Simhadri V. S. D. N. A. Nagesh1* , Kannan I2
, Kannan I2  and Bairagi K. K.3
and Bairagi K. K.3
1Department of Pharmacology, Shri Ram Murti Smarak Institute of Medical Sciences, Bareilly.
2Department of Microbiology, Tagore Medical College and Hospital, Chennai.
3Department of Forensic Medicine, Shri Ram Murti Smarak Institute of Medical Sciences, Bareilly.
Corresponding Author E-mail: nageshsai117@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/2626
Abstract
The majority of current pharmaceuticals are derived from traditional plants; one of these, Azadirachta indica, also known as neem, has a variety of therapeutic applications ranging from simple infections to cancer. All of these pharmacological effects are due to the secondary metabolites present in the various plant parts. Diverse researchers made numerous attempts to identify the active ingredients using techniques such as Gas Chromatography-Mass Spectrometry (GC-MS), High-performance liquid chromatography (HPLC), and High performance thin-layer chromatography (HPTLC), among others. The GC-MS technique is used to isolate various secondary metabolites from the leaves of an aqueous extract of A.indica. The isolated compounds were analysed for their pharmacokinetics and pharmacodynamics properties using software such as SWISSADME, OPENBABEL, Swiss target prediction, etc. The aqueous extract of A.indica yielded 13 compounds, but only 5 compounds showed the highest number of hits; those with the highest concentration were chosen to obtain the pharmacodynamic, pharmacokinetic, and toxicological profiles. All five compounds are non-toxic and can be administered orally, and molecules with specific properties are capable of modulating a variety of proteins, including some enzymes. Based on this information, we can assume that these molecules can be used as "hit" or "lead" molecules in preclinical studies.
Keywords
Azadirachta indica; GC-MS; Pharmacokinetics; Pharmacodynamic; Secondary Metabolites; SWISSADME; Swiss target prediction
Download this article as:| Copy the following to cite this article: Nagesh S. V. S. D. N. A, Kannan I, Bairagi K. K. Pharmacophoric Evaluation of Compounds Isolated from GC-MS Analytical Method of Aqueous Extract of Azadirachta indica Leaves. Biomed Pharmacol J 2023;16(1). |
| Copy the following to cite this URL: Nagesh S. V. S. D. N. A, Kannan I, Bairagi K. K. Pharmacophoric Evaluation of Compounds Isolated from GC-MS Analytical Method of Aqueous Extract of Azadirachta indica Leaves. Biomed Pharmacol J 2023;16(1). Available from: https://bit.ly/3GRxMlr |
Introduction
Since ancient times, the use of medicinal plants to treat both common and uncommon ailments has been documented. Azadirachta indica, commonly known as Neem, is a plant that has been used traditionally to treat a variety of human diseases. It is a member of the Meliaceae family and is native to Burma and the Indian subcontinent. Melia azadirachta Linn is an alternative name for this plant. Indian lilac (English), neeb (Arabic), Azadirakhta (Persian), Margosa, Dogon yaro (certain Nigerian languages), Pokoksemambu (Malaysia), Kohomba (Sinhala), Tamar (Burmese), Nimba (Sanskrit), Vepa (Telugu), and neem are all names for the neem tree (Hindi and Bangla). It is known as Mwarobaini (Swahili) in east Africa, which literally translates to “tree of the 40” due to its ability to treat 40 different diseases1. Active components of the neem plant have been used medicinally by the AYUSH department, and modern medicine is currently employing this “divine tree” to treat a wide range of ailments, including infections, metabolic disorders, and cancer2. As evidenced by numerous research studies, every part of the plant has been examined for its pharmacological activity3, and it is well-established that this plant is used to treat a variety of diseases in numerous countries, including the Indian subcontinent4. In 1992, the United States National Academy of Sciences published a paper on “Neem”5. The chemical and biological analysis of neem discovered the existence of more than 300 bioactive substances in various plant parts, including at least 50 limonoids6. Bark, leaves, and roots contain antimicrobial, antifungal, insecticidal, antiviral, anti-malarial, antiperiodic, mosquito larvicidal, anti-inflammatory, antifertility, spermicidal, and hypoglycemic properties; they are also effective against periodontitis, gingivitis, boils, sores, splenomegaly, malaria, hyperpyrexia at childbirth, smallpox, and measles. Neem oil is employed as an intravaginal contraceptive, a treatment for vaginal infections, and a mosquito repellent7. Some of the well-established secondary metabolites, including nimbin, azadirachtin, nimbidiol, quercetin, and nimbidin1, are responsible for their pharmacological actions8, which is why, according to the World Health Organization, 80% of people rely on ethnomedicine (WHO)9. The purpose of Gas Chromatography-Mass Spectrometry (GC-MS) is to isolate various substances within a given sample, which is then used to retrieve the accessible compounds from the plant extract10. Previous research has documented the presence of countless secondary metabolites with potent antibacterial, antifungal, insecticidal, anti-inflammatory, antiviral, antioxidant, anti-cancer, and antimutagenic properties11. The objective of this study is to investigate the pharmacophoric properties of an aqueous extract of A. indica leaves.
