Alamu Juliana , K. V. Leela.
, K. V. Leela. , Anusha Gopinathan
, Anusha Gopinathan and T. Jayaprakash
and T. Jayaprakash
Department of Microbiology, SRM Medical College Hospital and Research Centre. SRM Institute of Science and Technology, SRM Nagar, Kattankulathur, Tamil Nadu, India.
Corresponding Author E-mail: alamukemii@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/2546
Abstract
Background and objectives: Biofilm formation is an important virulence factor that protects an organism from antimicrobial agents as well as host immune effectors, thus allowing organisms to invade, survive, and cause persistent-reoccurring infection in host cells. The aim of this study was to investigate the ability of sepsis-causing gram-negative bacteria to form biofilms, evaluate the association between antibiotic resistance pattern and biofilm formation, determine the role and influence of biofilm formation on pathogenicity and clinical outcome of sepsis. Methods: A prospective study conducted from October 2020 to August 2021, non-replicated gram-negative bacteria isolates were recovered from blood samples of patients with suspected bacteremia, sepsis, and sepsis shock and identified using biochemical procedures. Antimicrobial susceptibility patterns of GNB isolates were determined using the Kirby-Bauer disc diffusion method and interpreted using CLSI guidelines. The ability of GNB isolates to form biofilm was assessed using Congo red agar and the tissue culture plate method. Results: Of the 160 Gram-negative bacteria tested, biofilm formation was seen in 73 (45.63%) isolates. Isolates are Klebsiella pneumoniae (39.73%), Acinetobacter spp. (34.25%), Escherichia coli (23.29%), Pseudomonas aeruginosa (1.37%), and other non-fermenters (1.37%). Isolates were highly resistant to cephalosporins, fluoroquinolones, and the penicillin group of antibiotics. No statistical relationship was found between resistance pattern, clinical outcome, and biofilm formation. Conclusion: In the current study, we found that 45.63% of gram-negative bacteria causing sepsis were biofilm producers. Klebsiella pneumonia isolates exhibited the highest levels of biofilm formation and antimicrobial resistance. Based on the strength of biofilm formation, most isolates were weak biofilm producers, and there was no statistical correlation between the formation of biofilms and antimicrobial resistance, indicating that the formation of biofilms was not a determining factor for resistance.
Keywords
Antibacterial agent; Bacteria; Biofilm; Persistence infections; Virulence
Download this article as:| Copy the following to cite this article: Juliana A, Leela K. V, Gopinathan A, Jayaprakash T. Biofilm Formation and its Association with Gram Negative Sepsis Pathogenicity. Biomed Pharmacol J 2022;15(4). |
| Copy the following to cite this URL: Juliana A, Leela K. V, Gopinathan A, Jayaprakash T. Biofilm Formation and its Association with Gram Negative Sepsis Pathogenicity. Biomed Pharmacol J 2022;15(4). Available from: https://bit.ly/3slFu0z |
Introduction
Biofilms are microbial-derived surface-associated cells that are enclosed in an extracellular polymeric substance matrix (EPS) and attached to a substratum or to each other in an irreversible way.1,2 They are important virulence factors that are produced through a multi-step process that begins with a single species of bacteria with fimbriae, pilli, or flagella attaching to conditioning film and progresses to micro-colonies after longer exposures, eventually forming a three-dimensional structure that detaches after maturation. 2,3
Biofilm serves as a protected mode of growth and an efficient barrier in hostile environments, allowing cells to survive while also dispersing to colonise new niches.4,5 Research shows that 80% of chronic persistent bacterial infections are linked to biofilms, which are usually formed at the primary focus of infection, such as meningitis, UTI, cystic fibrosis, or infective endocarditis, and then disseminated into the bloodstream via the penetration of injured tissues.3,6,7 The presence of an organism in the bloodstream, particularly planktonic bacteria, triggers an immune response that typically destroys pathogens. However, a dysregulated response by host immune cells to infection results in sepsis, which further leads to organ dysfunction, septic shock, or death if left untreated.8
Biofilm formation is a serious clinical issue that promotes antimicrobial resistance by slowing antimicrobial diffusion and facilitating plasmid exchange, which requires aggressive treatment.9,10 Several studies show that biofilm formation is associated with infection severity, persistence, and relapse. 2,10,11,12,13. However, only a few studies looked at the biofilm-forming ability of gram-negative bacteria that cause sepsis and its correlation to antimicrobial resistance and sepsis outcome. Studies on biofilm formation in sepsis-causing gram-negative bacteria are very important as they will play a key role in understanding the virulence of GNB causing sepsis, provide information on the role of biofilm in resistance, as well as give a deeper insight into treatment strategies, which might help reduce the mortality and morbidity rate associated with sepsis. The aim of this study was to investigate the ability of sepsis-causing gram-negative bacteria to form biofilms, evaluate the association between antibiotic resistance pattern and biofilm formation, determine the role and influence of biofilm formation on pathogenicity and clinical outcome of sepsis.
