Angelica Kresnamurti1* , Farizah Izazi2
, Farizah Izazi2 , Ersanda Nurma Praditapuspa3
, Ersanda Nurma Praditapuspa3 and Siswandono4
and Siswandono4
1Department of Clinical Pharmacy, Hang Tuah University, Surabaya, Indonesia
2Department of Biology Pharmacy, Hang Tuah University, Surabaya, Indonesia
3Department of Chemistry Pharmacy, Hang Tuah University, Surabaya, Indonesia
4Department of Pharmaceutical Chemistry Pharmacy, Airlangga University, Surabaya, Indonesia.
Corresponding Author E-mail: angelica.kresnamurti@hangtuah.ac.id
DOI : https://dx.doi.org/10.13005/bpj/2614
Abstract
SARS-CoV-2 is a kind of coronavirus that produces Covid-19 illness, which is still a public health concern in Indonesia. Meanwhile, an effective drug has not yet been found and although vaccination has been carried out, in several regions and neighboring countries there is still an increase in Covid-19 cases. This study aimed to obtain bioactive compounds from sea urchins (Echinometra mathaei) that have greater antiviral potential and lower toxicity than remdesivir. This research was started by predicting druglikeness with SwissADME, followed ADMET predicition with pkCSM online, and docking of molecule using the Molegro Virtual Docker (MVD) 5.5 software against the main protease (Mpro) target (PDB ID: 6W63). The results showed that six compounds from sea urchins (Echinometra mathaei) had antiviral activity, where the bioactive compound from sea urchins (Echinometra mathaei) with the highest affinity was shown by Spinochrome C a smaller rerank score compared with Remdesivir and native ligand (X77). So that Spinochrome C compounds are candidates as SARS-CoV-2 inhibitors potential developed drug.
Keywords
ADMET; Druglikeness; molecular docking; Sea urchin; SARS-COV-2
Download this article as:| Copy the following to cite this article: Kresnamurti A, Izazi F, Praditapuspa E. N, Siswandono S. In Silico Analysis of Bioactive Compounds from Sea Urchin (Echinometra mathaei) against SARS-COV-2. Biomed Pharmacol J 2023;16(1). |
| Copy the following to cite this URL: Kresnamurti A, Izazi F, Praditapuspa E. N, Siswandono S. In Silico Analysis of Bioactive Compounds from Sea Urchin (Echinometra mathaei) against SARS-COV-2. Biomed Pharmacol J 2023;16(1). Available from: https://bit.ly/3RTVFy5 |
Introduction
Coronavirus Disease-2019 (COVID-19) is a pneumonia-like illness caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)1. SARS CoV 2 is an RNA virus with a positive strand. Symptoms of this viral infection include fever, a dry cough, and shortness of breath. However, many patients are carriers and show no symptoms. This is one of the reasons for the widespread transmission of this virus2. Since 2019 until 2022 almost three years COVID-19 has surge around the world. Some things have been done for over come COVID-19 such us vaccine or treatment therapy. Treatment of Covid-19 patients is carried out by giving medicines that have been previously prescribed received FDA approval, such as Remdesivir, but assessment Remdesivir has little benefit on COVID-19 patients who are already being ventilated. It has a little influence on mortality or progression to ventilation in other hospitalised patients (or both)3. As time goes by, the variants COVID-19 become more variety.
This study focused on the main protease (Mpro), where Mpro is one of the CoV proteases that cleaves polyproteins. RdRp, helicase, RNA-binding protein, exoribonuclease, endoribonuclease, and 2-O-ribose methyltransferase are among the proteins that have been removed4. This makes compounds that inhibit Mpro have broad antiviral activity2. Protease activity disruption can result in a variety of illnesses. As a result, host proteases can be exploited as possible therapeutic targets in general. Proteases serve a key role in viral replication in many viruses. As a result, proteases are frequently employed as protein targets in the development of antiviral medicine5. Molecular modeling and structure-based drug design has shown to be an effective method for discovering new inhibitors and has become a well-known approach in contemporary drug development. Until the advent of computational drug discovery, medications were discovered by accident via try and error. Computer-aided drug design facilitates and accelerates the drug design process, which includes a number of approaches for identifying new molecules6.
