Manikantan Pappuswamy* , Aditi Chaudhary
, Aditi Chaudhary and Anushka Shitut
and Anushka Shitut
Department of Life Sciences, CHRIST (Deemed to be University), Bangalore, Karnataka, India.
Corresponding Author E-mail: manikantan.p@christuniversity.in
DOI : https://dx.doi.org/10.13005/bpj/2648
Abstract
As the new strains spread around the world, scientists have been trying to learn more about the different strains, especially Omicron, and how SARS-CoV2 acts in general. Studying historical trends of virus spread and the structure of the virus and its strains, as well as all the mechanisms it needs to survive, can help identify the symptoms and diagnose and treat the disease. The research has shown that the new strains, including Omicron, have a higher rate of mutation and transmissibility. Additionally, due to the rapid spread of the virus, there has not been a significant amount of time to understand the severity of the infection. To better understand the novel variants, a detailed analysis of the basic pathophysiology of the virus is needed. This includes transcriptome analysis for the recombination index to identify variation in the strand. This aided in the diagnostic process, and therapeutics for mutants of the virus could be treated. The Omicron strain is particularly threatening due to its rapid transmission rate and its property of immune evasion, which can make it less vulnerable to vaccination.
Keywords
Entry mechanism; Omicron; SARS-CoV-2 Variant
Download this article as:| Copy the following to cite this article: Pappuswamy M, Chaudhary A, Shitut A. A Systemic Review on Omicron Variant of SARS-CoV-2. Biomed Pharmacol J 2023;16(2). |
| Copy the following to cite this URL: Pappuswamy M, Chaudhary A, Shitut A. A Systemic Review on Omicron Variant of SARS-CoV-2. Biomed Pharmacol J 2023;16(2). Available from: https://bit.ly/3BoE0an |
Introduction
While in the past, coronaviruses have only been responsible for very mild respiratory infections, with the arrival of COVID-19 in 2019, the whole world was taken by surprise, culminating in a pandemic. The scientific research community and the medical community actively collaborated to develop a vaccine and a method to prevent the spread of the disease. This has resulted in several skills being developed to help deal with similar outbreaks. Having already gone through two to three major waves worldwide, researchers were certainly more prepared when the new strain, Omicron, came to light. There was rapid progress in the field in learning more about the patterns and behaviours of the coronavirus with the new strain. To better understand the variant, it is essential to compare it to the original strain1. This would involve studying the morphology and genomic structure of the virus, along with major processes like replication, transcription, recombination, and translation. Understanding how to deal with the new strain would involve studying the infection cycle and the process of diagnosis and treatment for the new strain. Meanwhile, a comparison with the other strains of the virus would also be essential to understanding the patterns of infection based on the mutations in the genome. Alongside this, taking a look at the biomarkers can help identify the disease based on the virus and all its strains to distinguish them and decide the course of treatment for the disease.
Background of coronavirus
Rapid spread of communicable airborne viruses is not an uncommon occurrence. However, coronavirus is among the few outbreaks that have progressed to pandemic levels. The virus has a characteristic crown-like morphology, hence its common and familial names. The first human coronavirus in records is human coronavirus strain B814 (HCoV-B814 and Coronavirus-OC43 (CoV-OC43). These were detected in the respiratory tracts of patients demonstrating symptoms of the common cold. The overlapping features of the pathogenesis are predominant pneumonia, asthma, and chronic bronchitis1, 2. The spread is more prevalent in humid and cold climatic conditions. The novel virus was first spread in China between the end of 2002 and the beginning of 2003, then spread worldwide. After isolation, it was planted on a cell line and studied more extensively than strains like 229E and OC43 in particular. It spread in 2002 and stayed an epidemic until 2004. The different strains of coronavirus spread zoonotically through bats while also remaining prevalent in human forms. The second outbreak was MERS-CoV (Middle Eastern respiratory syndrome), for which there was an outbreak in Saudi Arabia in 2012. While most of the coronavirus alpha and beta species proliferated through bats, humans acted as vectors. Human COVID infection rates were low till it emerged in Wuhan, China, resulting in a major outbreak leading to the pandemic3. There are a lot of different viral strains under COVID 19. In the 2019 pandemic, there are different strains due to mutations after infections, which will be discussed in detail later. There are several variants of COVID-19 in the world, with Omicron being one of the more recent and possibly more dangerous variants of the disease4.