Methodology
Isolates collection
This was a prospective study conducted at SRM Medical College and Research Centre’s Department of Medical Microbiology (October 2020–August 2021). 160 non-repetitive GNB were recovered from blood samples submitted by various outpatient and inpatient wards to the laboratory. All blood positive cultures identified as gram-negative bacteria between the study periods were included, while blood positive cultures identified as contaminants or gram-positive were excluded.
Colony characterization and bacterial identification
According to bacteriological guidelines, the specimens were cultured on blood, MacConkey, and chocolate agar, and incubated at 37oC for 24 hours. Following incubation, colony morphology was evaluated based on size, mucoid nature, pigment, odour, and lactose fermentation. Biochemical identification was done using unique tests such as Oxidase, Indole, Motility, Triple Sugar Iron, Citrate, Urease, Methyl Red Voges-proskauer, and Amino acid.
Susceptibility testing
Antimicrobial susceptibility testing was done using the Kirby Bauer disc diffusion technique and interpreted as per Clinical and Laboratory Standards Institute (CLSI) guidelines.14
Detection of Biofilms
Biofilm detection was performed using Congo red agar and the tissue culture plate method as previously described by Hassan et al. and Dhanalakshmi et al. 1,10
Congo Red Agar (CRA) Method
Congo red medium was prepared using brain heart infusion broth at 37gm/l, agar 10gm/l, sucrose 50gm/l, and Congo red stain 0.8gm/l. First, an aqueous concentrated solution of Congo red stain was prepared, autoclaved, and added to the other medium constituents at 55oC. Isolates were then cultured on CRA plates and incubated for 24 hours at 37oC. CRA was repeated in triplicates and the results were interpreted based on whether black colonies with a dry crystalline consistency were produced or red colonies. (Fig 1).1,10
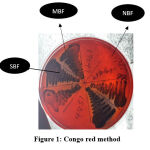 |
Figure 1: Congo red method. |
Tissue culture plate method (TCP)
In brief, 200 µl of bacterial suspension was aliquoted into 96-well flat-bottom tissue culture plates and incubated for 24 hours at 37°C. Following incubation, wells were rinsed with phosphate buffer saline (pH 7.2). Adhering bacteria were fixed with 2% sodium acetate and stained with 0.1% crystal violet. An ELISA auto reader (Model 680) was used to quantify stained adhering biofilm at an optical density of 450 nm, and the results were interpreted according to Stepnovic et al. criteria. Each experiment was carried out in triplicate, with an uninoculated medium serving as a control. 1,10,15.
 |
Figure 2: Tissue culture plate method. |
Statistical Evaluation
Statistical calculations were performed using SPSS (IBM SPSS V23). The relationship between resistance pattern and biofilm formation was assessed using Pearson correlation. The chi-square table was applied to compare variables (Table I). P value <0.05.
Table 1: Chi square table. P valve = 0.057
| Negative | Positive | Total | |
| TCP | 87 | 73 | 160 |
| CRA | 70 | 90 | 160 |
| Total | 157 | 163 | 320 |
Table 1 compares data obtained via the Congo red agar (CRA) method with the Tissue Culture Plate (TCP) method. P<0.05
Ethics
The study was approved by our institution’s ethical committee (2196/IEC/2020). Patient consent was not required for the study because isolates were collected directly from the laboratory.
Result
Out of the one hundred and sixty (160) non-repetitive gram-negative bacteria isolates studied for biofilm formation, 73 were identified as biofilm producers via TCP method and 90 as biofilm producers via CRA method (Table 2). A true biofilm producing organism is considered an organism that shows biofilm formation in both methods. The most common biofilm producing organism was Klebsiella pneumoniae 29 (39.73%), followed by Escherichia coli 17 (23.3%). Table 2 shows the percentages of biofilm and non-biofilm production identified by CRA and TCP.