Objective
To evaluate the various compounds present in the aqueous extract of A.indica by GC-MS analytical method.
To know the properties of Absorption, Distribution, Metabolism, Elimination and Toxicology of the major compounds obtained by analytical method
To obtain the pharmacodynamic properties for the major compounds obtained in GC-MS analysis.
Materials and Methods
The A.indica leaves were collected identified and authenticated by an expert botanist. The collected fresh leaves from the Rathinamangalam area of Chennai, Tamil Nadu, India were cleaned with fresh running tap water followed by distilled water, and dried in a shaded sunlight area after authentication which were later finely powdered. The powdered leaves were subjected to aqueous extraction by maceration. The obtained extract was subjected to quantitative chemical analysis with GC-MS to evaluate the compounds present. We further attempted to obtain from those compounds to know their pharmacokinetic and toxicological properties and their pharmacodynamic activity.
Gas chromatography-Mass Spectrometry
Analysis of A. indica‘s aqueous extract was carried out using GC-MS equipment. The GC-MS system used a TR 5MS capillary standard non-polar column with a diameter of 30 Mts, an ID of 0.25 mm, and a film thickness of 0.25 m. The flow rate of the mobile phase was set to 1.0 mL/min from the start. In the gas chromatography section, the temperature was raised from 40°C to 250°C at a rate of 5°C/min, with an injection volume of 1 microliter. The Wiley Spectral library search tool was used to analyze the outcomes of the samples immersed in chloroform over a mass spectrum of 50650 m/z12.
Preparation of ligand to know the pharmacological properties
The compounds which were retrieved from GC-MS analysis were taken up to find out their International Union of Pure and Applied Chemistry (IUPAC) names. Using the Chemicalize software and/or Pubmed compound NCBI website, we downloaded the .sdf file; by using the .sdf file, the Simplified Molecular Input Line Entry System (SMILES) for all the compounds were obtained by using an online SMILES translator. By using the same SMILES, with the help of SwissADME web tool, wherein we procured the data of physicochemical parameters, nature of solubility, pharmacokinetic parameters, druglikeliness, and medicinal properties. By using admetSAR which is an interface that is simple to utilize to search the ADME/T (Absorption, Distribution, Metabolism, Excretion, and/ Toxicity) properties of any molecule, we retrieved the toxicity profile. Predicting the most prospective macromolecular targets of a small molecule that is believed to be bioactive is done using the Swiss Target Prediction Interface, which compares small molecules to over 3000 distinct proteins from various species to find molecules that are comparable in 2D, and 3D structure.
Results
A total of 74 compounds were retrieved from the GC-MS analysis, out of which 13 compounds are showing significance (2 compounds having two peaks) and out of 13 compounds, 5 compounds had more hits, the obtained chromatogram was presented in the figure 1a and 1b. The compounds having a greater number of hits were subjected to evaluation of the pharmacodynamic properties.
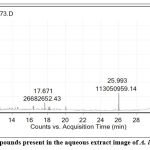 |
Figure 1a: Chromatogram of the compounds present in the aqueous extract image of A. indica obtained from GC-MS analysis. |
 |
Figure 1b: Chromatogram of the individual 13 compounds obtained from the aqueous extract image of A. indica obtained by GC-MS analysis. |
 |
Table 1: General Properties of the compounds retrieved from GC-MS analysis of aqueous extract of A.indica. |
Table 1 depicts the availability of various compounds in the aqueous extract of A. indica which may be important for the pharmacodynamic and pharmacokinetic potency and their general physicochemical properties. A total of 13 compounds are seen in the chromatogram but only 5 compounds are predominantly observed as productive based on the area and peak obtained in the chromatogram and may be responsible for pharmacological actions of aqueous extract of A.indica.
Table 2: Physicochemical Properties of the compounds retrieved from GC-MS analysis of aqueous extract of A.indica.
