Semuel Sandy1*  and Elisa Winanda2
and Elisa Winanda2
1Center for Public Health and Nutrition, National Research and Innovation Agency-Indonesia, Coworking Space, BRIN Archeolog Papua Jl Isele Waena Kampung, Waena Jayapura
2Research Center for Food Technology and Processing, Coworking Space, BRIN Archeology, Jl. Jl. Pajjaiang No.13, Sudiang Raya, Makassar,
Corresponding Author E-mail: mercury.sandy56@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/2515
Abstract
The odorant binding protein (OBP) has a role as a target protein for potential interaction mechanism activity for the development of repellent compounds. The purpose of this study was to analyze the physico-chemical properties of the protein, the stability of the three-dimensional structure of the OBP1 Anopheles farauti protein, and to predict the binding site pocket as the target of the active protein site against inhibitors. Analysis of physico-chemical properties was carried out by the ProtParam Expasy server. The theoretical calculated isoelectric point (pI) was found to be less than 7 indicating the acidic nature of this protein. The aliphatic index of 78 indicates the thermal stability of the protein. The Grand Average of Hydropathicity (GRAVY) is estimated at -0.355; This lower GRAVY value indicates a possible better interaction of this protein with water. Secondary structure analysis was carried out by SOPMA which revealed that Alpha helix (55.86%) predominated among the secondary structural elements followed by Random coil (32.41%), Extended strand (8.97%), and Beta turn (2.76%). Three-dimensional structure modeling of OBP1 Anopheles farauti was performed with the Swiss-Model server and the protein refine Galaxy server. Homology modeling results obtained PDB ID 2ERB template with sequence identity 94.4%. The model was validated for the three-dimensional structure of the protein using the MolProbity, ProSA, ProQ, ERRAT, Verfy3D, and PROCHECK servers. The prediction results of pocket binding sites using DoSiteScore obtained three pocket binding site locations, namely P_0 (Drug score 0.84); P_1 ((0.75); P_2 (Drug score 0.64). Conclusion Homology modeling of the protein OBP1 Anopheles farauti has been carried out and the three-dimensional structure of the Model_OBP1_04 protein has been obtained that meets the criteria for valid structural parameters so that this structure can be used In-silico molecular docking and molecular dynamic studies for the development of mosquitoes repellent.
Keywords
Anopheles farauti; Homology Modeling; In-silico; Malaria
Download this article as:| Copy the following to cite this article: Sandy S, Winanda E. Homology Modeling Odorant-binding Protein-1 (OBP1) Anopheles Farauti Protein Target for Mosquito Repellent. Biomed Pharmacol J 2022;15(3). |
| Copy the following to cite this URL: Sandy S, Winanda E. Homology Modeling Odorant-binding Protein-1 (OBP1) Anopheles Farauti Protein Target for Mosquito Repellent. Biomed Pharmacol J 2022;15(3). Available from: https://bit.ly/3S5GlxC |
Introduction
Mosquitoes are serious vectors of diseases that infect millions of people and animals around the world, such as malaria, filariasis, and important arboviruses such as dengue fever, yellow fever, chikungunya, West Nile virus, and Zika virus 1.
Malaria is a disease transmitted by mosquitoes of the genus Anopheles. Malaria in humans is caused by infection with one or several species of Plasmodium (Plasmodium falciparum, Plasmodium vivax, Plasmodium ovale, Plasmodium malariae, and Plasmodium knowlesi) which are transmitted by bite of the female Anopheles mosquito 2. Malaria remains an important health threat in rural Southeast Asia including Indonesia. Various efforts have been made to control, prevent, and even eliminate the disease. Studying the diversity and behavior of the Anopheles mosquito, which is a vector of malaria, plays an important role in developing effective and successful vector control program strategies to combat malaria transmission 3.
Many efforts to control malaria vectors have been carried out, including the distribution of insecticide-treated mosquito nets (Long Lasting Insecticide Treated Nets) and spraying of house walls with insecticides (Indoor Residual Spraying) 4. Malaria vector control through personal protection with the use of repellents, either derived from chemical sensitizers or derived from natural materials such as extracts and essential oils from plants, has been widely developed either in the form of sprays or in the form of lotions). The insect repellent works by providing a vapor barrier that prevents mosquitoes from contacting the skin surface. The development of essential oils and plant extract repellents has the potential to replace chemical synthetic materials. Plant materials are generally safe for the environment and also for living things 1.
