Meraj Banu1 , Akbar Ali Khan Pathan2
, Akbar Ali Khan Pathan2 and K.V. Chaitanya3*
and K.V. Chaitanya3*
1Department of Business Management, Osmania University, Hyderabad-500007, India
2MGI Tech. Co. Ltd, Shenzen-518083, China
3Department of Microbiology and FST, GIS, GITAM University, Visakhapatnam-530045, India
Corresponding Author E-mail: Viswanatha.chaitanya@gmail.comDOI : https://dx.doi.org/10.13005/bpj/2646
Abstract
The frequent occurrence of chromosomal abnormalities in humans is one of the main factors responsible for the birth of children with disabilities. More than 7.6 million infants per year are diagnosed with severe genetic abnormalities. An increase in genetic abnormalities among children may be attributed to women suffering from hormonal disorders. Genetic malformations can either be hereditary or spontaneous due to the exposure of germinal cells to toxins and mutagens or even oxidative stress. Most genetic disorders lack proper treatment. However, proper counseling, therapy, and medication can minimize its impact. Early diagnosis of abnormalities in the fetus will benefit the parents in options assessment. Fetal chromosomal analysis is the best option for an appropriate genetic disorder diagnosis. The latest and emerging technologies involved in detecting chromosomal abnormalities at the prenatal stage are discussed in this review. Significant developments in prenatal diagnostics and the best globally available economical options were also discussed.
Keywords
Cytogenetics; Fluorescent In situ Hybridization; Genetic disorders, Karyotyping; MLPA; Next Generation Sequencing; QF-PCR
Download this article as:| Copy the following to cite this article: Banu M, Pathan A. A. K, Chaitanya K. V. Diagnostics for Genetically Inherited Disorders: From Cytogenetics to Genomics Technologies- A Review. Biomed Pharmacol J 2023;16(2). |
| Copy the following to cite this URL: Banu M, Pathan A. A. K, Chaitanya K. V. Diagnostics for Genetically Inherited Disorders: From Cytogenetics to Genomics Technologies- A Review. Biomed Pharmacol J 2023;16(2). |
Introduction
Condensation of DNA inside the cellular nucleus will form chromatin. During cell division, the DNA further condenses into a rod-shaped chromosome. The entire DNA in the nucleus of human cells packs into 23 sets of chromosomes, comprising 22 homologous autosomes and one allosome set, XX and XY, respectively. Every nuclear chromosome has a characteristic size and shape. The chromosomes morphology and staining allow their identification and numbering, termed karyotype. Chromosomal disorders arise from excess or deficiency in the chromosomes or their part, known as abnormalities. Chromosomal abnormalities are numeric and structural. Chromosomal disorders are monogenic, digenic, oligogenic, and polygenic (multifactorial). A chromosomal abnormality might occur due to numerical or structural changes in the chromosome, resulting in the rearrangement of genetic information leading to abnormalities in humans’ growth, development, and functioning. Structural and numerical changes occur during spermatogenesis, oogenesis or shortly after embryo formation. Chromosomal disorders form a major category of human diseases responsible for more than 100 genetic disorders, most of which are due to single-gene disorders. The past four decades have witnessed a remarkable development of novel genomic technologies for improving the diagnosis of existing disorders in the health sector. Compared with the genomics technologies available for diagnosing genetic disorders, pre-natal tests are still preferred in developing countries due to the unavailability of the technology in rural places, the cost of the tests, and the appropriate reimbursement system. All these factors are responsible for the abnormal delay in the diagnosis resulting in severe damage to human health. Prenatal diagnostics genetic techniques for assessing chromosomal abnormalities and their possible outcomes are discussed in this review, which provides an in-depth discussion of associated costs, significant players operating in the market, and the implications and limitations.
Prenatal Diagnostics
Prenatal testing comprising traditional and non-invasive along with preimplantation genetic testing, traditional prenatal testing forms the three significant components in the prenatal diagnosis. Rapid advances in the diagnostics combined with availability of the human genome and increased accuracy in sequencing have contributed to a major change in the accuracy and the noninvasive testing of genetic abnormalities1. These tests utilize cell-free DNA of maternal plasma. Techniques include Karyotyping, Molecular DNA Testing, FISH, Comparative Genomic Hybridization (CGH), Microarray Analysis, Next-Generation Sequencing, fetal blood analysis, and screening for cytogenetic malformations are into clinical diagnosis2. These recent developments have created the technical and logistical challenges and magnified the ethical and public policy issues since its inception. The conductance of genetic tests during the prenatal and early postnatal periods is becoming more inevitable, and the testing is performed on any part of the reproductive lifecycle (Figure 1). Genetic testing might not apply to chromosomal abnormalities during pregnancy in women over 35 years3. A genetic test can be performed on an individual with a known family pedigree consisting of abnormalities or genetic disorders.
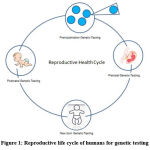 |
Figure 1: Reproductive life cycle of humans for genetic testing. |
Cytogenetics Tests
The global molecular cytogenetics market has been growing with an 11% compound annual growth rate (CAGR), doing a business total of USD 2,266.4 million since 2016. Cytogenetics studies the structure, properties, and changes in the chromosomes during mitosis and meiosis. It is also involved in understanding the influence of chromosome behavior during the cell division on the phenotype and the role of factors such as mutagens on the changes in chromosomes4. Chromosomes and chromatin are the dark staining region in the nucleus. During interphase, chromatin material is organized into extended, loosely coiled chromatin reticulum. During cell division, these structures condense to form chromosomes, carrying genetic information. Except for mosaic, this chromosomal complement is embedded in an individual’s diploid and haploid cells. Q-banding is one of the first techniques for banding a chromosome, discovered by Casperson and his colleagues,5 involving chromosomal staining with a fluorochrome and examining them under fluorescent microscopy. Low optimization of this technique has further led to the optimization of other banding patterns, including G, R, C, and NOR, using different staining techniques, with specific properties and applications. However, the resolution of the chromosomes was kept low during high condensation, making detecting the chromosomal rearrangements difficult. This situation was improved by developing high-resolution banding of lymphocyte cells obtained in pro and prometaphases6. High-resolution banding has helped to locate the chromosomal breakpoints and assignment of gene loci. High resolution banding and sub-banding of late prophase have provided twice the number of bands that are visualized during metaphase.