Background of Omicron
Omicron is more easily transmissible in comparison to the delta-variant, or “variant of concern”. There are three hypotheses about the evolutionary history of the O-micron variant of coronavirus. These include poor surveillance, antigenic drift, or zoonosis5, 6. Antigenic drift due to host sensitivity and high mutability is the most probable explanation. The mutations could have occurred due to ROS (reactive oxygen species) or the translocation of cytidine deaminases. However, according to RNA assays conducted, the Omicron variant had 31 mutations uncharacteristic of the human-human transition. This introduces the possibility of host shift from transitional vectors like mice. The variant demonstrated rapid transmission, which made it a public threat.
Table 1
| S No. | Strain | Type of variant | Country of first seen | Date of the first case | Characteristic feature |
| 1 | Alpha | Variant of concern | United Kingdom | September 2020 | High severity and rapid circulation |
| 2 | Beta | Variant of concern | South Africa | May 2020 | Spike protein mutation, increased transmissibility |
| 3 | Omicron | Variant of concern | Multiple countries | November
2021 |
Rapid replication rate, immune evasion |
| 4 | Delta | Variant of concern | India | October
2020 |
High infection rate |
| 5 | Gamma | Variant of concern | November
2020 |
3 mutations in spike proteins | |
| 6 | Mu | Variant of interest | Peru | December 2020 | Completely evades vaccines’ immunization, 3 mutations |
| 7 | Lambda | Variant of interest | Colombia | Jan 2021 | High mortality rate and high transmissibility |
There were several dangerous strains of the family Coronaviridae, which is part of the order Nidovirales. There are four genera under the family Coronaviridae, including alpha, beta, gamma, and delta. Seven strains of Co-Vs were detected in the respiratory tract. Viruses in general are classified on the basis of variants of interest, being monitored, of concern, and variants of consequence and interest. According to this system of classification, Omicron currently is a strain of concern, with a sharp increase in the number of cases across the world.
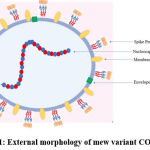 |
Figure 1: External morphology of mew variant COVID-19 |
Morphology of coronavirus strand
They are spherical, positive single-stranded RNA viruses enveloped in a lipid bilayer, containing a structural club-shaped glycoprotein, which is known as a spike protein. It is a triple-enveloped structure containing a membrane, a spike, and an envelope. The virion size varies between 70 and 90 nm. After entry through nasal pathways, the virus can be immobilised and inoculated with 2% paraformaldehyde and 2.5% glutaraldehyde. The virus attaches to the target cells with the help of the spike protein through interaction with the receptor. The virion is either spherical or polymorphic, vested with coronal fibrils on the envelope. The spike protein constructs a homotrimer emerging from the viral surface. The spike is composed of two functional subunits, S1 for binding to the host cell receptor and S2 for fusion of the viral cell membrane with the host cell membrane. The RBD (receptor binding domain) for the spike protein does not possess glycan shielding, resulting in immunodominance of the domain. ACE2 (Angiotensin-converting enzyme-2) was identified as the highest-functioning receptor that enables infection. The binding affinity of ACE2 often determines the severity of the infection. This is dependent on glycans and integrins present in host7. The virus genome encodes 9680 amino acids.
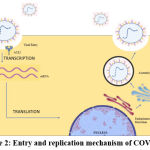 |
Figure 2: Entry and replication mechanism of COVID-19 |
Genomic organization
Being an RNA-based single-stranded genome; it can be transcribed and replicated within the capsid transcription to form messenger RNA. The genome contains 16 non-structural and 4 structural proteins. The genome is about 26–32 kb with 6–11 ORFs (open reading frames). The common features among all six known strands include:
First code in the ORF is AUG- initiated end along with several UTRs (Untranslated region) at the 5` end. The UTR at the 5` end is in some part involved in translation and is known as “leader”. Translation begins with the Kozak sequence.
The UTR at the 3` end is approximately 288- 506 nucleotides with a greater number of internal duplications. The termination sequence is approximately octamer sequence GAGA GAGA8.
The large gene, separated by ORFs 1a and 1b, covers over two-thirds of the genome. It contains the proteins needed for viral genome replication and sub genomic mRNA synthesis.
Expressed gene analysis includes RNA coding for spike glycoprotein, small envelope, membrane glycoprotein, and nucleocapsid proteins present in the genome.
A third of the genome starting from the 3` end forms a nested set of sub genomic mRNAs (sg mRNA). These sets with high degree of variability are responsible for infliction of host responses, determinant properties of viral pathogenicity8.