The synthetic chemical N,N-diethyl-3-methylbenzamide (DEET) is a widely used repellent and is used as the gold standard for the development of repellants. The compound N,N-diethyl-3-methylbenzamide (DEET) has a target action on olfactory receptor neurons (ORNs) 5. Olfactory receptor neurons (ORN) of odor-detecting insects occur mainly in olfactory sensilla cells and involve membrane-bound olfactory receptor interactions. Odorant binding proteins (OBPs) is the most important agent in the olfactory system. This protein plays a key role in the perception and transmission of odors to receptor sites, signal transduction, and triggers a series of behavioral responses. The activity of the interaction of N,N-diethyl-3-methylbenzamide (DEET) against the OBPs protein in the malaria vector Anopheles gambiae has been studied. Findings for Anopheles gambiae OBP7 (AgamOBP7) suggest a more evolved protein as a result of increased structural complexity with eight Cys residues, four disulfide bridges, and a slightly longer C-terminal than the Classic OBPs protein model 6. Odorant binding proteins (OBPs) are expressed in the lymph fluid surrounding the olfactory dendrites, where they can reach concentrations in the millimolar range. Odorant binding proteins have multiple roles including protecting odorants from degradation and transporting them to olfactory receptors. There is evidence that OBPs has two major roles in odor perception. In the first model, several groups have proposed that OBPs act as passive carriers for odors, and changes in pH around the dendritic membrane cause conformational changes that stimulate ligand release, freeing the ligands to activate receptors 7.
Anopheles farauti is one of the vectors of malaria in Eastern Indonesia, especially in the Papua region. This mosquito has a protein sequence data code OBP1 on the UniProt server with gene code ID number AFAF003898. Modeling the three-dimensional structure of the protein by using the Homology modeling of the protein OBP1 Anopheles farauti using a protein sequence to obtain a stable three-dimensional structure template so that it can be used as an in-silico molecular docking and molecular dynamic analysis in repellent design. Protein structure Odorant-binding protein-1 (OBP1) Anopheles farauti experimentally, the protein crystal structure is not yet available in the protein data bank, so homology modeling is one way to predict the three-dimensional structure of the protein 8. The purpose of homology modeling of the three-dimensional structure of the protein OBP1 Anopheles farauti is to analyze the physico-chemical properties of the protein, the stability of the model protein structure, and predict the binding site pocket as the target of the protein’s active site against inhibitors.
The computational protein structure prediction method provides a cost-effective and time-effective alternative in the absence of experimentally derived structures. The Odorant-binding protein-1 Anopheles farauti protein homology modeling study is useful for understanding the structural properties of Anopheles farauti OBP1 protein, and the physico-chemical properties so that it will increase the prospect of its potential use to target mosquitoes’ repellent interaction mechanisms from the design of synthetic chemicals, extracts and essential oils from plants 9.
Material and methods
Tools and softwares
The tool used for analysis is Acer Nitro Notebook AN515-55 Intel Core I7 10870H CPU @ 2.20 GHz, 8 GB Memory, Microsoft Windows 10 Pro 64-bit Operating System. The software used is the Swiss Model server (https://swissmodel.expasy.org/); Galaxy refine server (https://galaxy.seoklab.org/index.html); SOPMA server (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html); SAVES v6.0 server (https://saves.mbi.ucla.edu/); Protein Quality Predictor server (ProQ) (https://proq.bioinfo.se/cgi-bin/ProQ/ProQ.cgi); ProtParam server (https://web.expasy.org/protparam/); DogSiteScore server (https://proteins.plus/);
Squence protein OBP1 Anopheles farauti
Sequence Odoront binding protein-1 (OBP1) Anopheles farauti gene code AFAF003898 downloaded from UniProt ID A0A182Q6A2 (https://www.uniprot.org/uniprotkb/A0A182Q6A2/entry). The OBP1 Anopheles farauti sequences can be seen as follows 10 :
>tr|A0A182Q6A2|A0A182Q6A2_9DIPT Odorant-binding protein 1 OS= Anopheles farauti OX=69004 PE=4 SV=1
MSKLLGFMCVALTCCSIVVADKTPRRDAEYPPPELLDAMKPLHDICVGKTGVTEEAIKKFSDEEIHEDEKLKCYMNCLFHEAKVVDDNGDVHLEKLHDALPNSMHDIAMHMGKRCLYPEGENLCDKAFWLHKCWKQSDPKHYFLV
Homology modeling
Homology modeling of the three-dimensional structure of the OBP1 protein using the Swiss Model Server by entering the data of the Anopheles farauti OBP1 protein sequence in the Swiss Model server (https://swissmodel.expasy.org/).