| S. No. | Name of the compound | No. of heavy atoms | No. of arom. heavy atoms | Fraction CSP3 | No. rotatable bonds | No. of H-bond acceptors |
No. of H-bond donors |
Molar Refractivity | Topological Polar Surface Area (TPSA) 0A2 |
| 1. | Benzaldehyde, 4-methyl |
9 | 6 | 0.12 | 1 | 1 | 0 | 36.80 | 17.07 |
| 2. | Ethanone, 1-(2-hydroxy- 5-methylphenyl) |
11 | 6 | 0.22 | 1 | 2 | 1 | 43.63 | 37.30 |
| 3. | Phenol, 2,6-dimethoxy |
11 | 6 | 0.25 | 2 | 3 | 1 | 41.45 | 38.69 |
| 4. | Eugenol | 12 | 6 | 0.20 | 3 | 2 | 1 | 49.06 | 29.46 |
| 5. | gamma-Elemene | 15 | 0 | 0.60 | 2 | 0 | 0 | 70.42 | 0.00 |
| 6. | 2(4H)-Benzofuranone, 5,6,7,7atetrahydro -4,4,7a-trimethyl-, (R) |
13 | 0 | 0.73 | 0 | 2 | 0 | 51.35 | 26.30 |
| 7. | Cyclohexane, 1-ethenyl-1-methyl-2 -(1 methylethenyl) -4-(1-methylethylidene) |
15 | 0 | 0.60 | 2 | 0 | 0 | 70.42 | 0.00 |
| 8. | 1H-Cycloprop[e]
azulen-7-ol, decahydro-1,1,7 -trimethyl-4-methylene-, [1ar-(1a.alpha., 4a.alpha., 7.beta.,7a.beta., 7b.alpha.)]- |
16 | 0 | 0.87 | 0 | 1 | 1 | 68.34 | 20.23 |
| 9. | Caryophyllene oxide | 16 | 0 | 0.87 | 0 | 1 | 1 | 68.27 | 12.53 |
| 10. | Caryophyllene oxide | 16 | 0 | 0.87 | 0 | 1 | 1 | 68.27 | 12.53 |
| 11. | 3,7,11,15-Tetramethyl-2-hexadecen-1-ol | 21 | 0 | 0.90 | 13 | 1 | 1 | 98.94 | 20.23 |
| 12. | Hexadecanoic acid, methyl ester |
19 | 0 | 0.94 | 15 | 2 | 0 | 85.12 | 26.30 |
| 13. | Phytol | 21 | 0 | 0.90 | 13 | 1 | 1 | 98.94 | 20.23 |
Extracted compounds are shown in Table 2 with their, number of heavy atoms, aromatic heavy atoms (AHA), proportion CSP3, number of rotatable bonds, molar refractivity, and Topological Polar Surface Area (TPSA)13. The number of atoms is in the permissible range, molar refractivity is maintaining the range 40-130 except for compound one “Benzaldehyde, 4-methyl-” as 36.80, the polar surface area of all the compounds is also less than 140 Ao, which indicates that the compounds are lipid soluble.
The Log p Octanol-Water partition coefficient14 values of the small molecules/compounds obtained are in the range of permissible -0.4 to +5.6 range implies a good lipophilic compound except the “3,7,11,15-Tetramethyl-2-hexadecen-1-ol” and “Phytol”. All the compounds show solubility in water except the last three compounds which are moderately soluble according to their hydrophilicity.
Only “gamma-Elemene,” “Cyclohexane, 1-ethenyl-1-methyl-2-(1 methylethyl)-4-(1-methylethyliden),” and “Phytol” have a low oral bioavailability, based on the GC-MS analysis of an aqueous extract of A.indica pharmacokinetic property15
All these compounds cross BBB15 except 4 compounds which are “gamma-Elemene”, “Cyclohexane, 1-ethenyl-1-methyl-2-(1 methylethenyl)-4-(1-methylethylidene”, “3,7,11,15-Tetramethyl-2-hexadecen-1-ol” and “Phytol”.
These compounds are not a substrate for p-glycoprotein16 which means they do not act as an efflux pump except “3,7,11,15-Tetramethyl-2-hexadecen-1-ol” and “Phytol”.
These compounds are not inhibitors of OATP2B1, MATE1, OCT2, BSEP transporter but inhibitor to OATP1B1 and OATP1B3 transporter.
All the compounds present are not inhibitors of CYP2D6 and CYP3A4 but compounds “Benzaldehyde, 4-methyl-”, “Ethanone, 1-(2-hydroxy-5-methylphenyl)”, “Eugenol”, and “Hexadecanoic acid, methyl ester” are inhibitors of CYP1A2; compound “Benzaldehyde, 4-methyl-”, “gamma-Elemene”, “Cyclohexane, 1-ethenyl-1-methyl-2-(1 methylethenyl)-4-(1-methylethylidene)”, “1H-Cycloprop[e]azulen-7-ol, decahydro-1,1,7-trimethyl-4-methylene-, [1ar-(1a.alpha.,4a.alpha.,7.beta.,7a.beta.,7b.alpha.)]-”, and “Caryophyllene oxide” are inhibitors of CYP2C19; the compounds “gamma-Elemene”, “Cyclohexane, 1-ethenyl-1-methyl-2-(1 methylethenyl)-4-(1-methylethylidene)”, “Caryophyllene oxide”, “3,7,11,15-Tetramethyl-2-hexadecen-1-ol”, and “Phytol” are inhibitors of CYP2C9.
The druglikeliness and lead likeliness of the compounds retrieved from GC-MS analysis of an aqueous extract of A.indica is that compounds follow Lipinski’s rule17of 5; the Ghose rule is not followed for the compounds “Benzaldehyde, 4-methyl-”, “Ethanone, 1-(2-hydroxy-5-methylphenyl)”, “Phenol, 2,6-dimethoxy-”, “3,7,11,15-Tetramethyl-2-hexadecen-1-ol”, “Hexadecanoic acid, methyl ester”, and “Phytol”; Veberrule18is not followed for the compounds “3,7,11,15-Tetramethyl-2-hexadecen-1-ol”, “Hexadecanoic acid, methyl ester”, and “Phytol”; the egan rule is not followed for the compounds “3,7,11,15-Tetramethyl-2-hexadecen-1-ol”, and “Phytol”; but no compound is following the muegge rule19. The bioavailability score20 for all the compounds is 0.55 and the synthetic accessibility score is 1.00 to 4.35.