The ORF (Open reading frame) of Omicron is highly mutated which defines the mechanism of action, allowing entry of the variant into the membrane through ACE2.
NSP 4, NSP 5, and NSP10 possess cis acting elements, and the mutation cannot be neutralized by the trans-acting elements of another virus. The action of the mutants can be determined by temperature shift protocols. These cis acting elements act as regulators of the rate of transcription. The positive strand of the RNA is also responsible for the folding and formation of domains.
Translation
Coronaviruses bind to cellular proteins through the receptor (i.e., ACE2) and host factor (serine protease TMPRSS2), promoting viral uptake and fusion with the host cell membrane, which is a characteristic move of the virus9. After the entry of the virus, the first step is the translation of ORF1a and ORF1b, forming the polyproteins PPLA and PPlAB. The two proteins are precursors for the viral transcription and replication complexes. Meanwhile, a safe microenvironment for replication and transcription is maintained with the help of double-membrane vesicles, convoluted membranes, and small linear vesicles. Further cleavage of PP1A and PP1AB forms NSP1, NSP2, NSP3, and NSP4. Proteolysis releases NSPs5–16 and NSP4. After proteomic analysis of viral components, MPRO can be blocked by lead components without affecting the host microenvironment. RTC is also a target for antivirals, having been important for the process of viral replication.
The presence of a 5`-terminal methylated cap on genomic and sub-genomic mRNAs is indicative of a host-ribosome-mediated entry. Based on the pattern analysis of UTR regions, there is a scanning mechanism for the interaction of ribosomes with ORF1. Capping is essential for translation to occur. A leader is responsible for the regulation of translation rate10. Promoter with bulged stem-loop conformation acts as a molecular switch for cis-regulation; an alternative but uncommon pathway involves cyclic phosphates, which act on NendoU of NSP15.
Replication and transcription
Target recognition site (TRS) is present next to a leader sequence; 70 nucleotides upstream, sub-genomic mRNAs are produced in infected cells. NSP3, NSPNSP4, and NSP6 act as replication organelles. Replication units include NSP3, NSP5, and NSP8. The viral infection cycle progresses through organelles like the endoplasmic reticulum (ER) to form a primitive secretory complex. This is followed by the replicated virions leaving the cell in the form of vesicles 11. The subgeneric microRNA follows discontinuous transcription12. The transcription is the leader-primed transcription, which is then transformed into minus-stranded sub-genomic RNA. Upstream of the leader peptide, transcriptional regulatory sequences are present.
Omicron variants have a higher affinity for infection of the bronchus in comparison to other strains that infect the lungs. The replication process is also less dependent on TMPRSS2 receptor 13. It is however more dependent on endocytosis. Replication is less efficient in comparison to delta variant14. Plus and minus strands are both regulated by cis-acting elements. Enzymatic activity in the viral proteins is required for bioenergetics and duplicating the virus.
Papain-like proteases formed in NSP3 is responsible for the deubiquitinating group with finger pal, thumb, and Fingertips containing z spin binding domain.
ADP[(adenosine dinucleotide phosphate) ribose phosphatase with ADRP or X domain in NSP 3 responsible for folding, with three layers, is common in other plus strand RNA viruses.
3C’ like cysteine proteins in NSP5 with proteinase activity with twelve antiparallel beta-strand alpha helix with Carboxyl terminal domain III
RNA dependent RNA polymerase with RdRp domain from NSP 12 with finger palm thumb-like structure.
5` to 3` helicases with HEL in NSP 12 modelled to E.coli rep helicase, adjacent to zinc-binding domain.
3` to 5’exonucleases with ExoN domain in NSP 14, structure like hex helical bundle but not capable of cleaving ribose 2-O methylated RNA substrates.
Uridylate-specific endonuclease with NendoU domain within NSP 15 with a butterfly fold.
S- adenosylmethionine dependent methyl transferase from NSP 16 with a methyl transferase fold.