Several clinical syndromes with deletions in the chromosomal q and p arms, including Prader-Willi syndrome, Angelman syndrome (deletions on the chromosome 15 q arm), Smith-Magenis syndrome, and Miller-Dieker syndrome (deletions in the chromosome 17 p arm), DiGeorge/Velo Cardio Facial syndromes (deletions in the chromosome 22 q arm) occurs due to chromosomal rearrangements giving rise to a microdeletion or contiguous gene syndrome concept7. The invention of specialized cytogenetic tests such as the Sister Chromatid Exchange (SCE) assay has provided the visualization of interchanging regions of bright and dull sister chromatid segments8. There was an increase in the sister chromatid exchange segments in people suffering from spondylitis, carcinoma of the cervix uteri, and specific medical conditions such as fragile X-syndrome, in smokers, after exposure to mutagens, carcinogens, and biomass fuels9. Fragile sites and chromosome gaps are consistent in these patients. Chromosome breakage has increased in individuals exposed to cytotoxic agents. A reduction in the DNA repair system has led to chromosome damage, as identified in autosomal recessive disorders.
Advanced Molecular Analysis Methods for the Detection of Abnormal Changes in the Chromosomes
Difficulties in detecting the abnormal changes in chromosomes at the cytogenetics level despite using high-resolution banding techniques have paved the path for the invention of hybridization, polymerase chain reaction, and genome sequence-based molecular techniques.
Fluorescence in Situ Hybridization (Fish)
Fluorescence In Situ Hybridization (FISH) has shown the path for the modernization of cytogenetics. FISH has a market value of $650 million in 2021, with an expected $978.6 million by 2027 with 7.2% CAGR. Developed by Pinkel and his colleagues,10 FISH facilitates the microscopic visualization of chromosomal and nuclear locations of a cell. This technology aid in detecting specific DNA sequences of cells and tissues fixed either on interphase or metaphase11. Here, fluorescently labeled probes will be subjected to hybridization with highly specific complementary DNA or RNA sequences. Consequently, the cells will undergo washing for removing the unbound or loosely bound probe and are analysed for the presence of a fluorescent signal. Probes used for the FISH hybridization are categorized based on the location of their hybridization in the genome and the type of chromosomal aberration they are detecting12. The FISH technique can diagnose contiguous gene syndromes involving the loss of genes that are functionally unrelated but are contiguous along the chromosome. Angelman syndrome, Di George syndrome, Miller-Dicker syndrome, and Prader Willi syndromes are contiguous gene syndromes diagnosed by FISH13. The carrier status of Duchenne or Becker muscular dystrophy in females can also be detected using FISH. It also diagnoses various genetic disorders in conjunction with chromosomal aberrations. FISH can detect chromosomal abnormalities in small DNA segments. FISH can subject the non-dividing cellular nuclei to karyotyping.
The global market for FISH probes was $ 618.8 million in 2018, and $ 0.9 billion in 2020, with an estimated $1.3 billion by 2025 at a CAGR of 7.4%. Oxford Gene Technologies, Life Science Technologies, PerkinElmer Inc, Abnova Corporation, Biosearch Technologies Inc., Genemed Biotechnologies, Inc., and F. Hoffmann-La Roche AG are Some key players operating in the market. Companies are adopting collaborative strategies to gain an advantage and for regional expansions. A cost per test demonstrates that, although probe-based testing is cheaper in Asia compared to US and UK, it is still considered an expensive test among the masses, and only some woman has the luxury to opt for it (Table 1).
Table 1: FISH cost per sample in three different continents.
| S.No. | Continent | Value | Lower Limit | Upper Limit | Source |
| 1. | USA | $330 | $300 | $360 | Lab. survey |
| 2. | Europe | $197.72 | $169.09 | $244.20 | Lab. survey |
| 3. | Asia | $ 111.57 | $100.00 | $150.00 | Lab. survey |
Spectral Karyotyping and Multicoloured Fish
Hybridization of a FISH probe will lead to the fluorescence of a single gene copy. Multicoloured FISH will enable the visualization of each chromosome in the human cell with different colours by using a variety of fluorescent dye combinations and different concentrations14. Multicoloured FISH provides every chromosome with a specific colour for identifying tumor-associated complex abnormalities. Spectral karyotyping provides the staining of the whole chromosome with a single colour. Multicoloured FISH combines fluorescent dyes representing different probes simultaneously, offering visibility of all 24 chromosomes in different colours15. Spectral Karyotyping and multicoloured FISH are resourceful while detecting complex chromosomal abnormalities such as translocations. However, the data generated by using the FISH technique is of specificity, restricted to a specific region in the genome of an organism, that can be hybridized with a probe(s) of high specificity, and are highly complementary. FISH cannot detect any modifications to the DNA that are less than 30 Kb, including point mutations in a single gene. The probe for the FISH hybridization is to be subjected to microscopic analysis, which is tedious and time-consuming. This process demands automation and technological advancement.
Prenatal Diagnosis of Chromosomal Aberrations Through Cytogenetics
The global prenatal testing market was $3.23 billion in 2019, growing with a 12.9% CAGR totaling $ 8.08 billion in business by 2027. Prenatal diagnosis refers to the techniques used to determine a developing fetus’s health. It forms a basis of the factors, including any disorder in the family pedigree, stage of the pregnancy, etc. Four prenatal genetic screening mechanisms, 1. Non-invasive prenatal testing, 2. Prenatal diagnosis (sometimes testing also), 3. In vitro fertilization with preimplantation genetic testing, and 4. Termination of pregnancy due to the presence of genetic disorders in the fetus. Non-invasive prenatal testing is performed for cases with either a history or presence of high-risk pregnancies, abnormal fetal ultrasound, and a family pedigree with affirmative history. Prenatal diagnosis and testing are performed after identifying abnormalities during initial testing. In vitro fertilization with preimplantation genetic testing is considered only if one or both parents are confirmed to possess or are carriers of a genetic disorder. Termination of pregnancy is suggestible if the current pregnancy is affected by a genetic disorder, regardless of the parents’ status16.