Using the free admetSAR21 programme, the GC-MS analysis of an aqueous extract of A.indica yielded information on the compounds’ toxicity profiles, the compounds “gamma-Elemene” and “Cyclohexane, 1-ethenyl-1-methyl-2-(1 methylethenyl)-4-(1-methylethylidene)” shows hERG inhibition which leads to Q-T prolongation; “Cyclohexane, 1-ethenyl-1-methyl-2-(1 methylethenyl)-4-(1-methylethylidene)” has a high risk of carcinogenicity; all the compounds except “1H-Cycloprop[e]azulen-7-ol, decahydro-1,1,7-trimethyl-4-methylene-, [1ar-(1a.alpha.,4a.alpha.,7.beta.,7a.beta.,7b.alpha.)]-” cause eye irritation; and no compound showed hepatotoxicity and AMES toxicity.
Table 3: Table showing the target proteins or macromolecules for a small compound “Benzaldehyde, 4-methyl-”
| S. No. | Target | Uniprot ID | ChEMBL ID | Target Class | Probability |
| 1. | D-amino-acid oxidase | P14920 | CHEMBL5485 | Enzyme | 0.044308319 |
| 2. | Alcohol dehydrogenase alpha chain | P07327 | CHEMBL1970 | Oxidoreductase | 0.023832743 |
| 3. | Muscarinic acetylcholine receptor M4 | P08173 | CHEMBL1821 | Family A G protein-coupled receptor | 0.023832743 |
| 4. | Neuronal acetylcholine receptor; alpha4/beta2 | P43681 P17787 | CHEMBL1907589 | Ligand-gated ion channel | 0.023832743 |
| 5. | Muscarinic acetylcholine receptor M5 | P08912 | CHEMBL2035 | Family A G protein-coupled receptor | 0.023832743 |
| 6. | Muscarinic acetylcholine receptor M2 | P08172 | CHEMBL211 | Family A G protein-coupled receptor | 0.023832743 |
| 7. | Muscarinic acetylcholine receptor M1 | P11229 | CHEMBL216 | Family A G protein-coupled receptor | 0.023832743 |
| 8. | Muscarinic acetylcholine receptor M3 | P20309 | CHEMBL245 | Family A G protein-coupled receptor | 0.023832743 |
| 9. | Neuronal acetylcholine receptor protein alpha-7 subunit | P36544 | CHEMBL2492 | Ligand-gated ion channel | 0.023832743 |
| 10. | Alcohol dehydrogenase beta chain | P00325 | CHEMBL3284 | Oxidoreductase | 0.023832743 |
A swiss target prediction is a web tool, which was used to predict the protein that modulates by the compound “4-methylbenzaldehyde”. It is a small molecule that acts as a ligand shows its activity on a totally of 94 proteins/targets which are 26.7 % on oxidoreductase, 13.3 % on enzymes and 33.3 % on G-protein coupled receptor, 13.3 % on ligand-gated ion channel, 6.7 % on voltage-gated ion-channel and 6.7 % on other cytosolic proteins. Table 3 shows the first 10 proteins/receptor with high probability to target the ligand, obtained from the Uniprot database.
Table 4: Table showing the target proteins or macromolecules for a small compound “Ethanone, 1-(2-hydroxy-5-methylphenyl)”
| S. No. | Target | Uniprot ID | ChEMBL ID | Target Class | Probability |
| 1. | Serine/threonine-protein kinase/endoribonuclease IRE1 | O75460 | CHEMBL1163101 | Enzyme | 0.063026 |
| 2. | Adenosine A1 receptor | P30542 | CHEMBL226 | Family A G protein-coupled receptor | 0.053518 |
| 3. | Adenosine A2a receptor | P29274 | CHEMBL251 | Family A G protein-coupled receptor | 0.053518 |
| 4. | Serum albumin | P02768 | CHEMBL3253 | Secreted protein | 0.053518 |
| 5. | Carbonic anhydrase II | P00918 | CHEMBL205 | Lyase | 0.053518 |
| 6. | Macrophage migration inhibitory factor | P14174 | CHEMBL2085 | Enzyme | 0.043919 |
| 7. | Tyrosinase | P14679 | CHEMBL1973 | Oxidoreductase | 0.043919 |
| 8. | Mannose-6-phosphate isomerase | P34949 | CHEMBL2758 | Isomerase | 0.043919 |
| 9. | Estradiol 17-beta-dehydrogenase 1 | P14061 | CHEMBL3181 | Enzyme | 0.043919 |
| 10. | Serotonin 2b (5-HT2b) receptor | P41595 | CHEMBL1833 | Family A G protein-coupled receptor | 0.043919 |
A swiss target prediction is a web tool used to predict the protein that causes to modulate, the compound “Ethanone, 1-(2-hydroxy-5-methylphenyl)” which is a small molecule that acts as a ligand and shows its activity on a totally of 100 proteins which 20 % on enzymes, 26.7 % on G-protein coupled receptor, 6.7% on secreted protein, 6.7 % on lyase, 13.3 % on oxidoreductase, 6.7% on ligand-gated ion channel, 6.7 % on isomerase, 6.7% on transferase and 6.7 % on other miscellaneous proteins. Table 4 shows the first 10 proteins with high probability to target the protein which is obtained from the Uniprot database.