Recombination in coronavirus
Homologous recombination, more prominent in positive-strand RNA viruses, occurs at a rapid rate. It has been discovered in several RNA viruses, which, through natural selection, give them a high genetic advantage to survive. This property also increases the difficulty for virologists to find vaccines for different strains of the virus. Normally, recombination is possible at any point in the viral genome. However, it is known that there are some preferred crossover sites where genetic recombination is far more prominent15. Genetic drift refers to a dramatic change in the frequency of an existing allele. This phenomenon occurs significantly with spike protein-coding gene16. The genomic instability of coronaviruses can however be used to our advantage through vaccination to cause RNA interference and silence specific genes in the viral genome. Additionally, because of pleiotropy in the modification of spike proteins, selective recombinants can hence be targeted to fight specific strains of the virus. Recombination occurs through reverse genetics with the involvement of NSP-mediated synthesis of RNA. Deletion is also done with the help of virus growth and RNA synthesis. Meanwhile, the N protein [1234556] is responsible for the rescue of recombinant coronaviruses17.
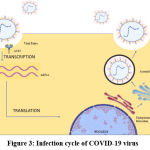 |
Figure 3: Infection cycle of Covid-19 virus |
It has been discovered in several RNA viruses, which, through natural selection, give them a high genetic advantage to survive. This property also increases the difficulty for virologists to find vaccines for different strains of the virus. Normally, recombination is possible at any point in the viral genome. However, it is known that there are some preferred crossover sites where genetic recombination is far more prominent. Genetic drift refers to a dramatic change in the frequency of an existing allele. This phenomenon occurs significantly with spike protein-coding gene 16. The genomic instability of coronaviruses can however be used to our advantage through vaccination to cause RNA interference and silence specific genes in the viral genome. Additionally, because of pleiotropy in the modification of spike proteins, selective recombinants can hence be targeted to fight specific strains of the virus. Recombination occurs through reverse genetics with the involvement of NSP-mediated synthesis of RNA. Deletion is also done with the help of virus growth and RNA synthesis. Meanwhile, the N protein is responsible for the rescue of recombinant coronaviruses 17.
Immune response to coronavirus
The body has its own mechanism of natural immunity through the lymphatic system. In order to keep track of microbes entering the body, there is a vesicular exchange between the lymphatic system and the blood. The most important part of the immune system is the WBCs (white blood cells)18. As the lymph moves to a lymph node, there are compartments where known viral antigens can be encountered. If an antigen is recognised, vessels filled with immune cells and antibodies flow through the body19, 20. In the case of SARS-CoV 2, pathogen recognition receptors present in the immune cells, for instance, toll-like receptors 3, 7, and 8, lead to interferon production21, 22. The humoral response meanwhile involves the production of IgG and IgM, which are responsible for the neutralisation of the immune system23. This is also accompanied by B cells targeting N proteins, as well as some targeting S proteins. Meanwhile, when it comes to the active response, there is a cytokine storm due to the hyperactive immune response, resulting in excessive inflammation.
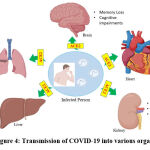 |
Figure 4: Transmission of COVID-19 into various organs |
Biomarkers of COVID-19
Biomarkers are essential for the diagnosis and prognostic treatment of the disease. An improved immune system and biochemicals injected to treat the disease can target it with the aid of biomarkers. Studying virus propagation techniques can help find biomarkers. Biomarkers can be haematological, biochemical, as well as inflammatory and coagulation biomarkers.
Haematological
These markers include the changes derived in blood content following infection with the virus. This includes eosinophil count, platelet count, neutrophil count, lymphocyte count, lymphocyte/neutrophil ratio count, as well as haemoglobin content24. Lymphopenia, which is a drastic decrease in the number of lymphocytes, is associated with a large number of SARS-CoV-2-positive patients. There is a higher leukocyte and neutrophil count, and in severe cases, a lower count of monocytes, eosinophils, and basophils25. The same applies to platelet count, which only drops in cases of severe infection. Both T cell and helper T cell counts drop significantly on infection by the virus 26.
Biochemical
The biochemical biomarkers were identified between fatal COVID cases and survivors. In non-survivors, there was a notable surge in bilirubin and creatine kinase (CK), along with an increase in iron concentration in the serum, WBCs, and IL-6. In cases of cardiac injury, this was also accompanied by a surge in cardiac troponin, which can progress into myocarditis and result in multiple organ failure. Multiple organ failure is brought about by the rise in liver enzymes, including aspartate aminotransferase and alanine aminotransferase27. Problems with liver function are a definite indicator of infection, as SARS-CoV2 binds to ACE2-positive cholangiocytes in the liver. Inadequate functioning of the liver related to cholangiocytes is associated with COVID-19 infection.