Non-Invasive Prenatal Testing (Nipt)
NIPT brings an efficient mechanism for testing the fetus for genetic abnormalities, with the significant advantage of minimizing the risk of miscarriage associated with invasive diagnostic procedures. During NIPT, the DNA of the fetal cells is isolated from the maternal blood samples at nine weeks of pregnancy for genetic screening. This prenatal test has a 99 % sensitivity and specificity for detecting chromosomal abnormalities and 100 % specificity in detecting aneuploidies such as trisomy 13,18, and 2117. NIPT testing was the objective for women with a high risk of aneuploidy pregnancies of age over 35 years. It is also for women with a trisomy baby during their first pregnancy. ACOG, in 2013, had recommended offering NIPT to females with immense risk after performing preliminary examinations. This statement has increased the conductance for NIPT in almost all corners of the world. In most cases, central labs have predominantly carried out these tests, while others have adopted service labs and smaller clinics for the conductance of NIPT. One ethical issue surrounding non-invasive prenatal testing is the high false positive rate, possibly due to collecting samples from the placenta rather than the fetus18. NIPT helps in the early diagnosis of genetic disorders much before ensoulment.
The cost of a single NIPT test in 2014 ranged from $800-2000 in the USA to $500 -1500 in other parts of the world. Due to its initial success, most companies have shown interest in developing NIPT-based tests. In two years, seven NIPT tests have been launched into the market (Table 2). In the next few years, many NIPT-based tests were on the market, reducing the cost to $600-800. The average cost of NIPT in India is reduced from Rs. 50,000 – 60,000 ($700 – $1000) to Rs. 15,000-25,000. Through the input of technically competent labs in the country, many new tests are developing, and old tests are improving with a diagnostic CE marking. Consequently, there is a gradual increase in the number of labs offering NIPT tests through the license from manufacturing companies or developing as LDTs.
Prenatal diagnosis of the fetus is performed through cytogenetic, array-based, PCR-based, and sequencing-based testing techniques. Cytogenetic analysis of the amniotic fetal cells is known as amniocentesis. This test is well-established, safe, and accurate for detecting chromosomal abnormalities and other genetic disorders19. It involves the removal of amniotic fluid from the sac around the fetus for identifying congenital disabilities. Prenatal genetic diagnosis will be of high source when sonographic findings cannot accurately predict the trisomic syndromes20. Amniotic fluid cells are derived from the skin, kidney, urinary bladder, and other fetal tissues. Two methods are under practice to culture the amniotic fluid cells;1. Culturing and processing the amniotic fluid cells on a coverslip for retaining the individual colonies, and 2. Culturing cells in flasks using trypsin in the medium for mixing of cells. Transcervical and transabdominal CVs and fetal blood sampling obtain cells from chorionic villi. Harvested cells from both methods can analyze chromosomes for aberrations. Structural chromosomal abnormalities such as deletion, translocation, and chromosomal aneuploidies can be prenatally detected21. Prenatal chromosome rearrangements and fetal abnormalities are detected with ultrasound. However, repeated amniotic fluid and chorionic villi collection from pregnant women might lead to fetal loss.
The prenatal diagnostic tests market is booming, with 6.1 billion $ in 2020, with an expected 8.2 $ billion by 2025, at a CAGR of 6.1% globally. Of this, 3.7% is from the maternity centres segment. The levels of maternal AFP determined during the initial three months of pregnancy were positively correlated with aneuploidy22. The false-negative rate of the existing methods ranges from 12-23% 23. The utilization of fetal cells and cell-free fetal DNA present in maternal circulation became resourceful in 2011 after decades of research in the form of NIPT24. According to fortune business insights, the global NIPT market was 2.95 billion $ in 2019, predicted to reach $ 10.88 billion by 2027. The NIPT market is exhibiting a CAGR of 17.8%. In addition to the non-invasive methods, invasive procedures like PUBS also are used for prenatal screening in most developing countries. However, these procedures carry a risk of miscarriage25.
From the sampling date, it might take several weeks or even months to obtain test results. Prenatal test results are obtained quickly due to the essentiality of the time for making decisions regarding the continuation or termination of pregnancy. The doctor or a genetic counsellor provides specific information about the cost of the prescribed test and the time taken to obtain the result. The prenatal diagnostics market is being redefined and redesigned, which might be normal after the impact of COVID-19 on the market. As part of the emerging scenario, the United States has a readjustment of up to 6.7% CAGR. In Europe, the prenatal diagnostics market of Germany is expected to be $ 85.2 Million over the next 7-8 years. Over $ 81.7 Million is projected from other European countries. Prenatal diagnostics might reach up to $319.2 million in Japan. China faces stiff political and economic challenges, primarily due to the pandemic. Its decoupling and economic distancing approaches might change its relationship with the rest of the world, triggering an antagonism and generating openings for the prenatal diagnostics market. India has one of the world’s highest birth rates, about 27 million annually. Regrettably, it also has an infant mortality rate of 9 million annually. The high infant mortality rate could be attributed to genetic and congenital abnormalities, occurring at a frequency of 25-60 per every1000 births26, which can be significantly decreased by regular early pregnancy screening. Even though the prenatal screening and diagnosis market is relatively new to India, it can make its impact by 2024. Depending on the sample and the disorder, the approximate cost per genetic test ranges from $100-2,000. The cost might further increase if the conduction of multiple tests is inevitable or it should be conducted on multiple family members. The test cost varies by state for new born infants, with some covering a part and others charging up to $15-60 per infant.
Table 2: NIPT tests carried out by different companies and their countries.
| S.No. | Year | Test | Company | Country of Origin | CE-IVD Mark |
| 1. | 2011 | MaterniT21 Plus | Sequenome | USA | NO |
| 2. | 2011 | NIFTY | BGI | China | Yes |
| 3. | 2011 | Bambni Test | Berry Genomics | China | NO |
| 4. | 2012 | Verifi test | Verinata Health | USA | Yes |
| 5. | 2012 | Harmony test | Ariosa Diagnostics | USA | Yes |
| 6. | 2012 | PrenaTest | LifeCodexx | Germany | Yes |
| 7. | 2012 | Panorama test | Natera | USA | Software |
| 8. | 2014 | InformaSeq | LabCorp | USA | No |
| 9. | 2014 | VisibiliT | Sequenome | USA | No |
| 10. | 2015 | Q-natal Advanced | Quest | USA | No |
| 11. | 2015 | IONA test | Yourgene Health | UK | Yes |
| 12. | 2017 | VeriSeq | Illumina | USA | Yes |
| 13. | 2018 | Vanadis | PerkinElmer | USA | Yes |
Prenatal Diagnosis Using Array-Based Tools
Karyotype analysis for the visualization of copy number variations and chromosomal abnormalities has been the mainstream of detecting genetic abnormalities for quite a long time. Due to the advent of technology, traditional analysis has been replaced by molecular tools such as hybridization-based microarrays, PCR-based tools, and sequence-based methods. In a seminal study, a microarray of 4,406 pregnancies identified 264 (6%) clinically significant copy number variations with structural anomalies27. However, microarray analysis could not detect balanced chromosomal variations. For detecting these variations, arrays have undergone certain modifications in the form of comparative genomic hybridization and chromosomal microarray analysis28.