Table 2: Biofilm detection by two different phenotypic (CRA & TCP) methods.
| Methods | Biofilm producers No (%) | Non-biofilm producers No (%) | Total No (%) | |||
| Congo red agar method (CRA) | Strong | Moderate | Weak | Total |
70(43.75) |
160(100) |
| 12(13.33) | 6(6.67) | 72(80.00) | 90(56.25) | |||
| n=90 | n=90 | n=90 | n=160 | n=160 | ||
| Tissue culture plate (TCP) | 0(0.00) | 8(10.96) | 65(89.04) | 73(45.63) | 87(54.38) | 160(100) |
| n=73 | n=73 | n=73 | n=160 | n=160 | ||
Table 2 shows the percentages of biofilm producers and non-biofilm producers detected using the Congo Red method and the Tissue Culture method.
Table 3: Diagnostic efficacy of Congo red agar method.
| Biofilm detection | Specificity
(%) |
Sensitivity
(%) |
*PPV
(%) |
**NPV
(%) |
Accuracy
(%) |
| Congo red method (CRA) | 92 | 76 | 92 | 76 | 83 |
*PPV- Positive predictive valve. ** NPV- Negative predictive valve.
Table III shows the diagnostic efficacy of the Congo red agar method. Specificity, sensitivity, PPV, NPV, and accuracy as compared to the tissue culture plate method, which is the standard method used for this study.
Table 4: shows the bacteriological profile and percentage of biofilm producing isolates.
| Blood Isolates | Biofilms producers Number (%) |
| Klebsiella pneumonia | 29(39.73) |
| Escherichia coli | 17(23.29) |
| Acinetobacter spp | 25(34.25) |
| Pseudomonas aeroginosa | 1(1.37) |
| Other -Non fermenting GNB | 1(1.37) |
| Salmonella typhi | 0(0.00) |
| Total | 73(100) |
Table IV shows the distribution and percentages of gram-negative bacteria isolates producing biofilm in our study. Klebsiella pneumonia was found to be the highest biofilm producer, followed by Escherichia coli. Salmonella typhi was found to be a non-biofilm producer.
Table 5: showing the resistance pattern of gram-negative bacteria isolates.
| Resistance in percentage (%) | ||||||
| Antibiotics | Escherichia coli | Klebsiella pneumonia | Acinetobacter spp | Pseudomonas aeruginosa | Citrobacter | Proteus |
| Aminoglycosides | ||||||
| Amikacin | 4.00 | 37.50 | 42.22 | 8.33 | 75.00 | 50.00 |
| Gentamicin | 38.00 | 41.67 | 53.33 | 8.33 | – | 50.00 |
| Cephalosporins | ||||||
| Cefazolin | 71.00 | 79.17 | 25.00 | 50.00 | ||
| Cefepime | 58.00 | 66.67 | 26.67 | 9.09 | 25.00 | 50.00 |
| Cefotaxime | 67.00 | 70.83 | 37.78 | – | 33.33 | |
| Ceftriaxone | 67.00 | 66.67 | 35.56 | – | 25.00 | 50.00 |
| Ceftazidime | 60.00 | 68.75 | 35.56 | 8.33 | 25.00 | 50.00 |
| Cefoxitin | 20.00 | 64.58 | – | – | 25.00 | |
| Cefuroxime | 69.00 | 72.92 | – | – | 25.00 | 50.00 |
| Carbapenem | ||||||
| Imipenem | 4.00 | 45.83 | 42.22 | – | – | – |
| Meropenem | 4.00 | 47.92 | 33.33 | 8.33 | – | – |
| Ertapenem | 5.00 | 47.92 | – | – | – | – |
| Fluroquinolones | ||||||
| Ciprofloxacin | 73.00 | 68.75 | 28.89 | – | – | 50.00 |
| Levofloxacin | – | – | – | 9.09 | – | – |
| Pencillins | ||||||
| Ampicillin | 91.00 | 100 | – | – | – | – |
| B lactam combination | ||||||
| Amoxacillin- clavulanate | 69.00 | 85.42 | 81.82 | 81.82 | 25.55 | 25.00 |
| Piperacillin tazobactam | 11.00 | 50.00 | 31.11 | 0.00 | 0.00 | 0.00 |
| Ceftazidime clavanic acid | 11.00 | 52.08 | 0.00 | 0.00 | 0.00 | |
| Tetracyclines | ||||||
| Tetracycline | 58.00 | 36.17 | 35.56 | 0.00 | 0.00 | 100.00 |
NA-Non applicable
Table 5 shows the resistance pattern of gram-negative bacteria isolates causing sepsis.