Inflammatory
Inflammation of blood vessels is associated with a certain stage of the infection. This is accompanied by an increase in CRP (C-reactive protein), which is a characteristic early feature of COVID 1928. Procalcitonin is released by the C cells in the thyroid. This is upregulated by endotoxin cytokines like IL-6, while it is reduced by the secretion of other cytokines (CT) like TNF-. The CT curve is more prominent in comparison to the curve for WBC. Release of IL-6 is associated with a phenomenon known as a cytokine storm, which is responsible for ARDS and acute lung injury. This, along with biochemical markers, is associated with more severe infections among patients29. Coagulation factors are associated with an increase in D-dimer and fibrin degradation products (FDP). The concentration of these products and the rate of coagulation peak when the patient is close to death. With freely circulating thrombin and no anti-coagulants to stop it, fibrinolysis occurs with the help of platelets30. This can lead to thrombocytopenia, which is a potentially fatal disseminated intravascular coagulation31.
Diagnosis
COVID tests are done after the detection of some basic symptoms, including those of the flu, including sore throat, cough, fatigue, and fever. This, however, is not applicable to all patients, as some people are asymptomatic. Nevertheless, the tests used for the diagnosis of SARS-CoV2 are the same, including RT-PCR (reverse transcription polymerase chain reaction), qRT-PCR (real-time quantitative reverse transcription PCR), and RT-LAMP (reverse transcription loop-mediated isothermal amplification) 32. The incubation time for COVID-19 is approximately 5.2 days. qRT-PCR, which detects ORF1b and N. The RT-PCR test displayed the results after several days (2–8). Meanwhile, the COVID-19 infection can be diagnosed with certainty with positive CT scans 33. CT scans have a higher accuracy in comparison to RT-PCR tests. After diagnosis, severe cases are admitted to follow the prognosis, while all other patients are responsible for practising isolation at home. The O-micron variant is also detected with the help of RT-PCR. The process conducts PCR, followed by looking for the genes responsible for the formation of the parts of the virus, including the spike (S), nucleocapsid (N), or inner area, and envelope (E). If the S gene is not detected, it is possible for the strain to be Omicron 52. This may be followed by genomic sequencing. The symptoms of omicron include aches and pains without taste or smell 34.
Treatment
Antivirals can prove to be a successful treatment for flu-like infections in the future. These are prescribed medicines capable of targeting and diminishing a virus. While none of the antivirals have been approved for SARS-CoV2, they can prove to be an efficient remedy for flu-like viruses. Some of the antivirals designed for the coronavirus include lopinavir, which blocks the protease activity of the coronavirus 35. Ribavirin works for a variety of different viruses, targeting SARS-CoV-2 5s RNA dependent RNA polymerase. Meanwhile, Remdesivir and IFN-beta have a high antiviral capability, which reduces the damage caused to the lung cells by COVID 1936. Meanwhile, chloroquine and hydroxychloroquine are broad-spectrum antivirals as well as immune boosters which can inhibit the virus through a change in endosomal pH resulting in merging of the virion membrane 37, 38. Another class of drugs capable of reducing the extent of COVID-19 infections is corticosteroids, which are capable of reducing swelling in the lungs 39. Monoclonal antibodies are an important part of the COVID-19 treatments and are generally targeted towards the spike protein, which is capable of inducing the response host. SARS-CoV-2, however, has a human-specific monoclonal antibody that can help neutralise the infection40. This, however, is more challenging to design for new infections, given that it has to be very specific to the antigen 41. Another way to introduce antibodies to the patient is through plasma transfusion, provided that the donor has recovered from COVID and has premade antibodies in the serum. This, however, introduces the probability of graft host rejection 42.