Validation of the three-dimensional structure of proteins
The results of the homology modeling of the three-dimensional structure of the OBP1 protein were then analyzed for the validation of its geometric structure using the ProSA server, SOPMA server, SAVES v6.0 server (PROCHECK, ERRAT, Verfy3D), ProtParam server, and ProQ server. If the homology modeling does not meet the required parameters, then the homology modeling refinement is carried out using the Galaxy refine server 11. The results of the refine homology modeling of OBP1 protein were then analyzed again. The results of OBP1 protein homology modeling that meet the requirements for good protein structure parameters are then analyzed for binding site prediction using the DoSiteScore server 12.
Result and Discussion
The results of the analysis of the physico-chemical properties of the Anopheles farauti OBP1 protein using the ProtParam server can be seen in Table 1. The OBP1 protein has 2303 atoms with a molecular weight of 16635.30 Daltons and consists of 145 amino acids. The isoelectric point (pI) of the calculation result of 5.67 (pI<7) so that the OBP1 protein has acidic properties. Calculating the isoelectric point value (pI) is useful in the development of buffer solutions for the protein purification process. The instability index provides an estimate of the stability of the protein in the reaction environment. Proteins with an Instability index value of less than 40 are predicted to be stable, an Instability index value above 40 predicts that the protein may be unstable. The calculation results obtained that the instability index is 49.65 so the template protein is not stable 13. The calculation of the instability index is useful for determining the storage conditions of the protein in a suitable solvent. The Grand average of hydropathicity (GRAVY) value for a peptide or protein is calculated as the sum of the hydropathic values of all amino acids, divided by the number of residues in that sequence. GRAVY Index calculation result -0.355. This lower value of the GRAVY index indicates a better possibility of interaction with water 8, 14. The results of the analysis obtained the composition of the most amino acids, among others, Leu(L); Lys(K); Asp(D); ala(A); Cys(C); Glu(E)His(H); and Val(V) (Table 2).
Table 1: Physicochemical properties of OBP1 protein parameters computed by ProtParam server
| Protein UniProt ID | AFAF003898 |
| Formula | C735H1144N194O214S16 |
| Total number of atoms | 2303 |
| Number of amino acids | 145 |
| Molecular weight | 16635.30 |
| Isoelectric point (pI) | 5.67 |
| The total number of negatively charged residues (Asp + Glu): | 25 |
| The total number of positively charged residues (Arg + Lys): | 18 |
| Instability index: | 49.65 |
| Aliphatic index: | 78.00 |
| Grand average of hydropathicity (GRAVY): | -0.355 |
Table 2: Amino acids composition Obp1 Anopheles farauti
| No. | Amino acids | Numbers | Composition |
| 1 | Ala (A) | 9 | 6.2% |
| 2 | Arg (R) | 3 | 2.1% |
| 3 | Asn (N) | 4 | 2.8% |
| 4 | Asp (D) | 13 | 9.0% |
| 5 | Cys (C) | 9 | 6.2% |
| 6 | Gln (Q) | 1 | 0.7% |
| 7 | Glu (E) | 12 | 8.3% |
| 8 | Gly (G) | 6 | 4.1% |
| 9 | His (H) | 9 | 6.2% |
| 10 | Ile (I) | 5 | 3.4% |
| 11 | Leu (L) | 15 | 10.3% |
| 12 | Lys (K) | 15 | 10.3% |
| 13 | Met (M) | 7 | 4.8% |
| 14 | Phe (F) | 5 | 3.4% |
| 15 | Pro (P) | 8 | 5.5% |
| 16 | Ser (S) | 5 | 3.4% |
| 17 | Thr (T) | 4 | 2.8% |
| 18 | Trp (W) | 2 | 1.4% |
| 19 | Tyr (Y) | 4 | 2.8% |
| 20 | Val (V) | 9 | 6.2% |
| 21 | Pyl (O) | 0 | 0.0% |
| 22 | Sec. (U) | 0 | 0.0% |
The secondary structure predicted using SOPMA is presented in Table 3. The results show that Alpha helix (Hh) 55.86% dominates among the secondary structural elements followed by Random coil (Cc) 32.41%, Extended strand (Ee) 8.97% and Beta turn (Tt) ) 2.76% 14.