Sea urchin is one of the many marine biota created for medications7-12. More than 5000 chemical compounds with high pharmacological potential have been isolated and described from marine organisms8. The quinone pigments are products of the secondary metabolism of marine animals, can have complex structures and become the basis for the development of new natural products in echinoids that are modulators of chemical interactions and possible active ingredients in medicinal preparations8. The antiviral activity of sea urchin is probably due to the virus-inhibiting activity of the compounds and because of their natural potentially antioxidant properties7 such as spinochrome, echinochrome, and naphtoquinone pigments9,10. According to studies (Mishchenko et al., 2020), the Spinochrome amine present in sea urchins has the ability to suppress the attachment and penetration herpes simplex virus type 1 into the cell11. According to Krylova et al., (2019) and Fedoreyevet al., (2018) Oral administration of the antioxidant composition of echinochrome A, ascorbic acid and α-tocopherol (5:5:1) protected 90% of the infected mice against death and reduced vaginal viral loads7 and affect virus particle of tick-borne encephalitis virus (TBEV) and herpes simplex virus type 1 (HSV-1)12.
This study aims to further identify the antiviral activity of Sea urchin (Echinometra mathaei) against COVID-19. So it is hoped that it will produce drug candidates that have the potential to be developed as anti-COVID-19 drugs.
Methods and Material
Hardware
The computer specifications are as follows: Intel® CoreTM i7 8565U@ 1.80 GHz processor (CPU), Nvidia® GeForce MX230 graphics processing unit (GPU), and 8 GB Random Access Memory (RAM) running Windows 10.
Compound Test Preparation
Eleven bioactive compounds from the sea urchin (Echinometra mathaei) were created in 2D and 3D models, and then they were optimized using the MMFF94 technique using Chem3D 20.0. Utilizing the Online SMILES Translator (https://cactus.nci.nih.gov/translate/), the structure is then converted to SMILES format.
Drug-likeness prediction
SwissADME (http://www.swissadme.ch/) are used to predict drug-likeness of the test compound with SMILES file format.
ADMET prediction
Prediction of pharmacokinetics (ADME) and toxicity of the test compound was done by the pkCSM website (http://biosig.unimelb.edu.au/pkcsm/prediction) with the SMILES format.
Molecular docking
N-nitrosourea is a COVID-19 inhibitor (4-tert-butylphenyl) -N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl] In the protein data bank, -1H-imidazole-4-carboxamide (X77) was described as an inhibitor in the binding state with the Mpro (PDB ID: 6W63). Molegro Virtual Docker (MVD) 5.5 software was used to assign the structures’ missing bond ordering, charges, bonds, and hybridization states13. The viral protein’s 3D structure was obtained from the Protein Data Bank (PDB ID: 6W63), and the proteins were created by removing all water molecules, ligands, and cofactors and assigning bonds, bond order, hybridization, and charges with the MVD 5.5 program. Search algorithms and scoring functions docking In computation screening, the Piecewise Linear Potential (PLP) approach is used to score functions. Using the cavity prediction wizard, the cavities were predicted and limited to three; the cavity with the largest volume was chosen as the origin for the binding site. The ligands were docked with viral protein, and the best-generated postures were chosen using docking scores14. The outcome of the in-silico investigation is the Rerank Score (RS). The rerank score predicts drug-receptor interaction.
Results and Discussion
Drug-likeness prediction
The results of the drug-likeness prediction of bioactive compounds from sea urchin (Echinometra mathaei) are shown in Table 1.
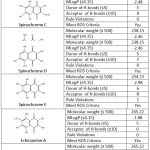 |
Table 1: Drug-likeness prediction. |
The prediction results of physicochemical properties based on Lipinski’s rule [Table 1] show that all bioactive compounds from sea urchin (Echinometra mathaei) meet Lipinski’s rule15.
ADMET prediction
The results of the ADMET prediction of bioactive compounds from sea urchin (Echinometra mathaei) are shown in Table 2.
Prediction of toxicity (ames toxicity, hepatotoxicity, and LD50) of bioactive compounds from sea urchin were done by the pkCSM online (https://biosig.lab.uq.edu.au/pkcsm/prediction) with the SMILES format.
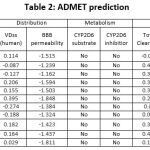 |
Table 2: ADMET prediction. |
Following oral administration, the small intestine is where medication molecules are primarily absorbed. Poor absorption is defined as less than 30%. Table 2 lists bioactive substances from sea urchins (Echinometra mathaei), which are projected to have good oral bioavailability since they have human intestine absorption rates of over 30%. The development of transdermal drugs is particularly linked to the problem of skin permeability. The log Kp (cm/h) measurements used to represent the skin permeation coefficient are capable of forecasting a medicinal compound’s skin permeability. When log Kp > -2.5, a chemical is predicted to have low skin permeability. As can be seen in Table 2, the bioactive substances from sea urchins have a lower log Kp value of < -3.0 (cm/h), indicating that they have a moderate level of skin permeability.