Table 5: Table showing the target proteins or macromolecules for a small compound “Eugenol”
| S. No. | Target | Uniprot ID | ChEMBL ID | Target Class | Probability |
| 1. | Adenosine A1 receptor | P30542 | CHEMBL226 | Family A G protein-coupled receptor | 0.133391 |
| 2. | Adenosine A2a receptor | P29274 | CHEMBL251 | Family A G protein-coupled receptor | 0.133391 |
| 3. | Vascular endothelial growth factor A | P15692 | CHEMBL1783 | Secreted protein | 0.133391 |
| 4. | Egl nine homolog 1 | Q9GZT9 | CHEMBL5697 | Oxidoreductase | 0.133391 |
| 5. | Fatty acid desaturase 1 | O60427 | CHEMBL5840 | Enzyme | 0.125076 |
| 6. | Histone deacetylase 6 | Q9UBN7 | CHEMBL1865 | Eraser | 0.125076 |
| 7. | Carbonyl reductase [NADPH] 1 | P16152 | CHEMBL5586 | Enzyme | 0.125076 |
| 8. | D-amino-acid oxidase | P14920 | CHEMBL5485 | Enzyme | 0.125076 |
| 9. | Cyclooxygenase-1 | P23219 | CHEMBL221 | Oxidoreductase | 0.125076 |
| 10. | Carbonic anhydrase II | P00918 | CHEMBL205 | Lyase | 0.125076 |
A swiss target prediction is a web tool used to predict the protein that causes to modulate, the compound “Eugenol” which is a small molecule that acts as a ligand shows its activity on total of 100 proteins which 20 % on Family A G-protein coupled receptor, 6.7 % on secreted protein, 20 % on oxidoreductase, 33.3 % on the enzyme, 6.7% on lyase, 6.7% on Family C G-protein coupled receptor and 6.7 % other miscellaneous proteins. Table 5 shows the first 10 proteins/receptors with a high probability to target the ligand obtained from the Uniprot database.
Table 6: Shows the target proteins or macromolecules for a small compound “gamma-Elemene”
| S. No. | Target | Uniprot ID | ChEMBL ID | Target Class | Probability |
| 1. | Peroxisome proliferator-activated receptor alpha | Q07869 | CHEMBL239 | Nuclear receptor | 0.052424 |
| 2. | Cannabinoid receptor 2 | P34972 | CHEMBL253 | Family A G protein-coupled receptor | 0.052424 |
| 3. | C-X-C chemokine receptor type 3 | P49682 | CHEMBL4441 | Family A G protein-coupled receptor | 0.042894 |
| 4. | LXR-alpha | Q13133 | CHEMBL2808 | Nuclear receptor | 0.042894 |
| 5. | Serotonin 2a (5-HT2a) receptor | P28223 | CHEMBL224 | Family A G protein-coupled receptor | 0 |
| 6. | Monoamine oxidase B | P27338 | CHEMBL2039 | Oxidoreductase | 0 |
| 7. | UDP-glucuronosyltransferase 2B7 | P16662 | CHEMBL4370 | Enzyme | 0 |
| 8. | Monoglyceride lipase | Q99685 | CHEMBL4191 | Enzyme | 0 |
| 9. | Cytochrome P450 19A1 | P11511 | CHEMBL1978 | Cytochrome P450 | 0 |
| 10. | Nuclear receptor subfamily 1 group I member 3 (by homology) | Q14994 | CHEMBL5503 | Nuclear receptor | 0 |
The swiss target prediction is a web tool used to predict the protein that causes to modulate, the compound “gamma-Elemene” which is a small molecule that acts as a ligand shows its activity on totally of 68 proteins which 33.3 % on nuclear receptor, 20 % on Family A G-protein coupled receptor, 6.7 % on oxidoreductase, 20 % on the enzyme, 13.3 % on Cytochrome P 450, and 6.7% on Voltage-gated ion channel. Table 6 shows the first 10 proteins/receptor with high probability to target the ligand which is obtained from the Uniprot database.
The last and fifth compound which shows maximum hits in GC-MS is “Cyclohexane, 1-ethenyl-1-methyl-2-(1 methylethenyl)-4-(1-methylethylidene)” which is also called “gamma-Elemene” this means that it has same properties as like “gamma-Elemene”.
Discussion
Azadirachta indica is acknowledged for a wide array of many medicinal properties for many years. The objective of this research was to detect the active bio-compounds, present in the extract which shows the pharmacological actions. In this study, we obtained 13 different compounds from the aqueous extract.