Comparative genomic hybridization (CGH) hybridizes numerous well-characterized probes with the DNA of a patient. Here, the fluorescence pattern of a hybridized spot is analyzed by the difference between the test and the reference. One CGH array is equivalent to the conductance of more than a thousand FISH experiments. The resolution provided by CGH is high and valuable for quantifying the copy number and identification of breakpoint segments that have been lost or generated29. CGH array can also say something about the differences and specific variations in the human genome. CGH is applied to identify genome-wide copy number variations of two individuals of size <100 kb. CGH is not used for the detection of triploidy, which is one of its major limitations30.
A high-resolution CGH for permitting the detection and exploration of the chromosomes is Chromosomal Microarray Analysis (CMA). This array-based technology is used to identify many abnormalities below sub-microscopic resolution31. Relative cost of CMA is one of the main hindrances to practicing CMA in prenatal diagnosis. This form of microarray can recognise copy number variations (CNVs) in the DNA responsible for an imbalance in chromosomal number32. CMA can provide more genome coverage than a CGH for better detection of CNVs. CMA is an alternative to the karyotype analysis due to its high sensitivity, condensed time, a requirement of low labour, and application of standard computational analysis. CMA concerned with prenatal diagnostics is highly specific, considerably reducing uncertain and insignificant results. This will minimise the parent anxiety and reduce the difficulty in making decisions33.
Despite its wide range of applications, CMA has a plethora of disadvantages. CMA cannot detect balanced chromosomal rearrangements, such as inversions or translocations, responsible for the deletion and duplication of genetic material. The chromosomal microarray is not sensitive in detecting low levels of tissue mosaicism in the fetus31.
Grand View Research, Inc. has reported raising the global microarray market with an expected 7.44 billion $ by 2026, at a CAGR of 8.7%. These tests are commercially available for $1500-3000, depending upon which assay is used (low density /high density. Similar to all medical tests, hospitals offer discounted costs to the reference laboratory or insurance carrier, lowering the test cost. CMA testing generates better genetic diagnosis at an incremental cost of US $2692 compared with karyotyping, with an average diagnosis cost $11,033. In addition, when a variant of unknown significance is obtained during CMA testing, CAM testing of both the parents can be obtained at an incremental cost of $4220. In this case, if both parents are unavailable for the analysis or sequencing, the patient’s DNA using next generation sequencing will be the right option at an incremental cost of $12,295. The cost of CMA per sample across the USA, UK, and Asia is outlined in Table 3.
Table 3: Cost of chromosome microarray per sample in three different continents
| S.No. | Continent | Value | Lower Limit | Upper Limit | Source
|
| 1. | USA | Variable | $1500 | $2000 | Lab. survey |
| 2. | Europe | Variable | $1300 | $1940 | Lab. survey |
| 3. | Asia | Variable | $247 | $630 | Lab. Survey |
PCR-Based Testing
Multiplex Ligation-Dependent Probe Amplification
Multiplex Ligation-Dependent Probe Amplification (MLPA) detects abnormal copy numbers in the sample through amplification. Application of MLPA will improve the copy number detection rate for genetic diseases due to partial or complete intragenic deletions and duplications34. More than 300 probes are available in the market for various disorders. Spinal muscular atrophy (SMA) is an autosomal recessive and progressive neurodegenerative disorder responsible for muscle weakness, atrophy, and partial or complete paralysis. SMA divided into the Type I, II, and III categories. Type I is severe, type II is in the intermediate stage of severity, and Type III is mild. 95% of the patients with SMA have a deleted region in the SMN1 gene exon 7. SMN2 gene, located next to SMN1, possesses a very high sequence homology. The number of SMN2 copies in the DNA will modulate the SMA phenotype.
MLPA diagnoses SMA in the following steps. 1. Genomic DNA is hybridized to specific probes. The specific probe for SMN1 exon 7 is 274 nucleotides long, and SMN2 exon 7 is 281 long. The critical single nucleotide difference in the ligation sites of SMN1 and SMN2 will distinguish them. 2. Ligation of the adjacent probes. 3. Ligation step is followed by a PCR amplification of ligated probes, whose products are the direct measure of the SMN1 and SMN2 copy numbers. 4. Following amplification, SMN1, and SMN2 amplified copies are separated through capillary gel electrophoresis. 5. Peaks are analysed by matching with the patterns of reference samples, respectively. MLPA is accurate in detecting SMA in 95% of patients. This technique cannot detect SMA in the remaining 5% of the patients due an inactive SMN1 gene, which remains its demerit. The inactiveness of the SMN1 gene in these patients is due to a point mutation35. There is also a possibility of polymorphic variations in the DNA, which might affect the probe binding.
Methylation PCR
DNA methylation is a phenomenon occurring in CpG islands for regulating the expression of imprinted genes in a genome. An extended stretch of either G or C nucleotides with methylated CpG dinucleotides is present in these islands. Their originating parent determines the expression of imprinted genes36. The failure in the imprinted gene expression could lead to neuro diseases Prader–Willi syndrome and Angelman syndrome37. Prader–Willi syndrome occurs due to paternal allele, and Angelman syndrome is due to maternal allele loss of functions, respectively.
Prader–Willi and Angelman syndrome can be diagnosed through molecular techniques such as Methylation-specific PCR with 99% accuracy38. The logic behind the utilization of methylation-specific PCR is the presence of more than 96% methylated cytosine residues in the SNRPN locus of the maternal allele. In contrast, no residue is methylated in the paternal allele. Initially, proband’s DNA is treated with sodium bisulfite to convert unmethylated cytosine residues to uracil. The resulting DNA comprising paternal and maternal SNRPN locus copies will be subjected to differential amplification, resulting in two bands of 174 bp and 100 bp for maternal and paternal alleles, respectively. Control DNA consists of both alleles amplified. Individuals with Prader–Willi syndrome will carry only the maternal allele, which gets amplified, and the patients of Angelman syndromes will have the amplification of the paternal band in their sample. The test cost for Prader–Willi / Angelman syndromes is around $300–360 per sample, depending on the bisulphite treatment kit used. The main drawback of the Methylation PCR is its incapability of detecting the Angelman syndrome occurring due to mutations in 10% of cases. Here, mutations occur due to deletions in the intragenic exon region of the UBE3A gene39. Further, this approach will only diagnose the disease but will not be able to provide insight into the disease mechanism.