Table 2: Name of vaccine type of vaccine, age group for vaccine, interval between actions
| Name of vaccine |
Type of vaccine |
Age group |
Interval between doses |
Efficacy |
| Pfizer/ BioNTech Comirnaty | mRNA | 65+ first priority
Not for people below 12 |
21-28 days | 95.3% |
| AstraZeneca/ Covishield | Chimpanzee adenovirus
vector |
Health workers and 56+ priority | 12-16 weeks interval | 78% |
| Janssen | Vector virus | Health workers and 56+ priority 18+ | 14 days | 86% |
| Moderna | mRNA vaccine | Health workers and 56+ priority 18+ | 28 days | 95% |
| Sinopharm | Inactivated vaccine with adjuvant | Health workers and 56+ priority 18+ | 3-4 weeks | 79% |
| Sinovac- CoronaVac | Inactivated vaccine with adjuvant | Health workers and 56+ priority 18+ | 2-4 week | 71% |
| Bharat biotech COVEXIN | Inactivated vaccine, indigenous | Health workers and 56+ priority 15+ | 28 weeks | 78% |
Meanwhile, vaccines are a well-known preventative measure against COVID. The list of vaccines approved by the WHO for COVID has been listed above. Unauthorised vaccines are only to be taken in case of a domestic emergency 43. The functions of the vaccine involve the introduction of a part of a virus, which can alert the immune system and probe lymphocytes to produce antibodies against the virus. When it comes to the treatment of the O-micro strain and others, it is not too different from that of the original strains44. The severity of the infection has not been confirmed yet, as the long-term reaction has not been monitored. Currently, symptoms of O-micron are managed with the help of corticosteroids by blocking the IL-6 receptor. It has also been confirmed that the strain can be neutralised with the help of monoclonal antibodies45-52.
Conclusion
The O-micron strain certainly caused concern for wave. Flipping the pages on the basic features of SARS-CoV-2 can certainly help us compare and contrast to create a clear picture for the diagnosis and treatment of the new strain. This includes studying the genome’s organisation, morphology, replication, transcription, and translation, along with the host’s response, the infection cycle, and general methods of diagnosis and treatment. While the coronavirus has had a negative impact on our lives, it is a point that will be beneficial for progress in the field of virology. With the unique recombination patterns and high transition rates observed in emerging viruses, it is crucial for the field to progress.
Abbreviation
-> Niche specific proteins
Covid 19-> SARS-CoV2
RTC-> replication-transcription complexes
ARDS-> acute respiratory distress syndrome
Acknowledgment
We would like to thank Fr. Jobi Xavier, Head of the Department of Lifesciences, Christ (Deemed to be University), Bangalore, Karnataka, India for providing us the with the opportunity and requirements needed for the accomplishment of the research project.
Conflict of Interest
Authors have declared that no competing interests exist.
Funding Source
No funding sources received for this review study
References
- Jahangir, M. A., Muheem, A., & Rizvi, M. F. Coronavirus (COVID-19): history, current knowledge and pipeline medications. Int J Pharm Pharmacol., 2020; 4: 140.
CrossRef - Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012 Apr;86(7):3995-4008.
CrossRef - Kahn JS, McIntosh K. History and recent advances in coronavirus discovery. Pediatr Infect Dis J. 2005 Nov;24(11 Suppl):S223-7, discussion S226.
CrossRef - Izurieta HS, Graham DJ, Jiao Y, Hu M, Lu Y, Wu Y, Chillarige Y, Wernecke M, Menis M, Pratt D, Kelman J, Forshee R. Natural History of Coronavirus Disease 2019: Risk Factors for Hospitalizations and Deaths Among >26 Million US Medicare Beneficiaries. J Infect Dis. 2021 Mar 29;223(6):945-956.
CrossRef - Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021 Dec;600(7887):21.
CrossRef - Kupferschmidt K. Where did ‘weird’ Omicron come from? Science. 2021 Dec 3;374(6572):1179. doi: 10.1126/science.acx9738. Epub 2021 Dec 2. PMID: 34855502.
CrossRef - Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S, Wong SK, Huang IC, Xu K, Vasilieva N, Murakami A, He Y, Marasco WA, Guan Y, Choe H, Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005 Apr 20;24(8):1634-43.
CrossRef - Kumar S, Thambiraja TS, Karuppanan K, Subramaniam G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J Med Virol. 2022 Apr;94(4):1641-1649.
CrossRef - Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 Apr 24;368(6489):409-412. doi: 10.1126/science.abb3405.
CrossRef - Nelson GW, Stohlman SA, Tahara SM. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J Gen Virol. 2000 Jan;81(Pt 1):181-8.
CrossRef - Hoffmann HH, Sánchez-Rivera FJ, Schneider WM, Luna JM, Soto-Feliciano YM, Ashbrook AW, Le Pen J, Leal AA, Ricardo-Lax I, Michailidis E, Hao Y, Stenzel AF, Peace A, Zuber J, Allis CD, Lowe SW, MacDonald MR, Poirier JT, Rice CM. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2021 Feb 10;29(2):267-280.e5.
CrossRef - Neumann G, Kawaoka Y. Reverse genetics systems for the generation of segmented negative-sense RNA viruses entirely from cloned cDNA. Curr Top Microbiol Immunol. 2004;283:43-60.