Table 3: Secondary structure prediction protein OBP1 Anopheles farauti by SOPMA server
| No. | The secondary structure | Numbers | Composition |
| 1 | Alpha helix (Hh) | 81 | 55.86% |
| 2 | 310 helix (Gg) | 0 | 0.00% |
| 3 | Pi helix (Ii) | 0 | 0.00% |
| 4 | Beta bridge (Bb) | 0 | 0.00% |
| 5 | Extended strand (Ee) | 13 | 8.97% |
| 6 | Beta turn (Tt) | 4 | 2.76% |
| 7 | Bend region (Ss) | 0 | 0.00% |
| 8 | Random coil (Cc) | 47 | 32.41% |
| 9 | Ambiguous states (?) | 0 | 0.00% |
| 10 | Other states | 0 | 0.00% |
Homology modeling of the protein structure of OBP1 Anopheles farauti using protein sequences data from the UniProt ID server A0A182Q6A2 then submitted to the SWISS-MODEL server (https://swissmodel.expasy.org/) 15. The results of homology modeling obtained the template for the protein structure of OBP1 Anopheles farauti with PDB ID code 2ERB (Figure 1). Parameters of homology modeling results can be seen in Table 4.
 |
Figure 1: Sequence template PDB ID 2ERB homology model protein structure OBP1 Anopheles farauti. |
Table 4: Swiss-Model parameters homology modeling OBP1 Anopheles farauti (PDB ID 2ERB).
| No. | Parameters | Value |
| 1 | Seq Identity | 94.40% |
| 2 | Resolution X-Ray | 1.50 A |
| 3 | Oligo state | monomer |
| 4 | GMQE | 0.81 |
| 5 | QMEANDisCo Global | 0.84 ± 0.07 |
| 6 | QMEAN | -0.67 |
| 7 | Cβ | -1.36 |
| 8 | All Atom | 0.13 |
| 9 | solvation | 0.60 |
| 10 | torsion | -0.64 |
Table 4 shows that the PDB ID 2ERB template resulting from homology modeling has a sequence identity of 94.4% so that the template is categorized as having good quality (larger identity > 67%). The 2ERB template also has a crystal structure with an X-Ray resolution of 1.5. The results of the evaluation of the parameter values for the Global Model Quality Estimate (GMQE) are 0.81 and QMEANDisCo Global is 0.84 ± 0.07. These results are categorized close to a value of 1 so that the resulting homology modeling template can be considered good (the scale for GMQE and QMEANDisCo Global is between 0 and 1). If the GMQE and QMEANDisCo Global values are less than 0.6 then the modeling results are not good. The evaluation of the QMEAN parameter value (-0.67) the results of the PDB ID 2ERB template homology modeling are still above -4.0
 |
Figure 2: (a) QMEANDisCo analysis homology modeling struktur protein OBP1 Anopheles farauti (Score 0.84 ± 0.07); |
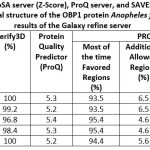
Validation of the three-dimensional structure of the homology result of Anopheles farauti protein modeling OBP1 using several applications including MolProbity server (Table 5), validation using the SAVES v.06 servers (PROCHECH, ERRAT, Vervy3D) 16, ProSA server and ProQ server (Table 6). The results of the validation of the three-dimensional structure of the Obp1 protein using MolProbity obtained a score of 0.69 and analysis using the Ramachandran Plot obtained 99.17% favored region 14. These results indicate that the results of homology modeling of the OBP1 protein have good quality. The results of the structural validation using the PROCHECH, ERRAT, Vervy3D, ProSA, and ProQ applications obtained quite different results from the validation using MolProbity, but the results of the structural analysis were still within the required parameter values (Table 6).