Quantitative fluorescence-polymerase chain reaction (QF-PCR)
Fluorescently labeled amplifications for the detection through capillary gel electrophoresis are produced by QF-PCR, which is ideal for stable and chromosome-specific STRs of 2-5 bp40. DNA isolated from amniotic fluid cells amplified using the fluorescently labeled primers for the simultaneous detection of aneuploidies in 13, 18, and 21 chromosomes, respectively41. The amplicons are separated on a capillary gel through electrophoresis. The peak areas of respective repeat lengths are analyzed by sequencing. QF-PCR facilitates a rapid diagnosis of chromosomal aneuploidies by analyzing the sample in a span of 24 hours with results available within a day as it need not have the culture of fetal cells42. However, this requires the environment and infrastructure to maintain sophisticated equipment and well-trained personnel. Expertise is in need for accurate interpretation of the cytogenetic analysis. Clinical trials in the USA have shown a very high accuracy of 100%, without any false negatives validating the efficiency of this technique. Prenatal QF-PCR testing and karyotyping on 7680 samples have consistently resulted in 99% of the cases, with abnormal karyotyping was not detected in 0.05% of samples43. QF-PCR can provide clinically validated results, but it cannot detect abnormalities related to the sex chromosomes.
Single-Gene Analysis by Sanger Sequencing
Sanger dideoxy terminator DNA sequencing, widely known as Sanger sequencing, is a laboratory technique used to interrogate genes or the entire coding sequence of minor disease-causing variants such as single-base changes, a few base-pair deletions, and duplications. Sanger’s sequencing is ideal to identify the minute changes in the DNA sequence whose applications are numerous. Sanger’s sequencing can detect variants in all cystic fibrosis gene exons44. These regions are translated into proteins and further into protein complexes. A few pathogenic variants might change this protein function, and alter its expression levels (dosage), adversely affecting its transportation to the destined location (for example, membrane proteins). PCR amplification of the exon DNA is the first step in exon analysis, followed by bidirectional sequencing through Sanger’s method. Here, the PCR amplicons are processed through capillary electrophoresis prior to their submission for sequencing. Fluorescently labeled ddNTPs will perform the sequencing and be analyzed using software for direct visualization and DNA identification. The result sequence can be compared to the reference human genome to determine changes in the base pairs.
The genetics community is generating databases for the disease variants by identifying several disease-causing genes, especially those manifested during infancy, early, and late childhood45. More than 1,700 variants for the CFTR gene have been identified, with some common variants in a population and others being rare variants reported only in individuals. Sanger sequencing confirms the genetic variants discovered using next-generation sequencing (NGS) assays. Sanger sequencing is expensive if it must be done in-house at ~$500/Mb compared to less than $0.50/Mb for NGS platforms. However, the cost per test for service labs, including purification and sequencing, ranges between $10-50, with many service companies like Macrogen and Bioserve offering the facility at a meager price. Sanger sequencing is highly resourceful in the identification of small mutations in DNA. However, it needs the enrichment of other technologies to detect large-scale genetic rearrangements and copy number variations. Low throughput, cost efficiency, and the requirement of skilled personnel are some of the limitations faced while sequencing and testing more than one gene. Sanger sequencing also suffers from low sensitivity and a 10–15% detection limit, which means the chances of mutation detection are zero if it is present in less than 10–15% of DNA molecules46.
Optical Genome Mapping (OGM)
Optical genome mapping (OGM) is one of the most sensitive and novel technologies for detecting all categories of structural variations, including copy number variations (CNVs)47. OGM is one of the next-generation cytogenomic techniques for detecting postnatal genetic disorders and hematological malignancies. The current potency of OGM in prenatal genetic testing is due to its ability to identify significant sequence variations (balanced and unbalanced). Currently, the karyotyping and metaphase analysis is done by FISH, CNVs are analyzed by CMA, repeat contraction disorders by Southern blotting, and multiple repeat expansion disorders by PCR-based methods or Southern blotting. The laboratories apply multiple molecular methods to discover the disorders related to repeat expansion and contraction. With a non-invasive prenatal screening test (NIPT) as the standard of careful screening assay for all global pregnancies, OGM is anticipated as a high-resolution cytogenomic diagnostic tool employed following a positive NIPT screen or for high-risk pregnancies with an abnormal ultrasound.
OGM can diagnose all genetic disorders. OGM, combined with Saphyr, has complete clinical concordance compared to traditional genetic analysis methods while diagnosing abnormalities in a cohort of 85 patients with constitutional48. Bionano’s Saphyr system is an extremely sensitive and highly specific research use-only platform for researchers and clinicians to accelerate novel diagnostics and therapeutic methodologies for understanding chromosome changes. Lineagen has provided genetic analysis services for families and healthcare providers for over nine years. So far, Lineagen has conducted more than 65,000 tests on patients with neurodevelopmental concerns. Depending on the expected coverage (400 to 1600X), the cost per sample is between $650 and $950 per genome.
Despite its high range of applications in cytogenomics, OGM needs a few limitations, such as 1. Single-molecule DNA sequencing requires high precision to match the confidence from the redundant read coverage provided by current next-generation sequencing technologies.2. Nicks on both strands at similar positions result in the low template during sequence-by-synthesis. 3. fluorochrome-labeled nucleotides are not removed after incorporation; because of these bulky labels, multiple incorporations might be difficult.