According to Saleem22et. al, A. indica contains a variety of phytochemicals for medicines, including alkaloids, steroids, flavonoids, terpenoids, fatty acids, and carbohydrates. The presence of azadirachtin and nimbin in the tree gives it fungicidal properties. In a study by Lu23 et. al, Salannin, 1-detigloyl-1-isobutylsalannin, salannol-3-acetate, salannol, spirosendan, 1-detigloyloxy-3-deacetylsalannin-1-en-3-one, nimbin, and 6-deacetylnimbin were identified from 95 percent ethanol extracts of neem (Azadirachta indica) seeds. In another study by Babatunde24et. al.,the major constituents were Eicosane (9.7662%), Diacenaphtho[1,2-j:1 _ ,2 _ -l]fluoranthene (11.301%), Phenol, 4-[[(4-methoxyphenyl)methylene]amino]- (11.84%) and (3Ar,6S,9ar)-1,2,3,4,5,6,7,9a- octahydro-8-methyl-3a,6-methano-3ah-cyclopentacycloocten-10-one (36.883%) in steam extracted oil; Eicosane (10.259%), Diacenaphtho[1,2-j:1 _ ,2 _ -l]fluoranthene (13.51%) and Butanamide, N-(2-methoxyphenyl)-3-oxo- (16.615%) in the ethanol extracted oil, and (3Ar,6S,9ar)-1,2,3,4,5,6,7,9a-octahydro-8-methyl-3a,6-methano-3ah-cyclopentacycloocten- 10-one (10.72%), n-Hexadecanoic acid (14.688%) and 9,12,15-Octadecatrienoic acid, (Z,Z,Z)- (34.719%) in the hexane extracted A. indica essential oil. Some studies done by Bolade25 et.al., disclosed many numbers of compounds that are bioactive phytoconstituents such as polyphenols and tannins by their GC-MS characterization. The study conducted by Akpuaka26, et.al., identified various phthalates like Diisobutyl phthalate, Dibutyl phthalate, Ethylhexyl phthalate, Heptyl methyl phthalate, Mono(n-octyl) phthalate, Mono(2-ethylhexyl) phthalate.In another research conducted by Shafie27et. al., revealed the presence of seven major compounds in the ethanolic leaf extract of A.excelsa which are 9, 12, 15-octadecatrienoic acid (42.34%), pentadecanoic acid, 14-methyl-, methyl ester (28.99%), phytol (10.63%), 9, 12, 15-octadecatrien-1-ol (5.37%), octadecanoic acid, methyl ester (4.36%), 9, 12-octadecadienoic acid, methyl ester (4.24%) and hexadecanoic acid, ethyl ester (4.06%).The other study done by Swapna sonale28 et. al., elicited twenty-five volatile compounds in hydro-distillate and in another extraction process called Supercritical fluid carbon dioxide (SCF) isolates forty volatile compounds with neem seed.
In another research conducted by Oshiobugie29 et. al., ten chemical constituents were identified in the leaf, six were found in the stem, and seven were identified in the root of A. indica. In a study conducted by Siddiqui11et. al., the researchers detected compounds like sixteen n-alkanes; three aromatics 2,6-bis-(1,1-dimethylethyl)-4-methyl phenol, 2-(phenylmethylene)-octanal, 1,2,4-trimethoxy-5-(1Z-propenyl)-benzene; three benzopyranoids 3,4-dihydro-4,4,5,8-tetramethylcoumarin, 3,4-dihydro-4,4,7,8-tetramethylcoumarin-6-ol, 1,3,4,6,7,8-hexahydro-4,6,6,7,8,8-hexamethyl-cyclopenta[g]-2-benzopyran; one sesquiterpene methyl-3,7,11-trimethyl-2E,6E,10-dodecatrienoate; three esters of fatty acids methyl 14-methyl-pentadecanoate, ethyl hexadecanoate, ethyl 9Z-octadecenoate and one monoterpene 3,7-dimethyl-1-octen-7-ol. According to Altayb30 et al., the GC-MS analysis of a methanolic extract of neem leaves revealed 30 peaks, which corresponded to 30 phytochemical compounds including 11 – fatty acids, 9- hydrocarbons, 2-pyridine derivatives, and 2 -aldehydes, 1- phenol group, 1-aromatic substances, 1-coumarins, and 1-monoterpenes, among these the compound 1,5-Anhydro-2-deoxy-L-arabino-hex-1-enitol was the most abundant. In another study by Hossain31 et.al.,, Hexane crude extract contains 33 different organic compounds, accounting for 7.12 % of the total extract; ethyl acetate extract contains 58 different organic compounds, accounting for 13.98 % of the total extract; chloroform extract contains 65 organic compounds, accounting for 29.12 % of the total extract; and butanol extract contains 49 different compounds, accounting for 19.56 % of the total extractbut in this study we observed 13 compounds (2 compounds are having two peaks) such as “4-methylbenzaldehyde”, “1-(2-hydroxy-5-methylphenyl)ethan-1-one”,“2,6-dimethoxyphenol”, “2-methoxy-4-(prop-2-en-1-yl)phenol”, “(1S,2S)-1-ethenyl-1-methyl-4-propan-2-ylidene-2-prop-1-en-2-ylcyclohexane”, “4,4,7a-trimethyl-6,7-dihydro-5H-1-benzofuran-2-one”, “1-ethenyl-1-methyl-2-(prop-1-en-2-yl)-4-(propan-2-ylidene)cyclohexane”, “(7S)-1,1,7-trimethyl-4-methylidene-1a,2,3,4a,5,6,7a,7b-octahydrocyclopropa[h]azulen-7-ol”, “4,12,12-trimethyl-9-methylidene-5-oxatricyclo[8.2.0.0.4,6]dodecane”, “3,7,11,15-tetramethylhexadec-2-en-1-ol”, “methyl hexadecanoate”, and “(2E)-3,7,11,15-tetramethylhexadec-2-en-1-ol” and we retrieved the potent drug targets that modify the pharmacological actions by these major compounds obtained through GC-MS.