CrossRef - Hui KPY, Ho JCW, Cheung MC, Ng KC, Ching RHH, Lai KL, Kam TT, Gu H, Sit KY, Hsin MKY, Au TWK, Poon LLM, Peiris M, Nicholls JM, Chan MCW. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022 Mar;603(7902):715-720.
CrossRef - Zhao H, Lu L, Peng Z, Chen LL, Meng X, Zhang C, Ip JD, Chan WM, Chu AW, Chan KH, Jin DY, Chen H, Yuen KY, To KK. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect. 2022 Dec;11(1):277-283.
CrossRef - Banner LR, Keck JG, Lai MM. A clustering of RNA recombination sites adjacent to a hypervariable region of the peplomer gene of murine coronavirus. Virology. 1990 Apr;175(2):548-55.
CrossRef - Kusters JG, Jager EJ, Niesters HG, van der Zeijst BA. Sequence evidence for RNA recombination in field isolates of avian coronavirus infectious bronchitis virus. Vaccine. 1990 Dec;8(6):605-8
CrossRef - Haijema BJ, Volders H, Rottier PJ. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J Virol. 2003 Apr;77(8):4528-38.
CrossRef - Keck JG, Makino S, Soe LH, Fleming JO, Stohlman SA, Lai MM. RNA recombination of coronavirus. Adv Exp Med Biol. 1987;218:99-107.
CrossRef - Kirchdoerfer RN, Cottrell CA, Wang N, Pallesen J, Yassine HM, Turner HL, Corbett KS, Graham BS, McLellan JS, Ward AB. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016 Mar 3;531(7592):118-21.
CrossRef - Gupta MK, Vemula S, Donde R, Gouda G, Behera L, Vadde R. In-silicoapproaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J Biomol Struct Dyn. 2021 Apr;39(7):2617-2627.
CrossRef - Su M, Chen Y, Qi S, Shi D, Feng L, Sun D. A Mini-Review on Cell Cycle Regulation of Coronavirus Infection. Front Vet Sci. 2020 Nov 5;7:586826.
CrossRef - Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020 Mar;579(7798):265-269.
CrossRef - Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol. 2020 Jun;20(6):339-341.
CrossRef - Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020 May;8(5):475-481.
CrossRef - Zhang B, Zhou X, Zhu C, Song Y, Feng F, Qiu Y, Feng J, Jia Q, Song Q, Zhu B, Wang J. Immune Phenotyping Based on the Neutrophil-to-Lymphocyte Ratio and IgG Level Predicts Disease Severity and Outcome for Patients With COVID-19. Front Mol Biosci. 2020 Jul 3;7:157.
CrossRef - Cossarizza A, De Biasi S, Guaraldi G, Girardis M, Mussini C; Modena Covid-19 Working Group (MoCo19)#. SARS-CoV-2, the Virus that Causes COVID-19: Cytometry and the New Challenge for Global Health. Cytometry A. 2020 Apr;97(4):340-343.
CrossRef - Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020 Jun 25;58(7):1021-1028.
CrossRef - Schons MJ, Caliebe A, Spinner CD, Classen AY, Pilgram L, Ruethrich MM, Rupp J, Nunes de Miranda SM, Römmele C, Vehreschild J, Jensen BE, Vehreschild M, Degenhardt C, Borgmann S, Hower M, Hanses F, Haselberger M, Friedrichs AK; LEOSS-study group. All-cause mortality and disease progression in SARS-CoV-2-infected patients with or without antibiotic therapy: an analysis of the LEOSS cohort. Infection. 2022 Apr;50(2):423-436.
CrossRef - Guan WJ, Liang WH, Zhao Y, Liang HR, Chen ZS, Li YM, Liu XQ, Chen RC, Tang CL, Wang T, Ou CQ, Li L, Chen PY, Sang L, Wang W, Li JF, Li CC, Ou LM, Cheng B, Xiong S, Ni ZY, Xiang J, Hu Y, Liu L, Shan H, Lei CL, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Cheng LL, Ye F, Li SY, Zheng JP, Zhang NF, Zhong NS, He JX; China Medical Treatment Expert Group for COVID-19. Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J. 2020 May 14;55(5):2000547.
CrossRef - Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 Mar 28;395(10229):1054-1062.