Next Generation Sequencing Technology (NGST)
Prenatal screening methods for detecting aneuploidy have undergone drastic development since the 1970s. Many Non-invasive diagnostic methods were developed during the late 1980s and 90s by combining maternal serum analytes and ultrasonography measurements. However, these methods suffer from a high false-negative rate of 23% and a high positive rate of 5%, with poor sensitivity, ranging up to 95% 49. The high frequency of uncertainty in the sample analysis has led to the development of invasive methods, including amniocentesis or chorionic villi sampling, for performing karyotyping on fetal samples. Both these procedures carry a risk of miscarriage50. The development of a process for isolating DNA from fetal cells has led to the development of a screening method using cell-free fetal DNA (cfDNA) for non-invasive prenatal testing (NIPT). Diagnosing trisomy in maternal blood using next-generation sequencing has further opened a race for developing the best NIPT tests using NGS, offering more accurate and safe methods. Clinical testing commonly uses the Illumina (Illumina, Inc., San Diego, CA, USA) or Thermo Fisher Scientific (Thermo Fisher Scientific, Waltham, MA, USA) platforms. The affordability of these tests is improving in developed countries after their coverage by health insurance. The clinical testing market is price sensitive in developing countries, and a few companies are actively involved in developing and launching cheaper alternatives to developed nations. Vanadis, a Swedish company associated with PerkinElmer, started using rolling circle amplification for its platform51. NIPD Genetics Ltd, a Cyprus-based company, has developed a methylation-based MeDIP qPCR technique for detecting aneuploidies. Bio-Rad is using it for NIPT testing using digital PCR52. Agilent OnePGT solution enables comprehensive insights for every IVF transfer with a single genome-wide NGS workflow integrating preimplantation genetic testing for single-gene disorders (PGT-M), translocations (PGT-SR), and aneuploidy testing (PGT-A) including verified automatic data analysis software with built-in QC metrics53. Launching these machines will increase the quality of the tests at a reduced time and cost. PerkinElmer PG-Seq™ Rapid Non-invasive PGT kit analyzes picogram quantities of DNA (low template DNA) from spent embryo culture media or blastocoelic fluid samples for non-invasive preimplantation genetic testing and A Novel, Combined Approach to PGT-A & PGT-M with One NGS Workflow54.
Even though next-generation sequencing is considered the most advanced technology available for genetic analysis laboratories, it still needs to be considered a comprehensive way of analyzing a sample and still has a significant number of limitations to overcome. Several regions in the genome, including long repetitive sequence elements, are challenging to sequence and analyze. Difficulty in interpreting novel or rare variants is mainly because of the insufficient knowledge or non-availability of relevant tests, which have put these variants with low or uncertain clinical significance. Genetic variants with structural gene and copy number variations are to be confirmed through additional tests, which increases the cost and increases anxiety among patients and their families. These limitations must be addressed to make NGS a single detection method for all genetic variants with clinical relevance.
Whole Exome Sequencing
There is a considerable improvement in prenatal diagnosis of the cases identified with structural differences through sonography. Whole exome sequencing has identified the pathogenic variants in 80% of cases whose karyotype is expected55. Whole exome sequencing focuses on the exons or protein-coding regions in the genome. The exons account for 1.5% of the human genome, equivalent to 22,000 genes. Most of the genes associated with the inheritance of genetic disorders consist of exons, making whole exome sequencing data more promising than whole genome sequencing56. Prenatal whole exome sequencing can diagnose fetal anomalies, increasing our understanding of the developmentally lethal variants. So far, more than 16 case series with 6 fetuses have used whole exome sequencing data with a diagnostic range of 57%. Whole exome sequencing has successfully diagnosed more than 6% of anomalies in 14% of fetuses57. There was an increase in the yield upon the performance of whole exome sequencing on maternal, paternal, and proband trios. Whole exome sequencing will prioritize the variants for their increased chances of succumbing to the disorders. Variants are found in the fetal cells but not in their parents. Variants of recessive inheritance are homozygous or compound heterozygous in the fetus and heterozygous in the parents. Whole exome sequencing is recommended for clinical indications such as multiple congenital disabilities and neurodevelopmental delay when other tests remain uninformative. Whole exome sequencing is economical, reduces the hospital costs and the number of postnatal tests to be performed, avoids the diagnostic odyssey, and decreases the hospital stay58. The American College of Medical Genetics and Genomics (ACMG) has recommended whole exome sequencing as a vital source of diagnosis when a phenotype determines a genetic disorder with genetic testing targeting the phenotype is unavailable along with its family history.
The cost of variants and their analysis is significant for performing the whole exome sequencing. Variants analysts comprise a team of molecular geneticists, cytogeneticists, clinical geneticists, and bioinformaticians to provide accurate results and genetic counselors to interpret the results. Re-analysis and re-evaluation of the variants are also needed to identify the genes whose function was previously unknown59. Whole exome sequencing poses many challenges while evaluating the SNPs in the coding regions of the genome, which can be overcome by in-depth sequencing of the region. Whole exome sequencing cannot detect the differences in the copy number variations, achieved using microarray technology. The whole exome sequence is not designed to detect aneuploidy, polyploidy, translocations, trinucleotide repeats, and mosaicism of low levels. GC-rich areas are not accurately sequenced using whole exome sequencing. The technology turnaround might meet these challenges, making whole exome sequencing faster and more cost-effective.
Conclusion
The available genetic tests were drastically different from prenatal and genetic tests that are available today, with more inventions, answers, and corrections promising in the future. Due to the rapid advancement in the utilization of technologies for prenatal testing, prenatal genomics will confer the ability to successfully interrogate the fetal genome and transcriptome noninvasively in the near future. Careful consideration and thorough analysis are needed while introducing these tools into the market. Most molecular tests are also performed in rural places due to reduced cost and shipment and regulatory issues raised when samples are sent abroad for diagnosis. Moreover, many manufacturing companies compete with diverse CE-IVD-marked products, reducing the cost per test. Many governments have adopted insurance policies that cover expensive diagnostic tests for women, helping diagnostics to select the right and successful therapy.
Conflicts of Interest
All authors report no financial or any other conflicts of interest related to this work.
References
- Hui L. Noninvasive approaches to prenatal diagnosis: historical perspective and future directions. Methods in Molecular Biology, 1885:45-58 (2019).
CrossRef - Waggoner D, Wain KE, Dubuc AM, et al. Yield of additional genetic testing after chromosomal microarray for diagnosis of a neurodevelopmental disability and congenital anomalies: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genetics in Medicine, 20: 1105-1113 (2019).
CrossRef - Neagos D, Cretu R, Sfetea RC, Bohiltea LC. The importance of screening and prenatal diagnosis in the identification of the numerical chromosomal abnormalities. Maedica,6:179-184 (2011).
- Yahaya TO, Oladele EO, Anyebe D, et al. Chromosomal abnormalities predisposing to infertility, testing, and management: a narrative review. Bulletinof the National Research Centre, 45:65 (2021).
CrossRef - Caspersson T, Zech L, Johansson C, et al. Identification of human chromosomes by DNA-binding fluorescent agents. Chromosoma, 30:215–27 (1970).
CrossRef - Yusuf M, Sajid A, Robinson IK, Lalani E.-N. 3D Ultrastructural imaging of chromosomes using Serial Block-Face Scanning Electron Microscopy (SBFSEM). DNA, 2:30–43 (2022).