The research findings observed in this study have few similarities and are vastly different from those observed by other researchers, which may be due to variation in plant ethnicity or variation in the GC-MS analytical method.
Conclusion
The medicinal plant A.indica is of prime importance since ancient days as it shows helpful in the treatment of a myriad of diseases but the real active principle, which is the basis of its pharmacological actions is not yet known completely. In this attempt of the research by GC-MS analytical technique, we isolated some 13 compounds (2 compounds having two peaks) that are present in this aqueous extract. The various compounds are “4-methylbenzaldehyde”, “1-(2-hydroxy-5-methylphenyl)ethan-1-one”, “2,6-dimethoxyphenol”, “2-methoxy-4-(prop-2-en-1-yl)phenol”, “(1S,2S)-1-ethenyl-1-methyl-4-propan-2-ylidene-2-prop-1-en-2-ylcyclohexane”, “4,4,7a-trimethyl-6,7-dihydro-5H-1-benzofuran-2-one”, “1-ethenyl-1-methyl-2-(prop-1-en-2-yl)-4-(propan-2-ylidene)cyclohexane”, “(7S)-1,1,7-trimethyl-4-methylidene-1a,2,3,4a,5,6,7a,7b-octahydrocyclopropa[h]azulen-7-ol”, “4,12,12-trimethyl-9-methylidene-5-oxatricyclo[8.2.0.0.4,⁶]dodecane”, “3,7,11,15-tetramethylhexadec-2-en-1-ol”, “methyl hexadecanoate”, and “(2E)-3,7,11,15-tetramethylhexadec-2-en-1-ol” which were successfully attempted to retrieve the drug targets for these compounds that can modulate the pharmacological actions that may conducive in re-establish the diseased condition after pre-clinical and clinical trials by using these as a “Hit” molecule or “Lead” molecule.
Limitations
In this research we were able to retrieve the compounds by GC-MS and able to know the pharmacokinetic and pharmacodynamic properties, but it is required to evaluate these for pre-clinical studies and clinical trials.
Acknowledgement
We, the authors of this article are very thankful to Dr. King Shuk Lahon, Professor & Head, Department of Pharmacology, VCSGG Institute of Medical & Research, Srinagar, Uttarkhand, India for his peer review at our request. Also we are thankful to Mr. M.J.Prasad a renowned botanist for helpful in identification and authenticated the neem leaves for this study.
Conflict of Interest
There are no conflict of interest.
Funding Sources
There is no funding sources.
References
- Paul R, Prasad M, Sah NK. Anticancer biology of Azadirachta indica L (neem): A mini review. Cancer Biol Ther. 2011;12(6):467–76.
CrossRef - Moga M, Bălan A, Anastasiu C, Dimienescu O, Neculoiu C, Gavriș C. An Overview on the Anticancer Activity of Azadirachta indica (Neem) in Gynecological Cancers. Int J Mol Sci. 2018;19(12):3898. doi: 10.3390/ijms19123898
CrossRef - Joy Sinha D, D.S. Nandha K, Jaiswal N, Vasudeva A, Prabha Tyagi S, Pratap Singh U. Antibacterial Effect of Azadirachta indica (Neem) or Curcuma longa (Turmeric) against Enterococcus faecalis Compared with That of 5% Sodium Hypochlorite or 2% Chlorhexidine in vitro. Bull Tokyo Dent Coll. 2017;58(2):103–9.
CrossRef - Alzohairy MA. Therapeutics Role of Azadirachta indica (Neem) and Their Active Constituents in Diseases Prevention and Treatment. Evid Based Complement Alternat Med. 2016;2016:1–11. Article ID 7382506. http://dx.doi.org/10.1155/2016/7382506
CrossRef - Kumar VS, Navaratnam V. Neem (Azadirachta indica): Prehistory to contemporary medicinal uses to humankind. Asian Pac J Trop Biomed. 2013;3(7):505–14.
CrossRef - Habluetzel A, Pinto B, Tapanelli S, Nkouangang J, Saviozzi M, Chianese G, et al. Effects of Azadirachta indica seed kernel extracts on early erythrocytic schizogony of Plasmodium berghei and pro-inflammatory response in inbred mice. Malar J. 2019;18(1):35. doi: 10.1186/s12936-019-2671-8
CrossRef - C.P. Khare. Indian Medicinal Plants An Illustrated Dictionary. Vol. 1st volume. Springer-Verlag Berlin/Heidelberg; 2007. 836 p.
CrossRef - Widiyana AP, Illian DN. Phytochemical analysis and total flavonoid content on ethanol and ethyl acetate extract from neem (Azadirachta indica JUSS.) leaves using UV–VIS spectrophotometric. 2022;8(1):71-77.