CrossRef - Terpos E, Ntanasis-Stathopoulos I, Elalamy I, Kastritis E, Sergentanis TN, Politou M, Psaltopoulou T, Gerotziafas G, Dimopoulos MA. Hematological findings and complications of COVID-19. Am J Hematol. 2020 Jul;95(7):834-847.
CrossRef - Bwire GM, Majigo MV, Njiro BJ, Mawazo A. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: A systematic review and meta-analysis. J Med Virol. 2021 Feb;93(2):719-725.
CrossRef - Pan F, Ye T, Sun P, Gui S, Liang B, Li L, Zheng D, Wang J, Hesketh RL, Yang L, Zheng C. Time Course of Lung Changes at Chest CT during Recovery from Coronavirus Disease 2019 (COVID-19). Radiology. 2020 Jun;295(3):715-721.
CrossRef - Callaway E, Ledford H. How bad is Omicron? What scientists know so far. Nature. 2021 Dec;600(7888):197-199.
CrossRef - Yao TT, Qian JD, Zhu WY, Wang Y, Wang GQ. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus-A possible reference for coronavirus disease-19 treatment option. J Med Virol. 2020 Jun;92(6):556-563.
CrossRef - Falzarano D, de Wit E, Rasmussen AL, Feldmann F, Okumura A, Scott DP, Brining D, Bushmaker T, Martellaro C, Baseler L, Benecke AG, Katze MG, Munster VJ, Feldmann H. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013 Oct;19(10):1313-7.
CrossRef - Savarino A, Di Trani L, Donatelli I, Cauda R, Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006 Feb;6(2):67-9.
CrossRef - Jallouli M, Galicier L, Zahr N, et al. Determinants of hydroxychloroquine blood concentration variations in systemic lupus erythematosus. Arthritis & Rheumatology (Hoboken, N.J.). 2015 May;67(8):2176-2184.
CrossRef - Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb 15;395(10223):497-506.
CrossRef - Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y, Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020 Feb 17;9(1):382-385.
CrossRef - Marovich M, Mascola JR, Cohen MS. Monoclonal Antibodies for Prevention and Treatment of COVID-19. JAMA. 2020 Jul 14;324(2):131-132.
CrossRef - Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 Mar;579(7798):270-273.
CrossRef - Ingraham NE, Ingbar DH. The omicron variant of SARS-CoV-2: Understanding the known and living with unknowns. Clin Transl Med. 2021 Dec;11(12):e685.
CrossRef - Mileto D, Micheli V, Fenizia C, Cutrera M, Gagliardi G, Mancon A, Bracchitta F, De Silvestri A, Rizzardini G, Lombardi A, Biasin M, Gismondo MR. Reduced neutralization of SARS-CoV-2 Omicron variant by BNT162b2 vaccinees’ sera: a preliminary evaluation. Emerg Microbes Infect. 2022 Dec;11(1):790-792.
CrossRef - Mohiuddin M, Kasahara K. Investigating the aggressiveness of the COVID-19 Omicron variant and suggestions for possible treatment options. Respir Med. 2022 Jan;191:106716.
CrossRef - Spaan W, Cavanagh D, Horzinek MC. Coronaviruses: structure and genome expression. J Gen Virol. 1988 Dec;69 ( Pt 12):2939-52.
CrossRef - Karim SSA, Karim QA. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021 Dec 11;398(10317):2126-2128. doi: 10.1016/S0140-6736(21)02758-6. Epub 2021 Dec 3. Erratum in: Lancet. 2022 Jan 8;399(10320):142.
CrossRef - Wei C, Shan KJ, Wang W, Zhang S, Huan Q, Qian W. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J Genet Genomics. 2021 Dec;48(12):1111-1121.
CrossRef - Mohapatra RK, Tiwari R, Sarangi AK, Islam MR, Chakraborty C, Dhama K. Omicron (B.1.1.529) variant of SARS-CoV-2: Concerns, challenges, and recent updates. J Med Virol. 2022 Jun;94(6):2336-2342.
CrossRef - Wang X, Powell CA. How to translate the knowledge of COVID-19 into the prevention of Omicron variants. Clin Transl Med. 2021 Dec;11(12):e680.
CrossRef - Wu YC, Chen CS, Chan YJ. The outbreak of COVID-19: An overview. J Chin Med Assoc. 2020 Mar;83(3):217-220.
CrossRef - Long C, Xu H, Shen Q, Zhang X, Fan B, Wang C, Zeng B, Li Z, Li X, Li H. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur J Radiol. 2020 May;126:108961.
CrossRef







