- Winkler J, Abisoye-Ogunniyan A, Metcalf K J, Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat.Commun. 2020; 111 11, 1–19
- Khawla H Z, Ahmed T E. Cytotoxic effect of Zno Nanoparticales on the viability of Lishmania donovani promastigotes in vitro, Iraqi Journal of science . 2016; vol.57,No.4C , pp: 2811-28117
- Stockert JC, Horobin RW, Colombo LL, Blázquez-Castro. “Tetrazolium salts and formazan products in Cell Biology: Viability assessment, fluorescence imaging, and labeling perspectives” (PDF). Acta Histochemica. 2018; A April, 120 (3): 159–167. doi:10.1016/j.acthis.2018.02.005. PMID 29496266. Archived (PDF) from the original on 2022-02-24. Retrieved 2020-04-15.
- Ibrahim J A, Alsafa N K evaluating the in vitro cytotoxicity of thymus vulgaris essential oil on Mcf.7 and Hela cancer cell line , Iraqi Journal of science. 2021; Vol.62. No, pp: 2862-2871.