Table 6: Relationship between Biofilm formation and Resistance pattern.
| P(<0.05) | ||||
| Antibiotics | Escherichia coli | Klebsiella pneumoniae | Acintobacter spp | Psedomonas aeroginosa |
| Aminoglycosides | ||||
| Amikacin | 0.886 | 0.834 | 0.962 | 0.753 |
| Gentamicin | 0.292 | 0.806 | 0.961 | 0.753 |
| Cephalosporins | ||||
| Cefazolin | 0.436 | 0.321 | 0.685 | |
| Cefepime | 0.529 | 0.822 | 0.418 | 0.753 |
| Cefotaxime | 0.642 | 0.442 | 0.390 | – |
| Ceftriaxone | 0.762 | 0.822 | 0.488 | – |
| Ceftazidime | 0.377 | 0.822 | 0.461 | – |
| Cefoxitin | 0.572 | 0.179 | 0.642 | – |
| Cefuroxime | 0.642 | 0.265 | 0.239 | – |
| Carbapenem | ||||
| Imipenem | 0.886 | 0.533 | 0.853 | – |
| Meropenem | 0.886 | 0.156 | 0.524 | 0.753 |
| Ertapenem | 0.886 | 0.533 | 0.560 | – |
| Fluroquinolones | ||||
| Ciprofloxacin | 0.790 | 0.293 | 0.423 | – |
| Levofloxacin | – | – | – | 0.753 |
| Pencillins | ||||
| Ampicillin | 0.277 | – | 0.520 | – |
| B lactam combination | ||||
| Amoxacillin- clavulanate | 0.528 | 0.231 | 0.991 | 0.546 |
| Piperacillin tazobactam | 0.445 | 0.657 | 0.418 | – |
| Ceftazidime clavanic acid | 0.917 | 0.744 | 0.303 | 0.7563 |
| Tetracyclines | ||||
|
Tetracycline |
0.622 | 0.661 | 0.580 |
– |
Table 6 shows the statistical relationship between antibiotic resistance and biofilm formation. The P value is <0.05
Discussion
The formation of biofilm is an efficient defence barrier used by microbes to invade hostile environments.5 It is associated with antimicrobial resistance, persistence, and severity of chronic infections and is a major cause of sepsis relapse. 8,13 Sepsis is a serious health threat with over 30 million causes recorded annually. Bacterial sepsis has been identified as a major cause of mortality and morbidity, even though its pathophysiology is not yet fully understood. In this study, the ability of gram-negative sepsis-causing bacteria recovered from the bloodstream to form biofilm was studied and correlated with the strength of biofilm formation, resistance pattern, and its influence on pathogenicity and clinical outcome.
Our finding indicates 73 (45.63%) of GNB organisms causing sepsis formed biofilm. This is nearly similar to Cepas et al.17 which reported 49.3% of biofilm formation in isolates and is contrary to studies by Swarna et al. 18 and Zubair et al.19 which receptively reported 91% and 80% of biofilm formation in their studies. The highest biofilm producing organism was Klebsiella pneumonia. Similar to our results, Klebsiella pneumonia was found to be the most common biofilm producing organism by Cepas et al.17, Karmi et al.20 while De et al.21 and Dumaru et al.13 identified Escherichia coli as the most common Gram-negative biofilm producing organism.
For biofilm detection, two (2) phenotypic methods were used; they are Congo red agar (CRA) and the Tissue Culture Plate method (TCP). Although there are many other biofilm detection methods, there is no standard procedure for the detection of biofilm formation. TCP was used as the gold standard for this study.10 Sensitivity and specificity of 75% and 92% were observed in the Congo red method. The percentage observed in sensitivity is similar to studies by Mathur et al. 22 (90.02%), Bose et al. 23 (96.23), and Chandana et al.2 (86.2%), while the sensitivity observed was lower in their study. The specificity percentage observed in this study is consistent with findings by Dhanalaskshmi et al.10 and Tayal et al.24 which had 80% and 94.59%, respectively.
On account of antimicrobial resistance, our findings revealed maximum percentage resistance to penicillin, cephalosporins, fluoroquinolones, and B-lactam combination agents (Table 5). Klebsiella pneumoniae was 100% resistant to Ampicillin, 83.64% to Amoxicillin-clavulanate, 80% to Cefazolin, 70.91% to Cefuroxime, and 63.64% to Ciprofloxacin. These findings are in line with those of Karimi et al.20 and Chandana et al.2, who found that Klebsiella pneumonia isolates had the highest resistance to cefotaxime, ampicillin, and ciprofloxacin in their studies. No significant statistical relationship was found between the resistance pattern and the strength of biofilm formation (Table VI), although it is important to note that in this study, no strong biofilm formation was observed by the TCP method, and the majority of strains were weak biofilm-formers.