CrossRef - Shubhangi K, Alka D, Prashant M. Chromosomal abnormalities – A review. Central India Journal of Dental Sciences, 4: 35-40 (2013).
- Tumini E, Aguilera A. The sister-chromatid exchange assay in human cells. Methods in Molecular Biology, 2153: 383-393 (2021).
CrossRef - Kannan TP, Zilfalil BA. Cytogenetics: past, present and future. The Malaysian Journal of Medical Sciences, 16: 4-9 (2009).
CrossRef - Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proceedings of National Academy of Sciences USA, 83:2934–2938 (1986).
CrossRef - Bishop R. Applications of fluorescence in situ hybridization (FISH) in detecting genetic aberrations of medical significance. Bioscience Horizons, 3: 85-95 (2010).
CrossRef - Shah J, Weltman H, Narciso P, Murphy C, Poruri A, et al. Dual color fluorescence in situhybridization (FISH) assays for detecting Mycobacterium tuberculosis and Mycobacterium avium complexes and related pathogens in cultures. PLOS ONE,12: e0174989 (2017).
CrossRef - Riegel M. Human molecular cytogenetics: from cells to nucleotides. Genetics and Molecular Biology, 37:194-209 (2014).
CrossRef - Liehr T, Starke H, Weise A, Lehrer H, Claussen U. Multicolor FISH probe sets and their applications. Histology and Histopathology,19: 229-237 (2009).
- Liehr T, Starke H, Heller A, Kosyakova N, Mrasek K, Gross M, Karst C, et al. Multicolor fluorescence in situ hybridization (FISH) applied to FISH-banding. Cytogenetics and Genome Research, 114: 240-244 (2006).
CrossRef - Malik SD, Al-Shafai M, Abdallah AM. The special features of prenatal and preimplantation genetic counseling in Arab countries. Genes,13: 167 (2022).
CrossRef - Stokowski R, Wang E, White K, Batey A, Jacobsson B, Brar H, Balanarasimha M, Hollemon D, Sparks A, Nicolaides K. Clinical performance of non-invasive prenatal testing (NIPT) using targeted cell-free DNA analysis in maternal plasma with microarrays or next generation sequencing (NGS) is consistent across multiple controlled clinical studies. Prenatal Diagnostics, 35: 1243-1246 (2015).
CrossRef - Van Opstal D, Srebniak MI, Polak J, de Vries F, Govaerts LC, Joosten M, Go AT, Knapen MF, Van den Berg C, Diderich KE, et al. False negative NIPT results: Risk figures for chromosomes 13, 18 and 21 based on chorionic villi results in 5967 cases and literature review. PLoS ONE, 11: e0146794 (2016).
CrossRef - Stavljenić-Rukavina A. Prenatal diagnosis of chromosomal disorders-molecular aspects. Electronic Journal of the International Federation of Clinical Chemistry and Laboratory Medicine,19: 2-6 (2008).
- Carlson LM, Neeta LV. Prenatal diagnosis: screening and diagnostic tools. Obstetrics and gynecology clinics of North America 2017; 44: 245-256.
CrossRef - Gregg AR, Skotko BG, Benkendorf JL, Monaghan KG, Bajaj K, Best RG, Klugman S, Watson MS. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genetics in Medicine, 18:1056-1065 (2016).
CrossRef - Russo ML, Karin JB. A historical and practical review of first trimester aneuploidy screening. Seminars in fetal & neonatal medicine, 19: 183-187 (2014).
CrossRef - Angie CJ, Neeta V. Whole exome sequencing: Applications in Prenatal Genetics. Obstetrics and Gynecology Clinics of North America, 45: 69–81(2018).
CrossRef - Rafi I, Hill M, Hayward J, Chitty LS, Non-invasive prenatal testing: use of cell-free fetal DNA in Down syndrome screening. British Journal of General Practice, 67: 298-299 (2017).
CrossRef - Alfirevic Z, Navaratnam K, Mujezinovic F. Amniocentesis and chorionic villus sampling for prenatal diagnosis. Cochrane Database Systemic Reviews, 9: CD003252 (2017).
CrossRef - Bhide P, Gund P, Kar A. Prevalence of congenital anomalies in an Indian maternal cohort: healthcare, prevention, and surveillance implications. PLoS One,11: e0166408 (2016).
CrossRef - Wapner RJ, Martin CL, Levy B, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. Engl. J. Med., 367: 2175–2184 (2012).
CrossRef - Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. J. Hum. Genet., 86: 749–764 (2010).
- Lee CL, Chuang CK, Tu RY, Chiu HC, Lo YT, et al. Increased diagnostic yield of array comparative genomic hybridization for autism spectrum disorder in one institution in Taiwan. Medicina, 58: 15 (2022).
CrossRef - Evangelidou P, Alexandrou A, Moutafi M, Ioannides M, Antoniou P, Koumbaris G, Kallikas I, Velissariou V, Sismani C, Patsalis PC. Implementation of high resolution whole genome array CGH in the prenatal clinical setting: advantages, challenges, and review of the literature. Biomedical Research International, 2013: 346762 (2013).
CrossRef - Levy B, Wapner R. Prenatal diagnosis by chromosomal microarray analysis. Fertility and sterility,109: 201-212 (2018).
CrossRef - Brynn L, Ronald W. Prenatal diagnosis by chromosomal microarray analysis. Fertility and Sterility, 109:201-212 (2018).
CrossRef - Çebi AH, Altıner S. Investigation of developmental disabilities and congenital anomalies: single center experience and review of NRXN3and NEDD4L Molecular Syndromology, 11:197-206 (2020).
CrossRef - Stuppia, L, Antonucci I, Palka G, Gatta V. Use of the MLPA assay in the molecular diagnosis of gene copy number alterations in human genetic diseases. International Journal of Molecular Sciences,13: 3245-3276 (2012).
CrossRef - Sun Y, Kong X, Zhao Z, et al. Mutation analysis of 419 family and prenatal diagnosis of 339 cases of spinal muscular atrophy in China. BMC Medical Genetics,21: 133 (2020).
CrossRef - Joshi RS, Garg P, Zaitlen N, Lappalainen T, Watson CT, Azam N, Ho D, et al. DNA methylation profiling of uniparental disomy subjects provides a map of parental epigenetic bias in the human genome. American Journal of Human Genetics, 99: 555-566 (2016).