Table 5: Results of the validation of the three-dimensional structure of the OBP1 Anopheles farauti protein model by MolProbity server.
| Model | Clash score, all atoms | Poor rotamers (%) | Favored Rotamers
(%) |
Ramachandran plot | Rama
(Z-score) |
MolProbity | Cβ deviation >0.25A
(%) |
Bad
bonds (%) |
Bad
angels (%) |
|
| Outliers
(%) |
Favored
(%) |
|||||||||
| Swiss Model (2ERB) | 0 | 1.8 | 96.4 | 0.00 | 99.17 | 0.76 ± 0.68 | 0.69 | 0.00 | 0.19 | 1.01 |
Table 6: Validation results of the three-dimensional structure of the OBP1 Anopheles farauti protein model by ProSA server, SAVES v6.0 server, and ProQ server.
| Model | ProSA
(Z-score) |
ERRAT Quality Factor (%) | Verify3D (%) | Protein Quality Predictor (ProQ) | PROCHECK Ramachandran plot | ||||
| Most of the time
Favored Regions (%) |
Additional
Allowed Regions (%) |
Generously
Allowed (%) |
Disallowed
Regions (%) |
Overall, the
G-factor |
|||||
| Swiss-Model (2ERB) | -6.9 | 94.6 | 95.9 | 5.4 | 92.6 | 7.4 | 0.0 | 0.0 | 0.01 |
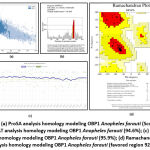 |
Figure 3: (a) ProSA analysis homology modeling OBP1 Anopheles farauti (Score -6.9); (b) ERRAT analysis homology modeling OBP1 Anopheles farauti (94.6%); |
Optimization of the three-dimensional structure of the OBP1 protein was carried out to increase the parameter values to approach the required ones. Optimization of the results of homology modeling of the three-dimensional structure of the OBP1 protein Anopheles farauti using Galaxy refine server 17. The optimization results obtained five models of the three-dimensional structure of the OBP1 protein. Then the OBP1 protein model was validated for its structure. The results of the validation of the three-dimensional structure of the OBP1 protein can be seen in Table 7 and Table 8. The validation results obtained that the results of the refinement of the homology modeling structure of the Anopheles farauti protein OBP1 with the code Model_OBP1_04 have good structural quality.
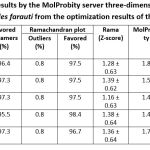 |
Table 7: Validation results by the MolProbity server three-dimensional structure of the OBP1 protein Anopheles farauti from the optimization results of the Galaxy refine server. |
The prediction results of pocket binding using the DoGSiteScorer (DGSS) Server obtained a drug score value > 0.5 so the prediction of the pocket binding site obtained 3 pocket models that meet these criteria (Table 5). The amino acid residues that make up the binding site pocket can be seen in Figure 4. The DGSS measures the volume of the pocket of interest and the amino acid composition and calculates the drug ability score, which indicates the possibility that the inhibitor compound will interact with the binding site pocket. 18. The visualization of the binding site pocket can be seen in the figure 4.
Table 9: Prediction of protein binding site protein structure model Model_OBP1_04 by DoGSiteScorer server.
| Model Pocket | Volume | Surface | Accept | Donor | Hydrophobic
Interactions |
Hydrophobicity | Drug Score |
| P_0 | 708.4 | 832.7 | 28 | 6 | 54 | 0.6 | 0.84 |
| P_1 | 457.4 | 844.6 | 39 | 15 | 33 | 0.4 | 0.75 |
| P_2 | 199.2 | 361.7 | 18 | 3 | 18 | 0.5 | 0.64 |
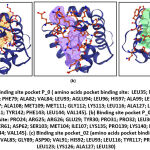 |
Figure 4: (a) Binding site pocket P_0 ( amino acids pocket binding site: |
Conclusion
The homology modeling of the OBP1 Anopheles farauti protein with physico-chemical properties was carried out by calculating the theoretical Isoelectric Point (pI 5.67), Molecular Weight 16635.30 Daltons, the number of positively charged residues (18) and negative (25), the Instability Index 49.65, the aliphatic index 78, and the Grand Average hydropathy (GRAVY -0.355). ). Secondary structure analysis was carried out by SOPMA which revealed that Alpha helix (55.86%) predominated among the secondary structural elements followed by Random coil (32.41%), Extended strand (8.97%), and Beta turn (2.76%). The three-dimensional structure modeling of Obp1 Anopheles farauti was carried out with the Swiss-Model server and the Galaxy refines protein server. Homology modeling results obtained PDB ID 2ERB template with sequence identity 94.4%. Three-dimensional modeling of the Anopheles farauti OBP1 protein using the Swiss-Model server. Refine the model using Galaxy refine protein Server to improve the quality of protein structure to produce a valid model. The OBP1 protein model Anopheles farauti was validated using PROCHECK and PROVE protein structure examiners to obtain the homology modeling protein Model_OBP1_04 and has three DGSS areas in the DGSS areas of the binding site pocket DGSS areas > 0.05 so that it can be used as a target for the development of mosquito repellents.