CrossRef - Sarkar S, Singh RP, Bhattacharya G. Exploring the role of Azadirachta indica (neem) and its active compounds in the regulation of biological pathways: an update on molecular approach. 3 Biotech. 2021;11(4):178. https://doi.org/10.1007/s13205-021-02745-4
CrossRef - David Sparkman O, Zelda P, Fulton K. Gas Chromatography and Mass Spectrometry: A Practical Guide. 2nd ed. Academic Press; 632 p.
- Siddiqui BS, Rasheed M, Ilyas F, Gulzar T, Tariq RM, Naqvi SN ul H. Analysis of Insecticidal Azadirachta indica A. Juss. Fractions. Z Für Naturforschung C. 2004;59(1–2):104–12.
CrossRef - Simhadri N, Muniappan M, Kannan I, Viswanathan S. Phytochemical analysis and docking study of compounds present in a polyherbal preparation used in the treatment of dermatophytosis. Curr Med Mycol. 2017;3(4):6–14.
CrossRef - Nagesh SV, M M, I K, S V. Antifungal properties of secondary metabolites of Azadirachta indica and Lawsonia inermis – An In-silico study. Asian J Pharm Clin Res. 2018;11(7):449-455.
CrossRef - Daina A, Michielin O, Zoete V. iLOGP: A Simple, Robust, and Efficient Description of n -Octanol/Water Partition Coefficient for Drug Design Using the GB/SA Approach. J Chem Inf Model. 2014 Dec 22;54(12):3284–301.
CrossRef - Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(1):42717. DOI: 10.1038/srep42717
CrossRef - Nagesh SV, M M, I K, S V. Antifungal activity of a secondary metabolite of Azadirachta indica and its derivatives – An In-Silico Study. Asian J Pharm Clin Res. 2018;11(1):175-184.
CrossRef - Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1–3):3–26.
CrossRef - Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J Med Chem. 2002;45(12):2615–23.
CrossRef - Muegge I, Heald SL, Brittelli D. Simple selection criteria for drug-like chemical matter. J Med Chem. 2001;44(12):1841–6.
CrossRef - Martin YC. A Bioavailability Score. J Med Chem. 2005;48(9):3164–70.
CrossRef - Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, et al. admetSAR: A Comprehensive Source and Free Tool for Assessment of Chemical ADMET Properties. J Chem Inf Model. 2012;52(11):3099–105.
CrossRef - Saleem S, Muhammad G, Hussain MA, Bukhari SNA. A comprehensive review of phytochemical profile, bioactives for pharmaceuticals, and pharmacological attributes of Azadirachta indica: A Comprehensive Review of Azadirachtaindica. Phytother Res. 2018;32(7):1241–72.
CrossRef - Lu XF, Dai DM, Yu RM, Song LY, Zhu JH, Fan XN, et al. Limonoids from seeds of Azadirachta indica and their cytotoxic activity. Zhongguo Zhong Yao Za Zhi Zhongguo Zhongyao Zazhi China J Chin Mater Medica. 2018;43(3):537–43.
- Babatunde DE, Otusemade GO, Efeovbokhan VE, Ojewumi ME, Bolade OP, Owoeye TF. Chemical composition of steam and solvent crude oil extracts from Azadirachta indica leaves. Chem Data Collect. 2019;20:100208. DOI:10.1016/j.cdc.2019.100208
CrossRef - Bolade OP, Akinsiku AA, Adeyemi AO, Williams AB, Benson NU. Dataset on phytochemical screening, FTIR and GC–MS characterisation of Azadirachta indica and Cymbopogon citratus as reducing and stabilising agents for nanoparticles synthesis. Data Brief. 2018;20:917–26.
CrossRef - Akpuaka A, Ekwenchi MM, Dashak DA, Dildar A. Biological Activities of Characterized Isolates of n-Hexane Extract of Azadirachta indica A.Juss (Neem) Leaves. N Y Sci J 2013;6(6):119-124
- Shafie I, Samsulrizal N, Sopian NA, Rajion MA, Meng GY, Ajat MM, et al. Qualitative phytochemical screening and GC-MSprofiling of Azadirachta excelsaleaf extract. Malays Appl Biol 2015;44(3):87-92.
- Swapna sonale R, Ramalakshmi K, Udaya Sankar K. Characterization of Neem (Azadirachta indica A. Juss) seed volatile compounds obtained by supercritical carbon dioxide process. J Food Sci Technol. 2018;55(4):1444–54.
CrossRef - Oshiobugie M, Olaniyi A, Raphael A. AAS and GC-MS Analysis of Phytocomponents in the Leaf, Stem and Root of Azadirachta indica A. Juss (Dongoyaro). Br J Pharm Res. 2017;15(4):1–12.
CrossRef - Altayb HN, Yassin NF, Hosawi S, Kazmi I. In-vitro and in-silico antibacterial activity of Azadirachta indica (Neem), methanolic extract, and identification of Beta.d-Mannofuranoside as a promising antibacterial agent. BMC Plant Biol. 2022;22(1):262.
CrossRef - Hossain MA, Shah MD, Sakari M. Gas chromatography–mass spectrometry analysis of various organic extracts of Merremia borneensis from Sabah. Asian Pac J Trop Med. 2011;4(8):637–41.
CrossRef