The theoretical volume required to hold the entire quantity of medicine delivered to disperse it while maintaining the same blood concentration is known as volume distribution (VDss). A substance that has a high VDss value is one that is dispersed more throughout the body than in blood plasma. The medication has a low distribution volume if the log VDss value is less than – 0.15. In contrast, it has a larger distribution volume when the log value of VDss is bigger than 0.4511. According to Table 2, the bioactive chemicals from sea urchins have modest distribution volumes, with log VDss values ranging from 0.395 to -0.274. Drug molecules with log BB > 0.3 can cross the blood-brain barrier but cannot be distributed well with log BB <-1. The log BB value for sea urchin bioactive chemicals is between -1.848 and -0.162, according to Table 2. These chemicals can cross the BBB somewhat with this value.
In the detoxification process, cytochrome P450 is a critical and key enzyme in the liver cells. The enzyme is important in the oxidation process and aids in the excretion of foreign organic substances such as pharmaceuticals. It is critical to understand that pharmacological molecules can decrease the activity of the cytochrome P450 enzyme. It is represented in this study by the cytochrome P3D6 isoform (CYP2D6). Table 2 shows that sea urchin bioactive chemicals have no effect on cytochrome P450 enzyme activity. The total clearance (Cltot) and the renal organic cation transporter 2 (OCT2) substrate can be used to predict drug compound excretion. The sum of hepatic clearance (liver and bile) and renal excretion is known as C1 total.
The compound speed of excretion of bioactive chemicals from sea urchin ranges from -0.063 to 0.413 in Table 2. OCT2 is a kidney transporter that plays critical functions in the disposition and clearance of drug and endogen chemicals in the body. Table 2 shows that bioactive chemicals from sea urchin have no impact on OCT2, allowing the substance to be readily secreted16. The Ames toxicity test is an assay used to measure the existing toxicity of a substance and has been widely used to analyze a drug’s mutagenesis potential using bacteria. A positive result implies that the substance has mutagenesis capabilities and may function as a carcinogen. Table 2 reveals that sea urchin bioactive chemicals are non-toxic. This implies that the chemicals are risk-free. They were subjected to Ames and hepatotoxicity testing. When the compounds were evaluated for LD50 in mice, the results ranged from 1.787 to 2.366 mol/kgBW. This demonstrates a low toxicity value, as to destroy 50% of the mice16. Furthermore, the results showed that all bioactive chemicals derived from sea urchin are non-hepatotoxic.
Molecular docking
Molecular docking is a type of computer modeling study that seeks to find ligand-receptor interactions. The RMSD number of 1.118 indicates the outcomes of docking validation, hence the docking procedure may be considered legitimate because the RMSD value is < 217. Table 3 displays the findings of molecular docking of bioactive chemicals from sea urchin (Echinometra mathaei).
Table 3: Molecular docking results ranked by the lowest Rerank Score and Interaction of ligands and amino acid residues.