Conflict of Interest
Not applicable
Funding Source
Not applicable
Reference
- Khater HF, Selim AM, Abouelella GA, et al. Commercial mosquito repellents and their safety concerns hanem. In: Intech., 2012, 13.
- Magdalena V, Wurisastuti T. Overview Of The Distribution of Anopheles species and their role as malaria vectors in the provinces of East Nusa Tenggara, Papua, and West Papua. Spiracles., 2021: 12(1); 46-59.
CrossRef - Senjarini K, Setiawan R, Wathon S, Oktarianti R. Species shifting composition of the Anopheles vector in Wongsorejo district – Banyuwangi, Indonesia. In: IOP Conference Series: Earth and Environmental Science., 2021: 913; 1-6.
CrossRef - Setiyaningsih R, Trapsilowati W, Mujiyono M, Lasmiati L. Malaria vector control in endemic areas of Purworejo regency, Indonesia. Balaba J R&D on Banjarnegara Animal-Sourced Disease Control. Published online., 2018; 1-12.
CrossRef - Dickens JC, Bohbot JD. Mini review: Mode of action of mosquito repellents. Pestic Biochem Physiol., 2013: 106(3); 149-155.
CrossRef - Venthur H, Mutis A, Zhou JJ, Quiroz A. Ligand binding and homology modelling of insect odorant-binding proteins. Physiol Entomol., 2014: 39(3); 183-198.
CrossRef - Davrazou F, Dong E, Murphy EJ, Johnson HT, Jones DNM. New insights into the mechanism of odorant detection by the malaria-transmitting mosquito Anopheles gambiae. J Biol Chem., 2011: 286(39); 34175-34183.
CrossRef - Sahay A, Piprodhe A, Pise M. In silico analysis and homology modeling of strictosidine synthase involved in alkaloid biosynthesis in catharanthus roseus. J Genet Eng , 2020: 18; 44
CrossRef - Aslanzadeh V, Ghaderia M. Homology modeling and functional characterization of pr-1a protein of Hordeum vulgare subsp. Vulgare. Bioinformation., 2012: 8( 17); 807-811.
CrossRef - Ghavami MB, Khoeini S, Djadid ND. Molecular characteristics of odorant-binding protein 1 in Anopheles maculipennis. Malar J., 2020: 19(1); 1-11.
CrossRef - Heo L, Park H, Seok C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013: 41; 384-388.
CrossRef - Volkamer A, Griewel A, Grombacher T, Rarey M. Analyzing the topology of active sites: On the prediction of pockets and subpockets. J Chem Inf Model., 2010: 50(11); 2041-2052.
CrossRef - Gupta MK, Vadde R, Donde R, et al. Insights into the structure–function relationship of brown plant hopper resistance protein, Bph14 of rice plant: a computational structural biology approach. J Biomol Struct Dyn., 2019: 37(7); 1649-1665.
CrossRef - Ratan V, Dixit S, Srivastava M, et al. Computational structure prediction and analyze active ligand binding site of defense and lytic enzymes of Trichodermaharzianum. Ann Phytomedicine An Int J., 2018: 7(2); 143-160.
CrossRef - Tran NT, Jakovlić I, Wang W-M. In-silico characterisation, homology modelling and structure-based functional annotation of blunt snout bream (Megalobrama amblycephala) Hsp70 and Hsc70 proteins. J Anim Sci Technol., 2015: 57(1); 1-9.
CrossRef - Roy S, Maheshwari N, Chauhan R, Sen NK, Sharma A. Structure prediction and functional characterization of secondary metabolite proteins of Ocimum. Bioinformation., 2011: 6(8); 315-319.
CrossRef - Vg V, Bhaskar A, Purushothaman P. In silico analysis and 3D modelling of SORD Protein in diabetic retinopathy. J Comput Methods Mol., 2011: 1(4); 22-27.
- Adams J, Thornton BP, Tabernero L. A new paradigm for kim-ptp drug discovery: Identification of allosteric sites with potential for selective inhibition using virtual screening and lei analysis. Int J Mol Sci., 2021: 22; 12206.
CrossRef