In most articles where a strong correlation was found between biofilm formation, pathogenicity, and resistance patterns, the biofilm formed was either strong or moderate. suggesting that biofilm formation strength may play an essential role in resistance. 16,20,25,26. Devanga Rugupathi et al.25 discovered a stronger correlation between strong biofilm formation and carbapenem resistance than between moderate and weak biofilm formation. This might imply that the susceptibility pattern of an organism is dependent on the strength of the biofilm formed by that organism. Several scientific studies have hypothesised that the formation of biofilm prevents the efficient diffusion of antibiotics, resulting in a significant decrease in bacteria’s exposure to antimicrobial agents and antibiotic activity.
Conclusion
In this study, resistance, pathogenicity, and clinical outcome of patients were found to be independently associated with weak biofilm formation. Klebsiella pneumoniae was the most resistant organism and had the highest biofilm production. No statistical relationship was found between biofilm formation and antibiotic resistance, and this could be because most isolates were weak biofilm producers. Further molecular investigation into biofilm associated genes and their role in sepsis severity is very important as findings will help in accurate treatment development, thus reducing mortality and morbidity associated with sepsis-associated Gram-negative bacteria.
Acknowledgement: None
Conflict of interest
The author(s) declare that there is no conflict of interest.
Funding Sources
There is no funding Source.
References
- Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J infect Dis. 2011 Jul-Aug., 15(4):305-11, PMID :21860999
CrossRef - Chandana D, Annapurna GS,. Comparison of three Different method for the detection of Biofilm in Gram Positive Cocci and Gram Negative Bacilli isolated from clinical specimens. J. Pharm. Sci & Res Vol. 7(11), 2015, 952-955.
- Jamal M, Ahmad W, Andleeb S, Jalil F, Imran M, Nawaz MA, et al. Bacterial biofilm and associated infections. J.Chin med Assoc. 2018 Jan;81(1):7-11. https://doi:10.1016/jcma.2017.07.012. Epub 2017 Oct 15. PMID: 2904218.
CrossRef - Hall-Stoodley. L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol.2004 Feb; 2(2):95-108, https://doi:10.1038/nrmicro821. PMID: 15040259.
CrossRef - Ghanbarzadeh Corehtash Z, Khorshidi A, Firoozeh F, Akbari H, Mahmoudi Aznaveh A. Biofilm Formation and Virulence Factors among Psedomonas aeroginosa isolated from burns patients. Jundishapur J Microbiol. 2015 Oct 2;8(10):e22345. https://doi.10.5812/jjm.22345. PMID: 26587205;PMCID: PMC4644346.
CrossRef - Pinto H, Simoes M,Borges A. Prevalance and impact of Biofilms on Bloodstream and Urinary Tract infections: A systematic Review Meta-Analysis. Antibiotics (Basel). 2021 Jul 8; 10(7):825. https://doi:10.3390/antibiotics10070825. PMID: 34356749; PMCID:PMC8300799.
CrossRef - Yousif A, Jamal MA, Raal L. Biofilm-based central line associated bloodstream infections. Adv. Exp Med Bio, 2015;830:157-79, https://doi:10.1007/978.3-319-11038-7-10. PMID: 25366227.
CrossRef - Minayan H. Sepsis: Mechanisms of bacterial injury to the patient. Scand J Trauma Resusc Emerg Med 27,19(2019). https://doi.org/10.1186/s13049-019-0596-4.
CrossRef - Ruhal R, Kataria R. Biofilm in gram-positive and gram-negative bacteria. Microbiol Res. 2021 Oct;251:126829. http://doi.10.1016/j.micres.2021. Epub 2021 Jul 23. PMID:34332222.
CrossRef - Dhanalakshmi TA, Venkatesha D, Aliya AN, Asharani N. Evaluation of phenotypic methods for detection of Biofilm formation in uropathogens. National journal of laboratory medicine 2018, Oct, vol-7(4):m006-m077.
- Devrari JC & Pai RV. Characterization of various virulence properties of Drug-resistant Klebsiella An Invitro Study among superbugs. Asian Journal of Pharmaceutical and Clinical Research. 2018;138-141.