| Compound | Rerank score (Kcal/mol) | Number of
H-bonds |
H-bond Interaction Site | Number of
Steric bonds |
Steric bonds Interaction Site |
| Spinochrome C | -73.3374 | 9 | Glu166, Cys145, Leu141, His163, Ser144, Phe140 | 8 | Glu166, Leu141, Phe140, His163, His164, Ser144, Cys145, Met165 |
| Echinamine B | -72.5032 | 7 | Glu166, Cys145, Leu141, His163, His164, Phe140 | 6 | His164, Cys145, His163, Leu141, Glu166, Phe140 |
| Spinochrome A | -71.6408
|
8 | Glu166, Ser144, Cys145, His163, Leu141, Phe140 | 7 | Glu166, Leu141, Phe140, His163, Ser144, Cys145, Met165 |
| Echinamine A
|
-71.4374 | 8 | Glu166, Ser144, Cys145, His163, His164, Asn142 | 5 | Leu141, Phe140, Cys145, His163, Asn142 |
| Echinochrome A | -70.4858 | 8 | Glu166, Ser144, Cys145, His163, His164, Leu141, Phe140 | 7 | Glu166, Leu141, Phe140, His163, Ser144, Cys145, His164 |
| Spinochrome D | -70.0122 | 8 | Glu166, Cys145, His163, His164, Leu141, Phe140 | 6 | Glu166, Leu141, Phe140, His163, His164, Cys145 |
| Remdesivir (standard drug) | -69.4683 | 9 | Glu166, Cys145, Cys44, Asn142, Gln189, Asp187, Tyr54 | 13 | Glu166, Cys145, Cys44, Asn142, Gln189, Asp187, Tyr54, His41, Met165, Met49, Gly143
|
| Spinochrome B | -68.1089 | 6 | Ser144, Cys145, His163, His164, Leu141 | 6 | Glu166, Leu141, Phe140, His163, His164, Cys145 |
| Spinochrome 282 | -67.4298 | 5 | Glu166, Ser144, Cys145, His163 | 8 | Glu166, Leu141, Phe140, His163, Ser144, Cys145, His164, Asn142 |
| Echinamine 253 | -66.3963 | 9 | Cys145, His164, His163, Glu166, Phe140, Ser144 | 7 | Glu166, Leu141, Phe140, His163, Ser144, Cys145, His164 |
| Spinochrome E | -65.5416 | 7 | Glu166, Cys145, His163, His164, Asn142, Phe140 | 7 | Glu166, Leu141, Phe140, His163, Ser144, Cys145, His164 |
| Spinochrome 252 | -65.0422 | 6 | Cys145, His164, His163, Glu166, Phe140 | 7 | Glu166, Leu141, Phe140, His163, Ser144, Cys145, His164 |
| X77
(native ligand) |
-64.0758 | 4 | Gly143, Cys145, Ser144, Glu166 | 5 | Asn142, Thr25, Ser144, His163, His164 |
The assumption of greater antiviral activity of the compounds is supported by the biggest number of bond couplings between bioactive compounds from sea urchins and amino acid receptors. Table 3 further shows that, in comparison to remdesivir and native ligand, all six sea urchin compounds (Spinochrome C, Echinamine B, Spinochrome A, Echinamine A, Echinochrome A, Spinochrome D) had a lower affinity and Rerank Score (X77). Rerank Score shows how the active substances, which are tiny molecules with molecular protein cell targets, are harmoniously bonded together. A limited affinity in the binding of molecular proteins and the chemicals indicates a stable bonding.
Moreno-gracia et al., (2022) discovered that the active components that were commonly found and had pharmacological properties in a review of articles on the content of sea urchins were echinocrome A, spinochrome A, spinochrome B, gallic acid, chlorogenic acid, and napthoquinone isolated from sea urchin shells and spine pigments from various types of sea urchins. These chemicals are expected to have significant antibacterial, antiviral, and antioxidant properties18.
As a result from this research, predictions for six sea urchin compounds’ antiviral activity (Spinochrome C, Echinamine B, Spinochrome A, Echinamine A, Echinochrome A, Spinochrome D) are higher than those for Remdesivir. Spinochrome C compounds had the lowest affinity/Rerank Score, indicating that they are likely to be the most effective as anti-Covid-19 agents.
Conclusion
Six bioactive substances from sea urchins have been shown to suppress SARS-CoV-2, according to studies. The outcomes of this in silico approach’s study yielded an idea of the precise and efficiency of antiviral drugs by interacting with Mpro so as to exhibit blockade and provide therapeutic therapy. In addition, among 11 bioactive compounds from sea urchins, spinochrome C is a potential compound. Spinochrome C has high affinity for 6W63 protein, spinochrome C has a smaller rerank score compare with Remdesivir and native ligand (X77). The results of this study are potentially useful, so Spinochrome C is a strong candidate for antiviral drugs as an inhibitor of SARS-CoV-2. However, further studies of bioactive compounds from sea urchins need to be done.
Acknowledgement
None
Conflict of Interest
There are no conflicts of interest.
Funding Sources
There is no funding source for this project.
References
- World Health Organization. Weekly Epidemiological Update – 10 Jan 2022. World Health Organization. 2022. Available from: https://www.who.int/publications/m/item/ weekly‑epidemiological‑update‑‑‑10‑January‑2022. [Last accessed on 2022 Jan 10].
- Goyal B., & Goyal D: Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS combinatorial science. 2020; 22(6): 297-305.
CrossRef - World Health Organization. Remdesivir and three other drugs for hospitalised patients with COVID-19: final results of the WHO Solidarity randomised trial and updated meta-analyses. Lancet 2022; 399: 1941–53. https://doi.org/10.1016/ S0140-6736(22)00519-0.