CrossRef - Eggermann T, Perez de Nanclares G, Maher ER, Temple IK, Tümer Z, Monk D, Mackay DJ, Grønskov K, Riccio A, Linglart A, Netchine I. Imprinting disorders: a group of congenital disorders with overlapping patterns of molecular changes affecting imprinted loci. Clinical Epigenetics 7:123 (2015).
CrossRef - Kotzot D. Complex and segmental uniparental disomy updated. Journal of Medical Genetics 45: 545–556 (2008).
CrossRef - Aguilera C, Viñas-Jornet M, Baena N, Gabau E, Fernández C, Capdevila N, Cirkovic S, Sarajlija A, Miskovic M, Radivojevic D, Ruiz A, Guitart M. Novel intragenic deletions within the UBE3A gene in two unrelated patients with Angelman syndrome: case report and review of the literature. BMC Medical Genetics 18: 137-143 (2017).
CrossRef - Dan DA, Maria G-H, Maria JT, Cristina G-G, Marta R.A, Carmen A, Carmen R-C, Isabel L-S. Application of quantitative fluorescent PCR with short tandem repeat markers to the study of aneuploidies in spontaneous miscarriages. Human Reproduction, 20:1235–1243 (2005).
CrossRef - Guo Q, Zhou Y, Wang X, Li Q. Simultaneous detection of trisomies 13, 18, and 21 with multiplex ligation-dependent probe amplification-based real-time PCR. Clinical Chemistry, 56: 1451-1459 (2010).
CrossRef - Badenas C, Rodríguez-Revenga L, Morales C, Mediano C, Plaja A, Pérez-Iribarne MM, Soler A, Clusellas N, Borrell A, Sánchez MÁ, Miró E, Sánchez A, Milà M, Jiménez W. Assessment of QF-PCR as the first approach in prenatal diagnosis. The Journal of Molecular Diagnostics, 12: 828–834 (2010).
CrossRef - Putzova M, Soldatova I, Pecnova L, Dvorakova L, Jencikova N, Goetz P, Stejskal D, QF-PCR-based prenatal detection of common aneuploidies in the Czech population: five years of experience. European Journal of Medical Genetics, 51: 209-218 (2008).
CrossRef - Cambraia A, Junior MC, Zembrzuski VM, Junqueira RM, Cabello PH, de Cabello G. Next-Generation sequencing for molecular diagnosis of cystic fibrosis in a Brazilian cohort. Disease Markers, 2021: 9812074 (2021).
CrossRef - Bean LJ, Hegde MR. Gene variant databases and sharing: creating a global genomic variant database for personalized medicine. Human Mutations, 37: 559-563 (2016).
CrossRef - Chin EL, da Silva C, Hegde M. Assessment of clinical analytical sensitivity and specificity of next-generation sequencing for detection of simple and complex mutations. BMC Genetics; 14:6 (2013).
CrossRef - Nikhil SS, Chi-Yu JL, Alex H, Ashis KM, Siavash D, et al. Optical genome mapping identifies rare structural variations as predisposition factors associated with severe COVID-19. iScience, 25: 103760 (2022).
CrossRef - Sahajpal NS, Barseghyan H, Kolhe R, Hastie A, Chaubey A. optical genome mapping as a next generation cytogenomic tool for detection of structural and copy number variations for prenatal genomic analyses. Genes, 12: 398 (2020).
CrossRef - Maxim LD, Niebo R, Utell MJ. Screening tests: a review with examples. Inhalation Toxicology, 26: 811-828 (2014).
CrossRef - Parikh R, Mathai A, Parikh S, Chandra Sekhar G, Thomas R. Understanding and using sensitivity, specificity and predictive values. Indian journal of ophthalmology, 56: 45–50 (2008).
CrossRef - Dahl F, Ericsson O, Karlberg O, et al.Imaging single DNA molecules for high precision NIPT. Science Reports, 8: 4549 (2018).
CrossRef - Papageorgiou EA, Koumbaris G, Kypri E, Hadjidaniel M, Patsalis PC, The epigenome view: an effort towards non-invasive prenatal diagnosis. Genes, 5: 310-329 (2014).
CrossRef - De Rycke M, De Vos A, Belva F, et al. Preimplantation genetic testing with HLA matching: from counseling to birth and beyond. Journal of Human Genetics 2020; 65: 445–454.
CrossRef - Martine DR, Veerle B. Preimplantation genetic testing for monogenic disorders. Genes 2020; 11: 871.
CrossRef - Yadava SM, Ashkinadze E. 125: Whole exome sequencing (WES) in prenatal diagnosis for carefully selected cases. The American Journal of Obstetrics and Gynecology 2017; 216: S87–S88.
CrossRef - Best S, Wou K, Vora N, Van den Veyver IB, Wapner R, Chitty LS. Promises, pitfalls and practicalities of prenatal whole exome sequencing. Prenatal Diagnostics 2017; 38:10-19.
CrossRef - Wapner RP, Brennan K, Bier L, Wou K, Goldstein D. Whole exome sequencing in the evaluation of fetal structural anomalies: A prospective study of sequential patients. The American Journal of Obstetrics and Gynecology 2017; 216: S5-S6.
CrossRef - Angie CJ, Neeta V. Whole exome sequencing: applications in prenatal genetics. Obstetrics and Gynecology Clinics of North America 2018; 45: 69–81.
CrossRef - Botkin JR, Belmont JW, Berg JS, Berkman BE, Bombard Y, Holm IA, et al. Points to consider: ethical, legal and psychosocial implications of genetic testing in children and adolescents. American Journal of Human Genetics 2015; 97: 6–21.
CrossRef
Expansions
NIPT: Non-Invasive Prenatal Testing
ACOG: American College of Obstetricians and Gynecologists
LDT: Large Distribution Transformers
SNP: Single Nucleotide Polymorphisms
FISH: Fluorescent In Situ Hybridization
CGH: Combined Genome Hybridization
CAGR: Compound Annual Growth Rate
SCE: sister Chromatid Exchange
AFP: Alpha Feto Protein
CE: Conformité Européene
CVS: Chorionic Villus Sampling
PUBS: Percutaneous Umbilical Blood Sampling
CMA: Chromosomal Microarray Analysis
CNVs: copy number variations
MLPA: Multiplex Ligation-Dependent Probe Amplification
SMA: Spinal Muscular Atrophy
OGM: Optical genome mapping
cf DNA: cell-free fetal DNA
ACMG: The American College of Medical Genetics and Genomics