CrossRef - Wang YC, Huang TW, Yang YS, Kuo SC, Chen CT, Liu CP, et al. Biofilm formation is not associated with worse outcome in Acinectobacter baumanni bacteraemic Pneumonia. Sci Rep. 2018 may 8:8(1): https://doi:10.1038/s41598-018-25661-9. PMID:29740176; PMICD: PMC5940913.
CrossRef - Dumaru R, Baral R, Shrestha LB. Study of biofilm formation and antibiotic resistance pattern of gram negative bacilli among the clinical isolates at BPKIHS Dharan. Bmc Res Notes. 2019 Jan 18:12(1):38. https://doi:10.1186/s13104-019-4084-8. PMID: 30658694; PMICID: PMC 6339267.
CrossRef - Clinical Laboratory Standards Institute, performance standards for Antimicrobial Susceptibility Testing. CLSI supplement M100, Clinical Laboratory Standard institute Wayne, PA, USA, 30th edition, 2020.
- Shirazi AS, Shafiei M,, Solgi H,Aslani MM, Azizi O, Badmasti F. Different Virulence Capabilities and ompA Expressions in ST2 and ST513 of Multi- Resistant Acinectobacter baumannii. Curr Microbiol. 2019 Jun;76(6):723-731. https://doi.10.1007/s00284-019-01686-9. Epub 2019 Apr 15. PMID:30989324.
CrossRef - Shadkam S, Goli HR, Mirzaei B et al. Correlation between antibiotic resistance and biofilm formation capacity among Klebsiella Pneumoniae strains isolated from hospitalized patents in Iran. Ann Clin Microbial Antimicrob 20, 13(2021) https://doi.org/10.1186/s12941-021-00418-X.
CrossRef - Cepas V, Lopez Y, Munoz E et al, Relationship between biofilm formation and antimicrobial resistance in gram negative bacterial. Microbial Drug Resistance, Vol 25, no 1, pp 72 -79.2019.
CrossRef - Swarna SR, Madhavan R, Gomathi S, Deveraj, Thamaraiselvi S. A study of biofilm on Diabetic foot ulcer. Int J Pharm Bio Sci 2012;4:1809-14.
- Zubair M, Malik A, Ahmad J, Rizri M, Farooqul KJ, Rizvi MW. A study of biofilm production by gram negative organisms isolated from diabetic foot ulcer patients. Biology and Medicine 2011; 3:147-157.
- Karimi K, Zarei O, Sedighi P, Taherei M, Doosti-Irani A, Shokoohizadeh. Investigation of antibiotics resistance and biofilm formation in clinical isolates of Klebsiella Pneumoniae. International Journal of Microbiology, Vol.2021, Article ID 5573388, 6 pages, 2021. https://doi.org/10.1155/2021/5573388.
CrossRef - De A, Deshpande D, Baveja SM, Taklikar S. Detection of biofilm formation in bacteria from cases of urinary tract infections, Septicemia, Skin and Soft tissues infections and post-operative infections by Congo Red Agar method. J Acad Med Sci 2021;2:46-47.
CrossRef - Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatima T, Rattan A. Detection of biofilm formation among the clinical isolates of staphylococci: an evaluation of three different screening methods. Indian J Med Microbiol 2006;24(1):25-9.
CrossRef - Bose S, Khodke M, Basak S, Mallick Sk. Detection of biofilm producing Staphylococci: need of the hour. J Clin Diagn Res 2009;3:1915-20.
- Tayal RA, Baveja SM, De AS. Comparison of phenotypic methods for the detection of biofilm production in uropathogens in a tertiary care hospital in India. Int J Curr microbial App Sci 2015;4(9):840-49.
CrossRef - Devanga Rugupathi NK, Muthuirulandi Sethuvel DP, Triplicane Dwarakanathan H, Murugan D, Urnashankar Y, Monk PN Karunakaran E, Veeraraghavan B. The Influence of Biofilms on Carbapenem Susceptibility and patient’s outcome in Device Associated Klebsiella pneumoniae Insights into Phenotype vs Genome – wide Analysis and Correction. Front. Microbiol. 11:591679. https://doi.10.3389/fmicb.2020.591679.
CrossRef - Lihua Q, Hao L, Chuanfu Z, Beibei L, Jie Li, Ligui W, et al. Relationship between Antibiotic Resistance, Biofilm Formation and Biofilm-Specific Resistance in Acinetobacter baumanni. Front Microbiol. 2016;7:483. https://doi:10.33889/Fmicb.2016.00483. PMID: 27148178.
CrossRef
Abbreviations
GNB-Gram negative bacteria
Spp – Species
UTI- Urinary tract infections