CrossRef - Zumla A., Chan JF., Azhar EI., Hui DS., & Yuen KY: Coronaviruses—drug discovery and therapeutic options. Nature reviews Drug discovery. 2016;15(5): 327-347.
CrossRef - Chang, K. O., Kim, Y., Lovell, S., Rathnayake, A. D., & Groutas, W. C: Antiviral drug discovery: norovirus proteases and development of inhibitors. Viruses. 2019; 11(2): 197.
CrossRef - Dubey, K., & Dubey, R : Computation screening of narcissoside a glycosyloxyflavone for potential novel coronavirus 2019 (COVID-19) inhibitor. Biomedical journal. 2020; 43(4): 363-367.
CrossRef - Krylova N.V., Leneva I.A., Fedoreev S.A., Ebralidze L.K., Mishchenko N.P., Vasileva E.V., Falynskova I.N., Iunikhina O.V., Lavrov V.F., Svitich O.A. Activity of compounds containing echinochrome A against herpes simplex virus type 2 in vitroand in vivo // Journal of microbiology, epidemiology and immunobiology. 2019; 96(6):56-64. doi: 36233/0372-9311-2019-6-56-64
CrossRef - Ageenko NV, Kiselev KV, Odintsova NA. Quinoid Pigments of Sea Urchins Scaphechinus mirabilisand Strongylocentrotus intermedius: Biological Activity and Potential Applications. Mar Drugs. 2022;20(10):611. doi:10.3390/md20100611
CrossRef - Kalinin VI. Echinoderms Metabolites: Structure, Functions, and Biomedical Perspectives. Mar Drugs. 2021;19(3):125. doi:10.3390/md19030125
CrossRef - Brasseur, L., Demeyer, M., Decroo, C., Caulier, G., Flammang, P., Gerbaux, P., & Eeckhaut, I: Identification and quantification of spinochromes in body compartments of Echinometra mathaei’s coloured types. Royal Society open science. 2018; 5(8): 171213.
CrossRef - Mishchenko NP, Krylova NV, Iunikhina OV, Vasileva EA, Likhatskaya GN, Pislyagin EA, Tarbeeva DV, Dmitrenok PS, Fedoreyev SA. Antiviral Potential of Sea Urchin Aminated Spinochromes against Herpes Simplex Virus Type 1. Marine Drugs. 2020; 18(11):550. doi: 10.3390/md18110550. PMID: 33167501; PMCID: PMC7694471.
CrossRef - Fedoreyev SA, Krylova NV, Mishchenko NP, Vasileva EA, Pislyagin EA, Iunikhina OV, Lavrov VF, Svitich OA, Ebralidze LK, Leonova GN. Antiviral and Antioxidant Properties of Echinochrome A. Marine Drugs. 2018; 16(12):509. https://doi.org/10.3390/md16120509
CrossRef - Dubey, K., Dubey, R., Gupta, R., & Gupta, A: In-silico Reverse Docking Studies for the identification of potential of Betanin on some enzymes involved in diabetes and its complications. Journal of Drug Delivery and Therapeutics. 2019; 9(2-A): 72-74.
- Heble, N. K., Mavillapalli, R. C., Selvaraj, R., & Jeyabalan, S. Molecular docking studies of phytoconstituents identified in Crocus sativus, Curcuma longa, Cassia occidentalis and Moringa oleifera on thymidylate synthase-an enzyme target for anti-cancer activity. Appl. Pharm. Sci. 2016; 6(12): 131-135.
CrossRef - Lipinski, C. A., Lombardo, F., Dominy, B. W., & Feeney, P. J : Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced drug delivery reviews. 1997; 23(1-3): 3-25.
CrossRef - Pires, D. E., Blundell, T. L., & Ascher, D. B: pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. Journal of Medicinal Chemistry. 2015; 58(9): 4066-4072.
CrossRef - Pagadala, N.S., Syed, K., & Tuszynski, J: Software for molecular docking: a review. Biophysical Reviews. 2017; 9(2): 91-102.
CrossRef - Moreno-García D.M, Salas-Rojas M, Fernández-Martínez E, López-Cuellar M.D.R, Sosa-Gutierrez C.G, Peláez-Acero A, Rivero-Perez N, Zaragoza-Bastida A, Ojeda-Ramírez D. : Sea urchins: an update on their pharmacological properties. PeerJ. 2022. 10:e13606. doi: 10.7717/peerj.13606. PMID: 35811815; PMCID: PMC9261939.
CrossRef









