Heggadadevanakote Kendaganna Pavan1,Bhargav Shreevatsa1, Chandan Dharmashekara1, Govindaraju Shruthi2 , Kollur Shiva Prasad3
, Kollur Shiva Prasad3 , Sharanagouda S Patil4
, Sharanagouda S Patil4 and Chandan Shivamallu1*
and Chandan Shivamallu1*
1Department of Biotechnology and Bioinformatics, School of Life Sciences, JSS Academy of Higher Education and Research, Mysore, Karnataka, India.
2Glimetomics Bioresolve Pvt Ltd, Mysuru, Karnataka-570 018, India.
3Department of Sciences, Amrita School of Arts and Sciences, Amrita Vishwa Vidyapeetham, Mysuru Campus, Mysuru – 570 026, Karnataka, India.
4ICAR-National Institute of Veterinary Epidemiology and Disease Informatics (NIVEDI), Bengaluru, Karnataka, India.
Corresponding Author E-mail: chandans@jssuni.edu.in
DOI : https://dx.doi.org/10.13005/bpj/2403
Abstract
Antibiotics are commonly used to treat bacterial respiratory infections, but they can exacerbate inflammation by releasing microbial components that overstimulate the immune system, leading to greater tissue damage. Klebsiella pneumoniae is a gram-negative, rod-shaped bacteria of the family Enterobacteriaceae. Knowing about Klebsiella pneumoniae is extremely important in the present situation, as it is one of the major causal organisms of pneumonia. Internal and external factors of K. pneumoniae are responsible for the entry and multiplication inside the host. Antibiotics against K. pneumoniae are a class of Penicillins, Cephalosporins, Monobactams, and Carbapenems which have the β-lactam ring in common with variable side chains. Combating the antibiotics by synthesizing the enzymes like beta-lactamases is the main reason for the survival of these organisms against newer generation antibiotics. In this review, we have tried to discuss about Klebsiella pneumoniae, antibiotics, and their mechanism of action.
Keywords
Beta-Lactamases; Biofilm; Klebsiella pneumoniae; Monobactam; Penicillin;
Download this article as:| Copy the following to cite this article: Pavan H. K, Shruthi G, Prasad K. S, Patil S. S, Shivamallu C. Review of Known and Unknown Facts of Klebsiella Pneumoniae and its Relationship with Antibiotics. Biomed Pharmacol J 2022;15(2). |
| Copy the following to cite this URL: Pavan H. K, Shruthi G, Prasad K. S, Patil S. S, Shivamallu C. Review of Known and Unknown Facts of Klebsiella Pneumoniae and its Relationship with Antibiotics. Biomed Pharmacol J 2022;15(2). Available from: https://bit.ly/3LVO11q |
Introduction
Klebsiella was named Friedlander bacillus initially when he had isolated the bacteria from the lungs of the patient who had been died because of pneumonia (1–3). The common habitat of Klebsiella species in the environment is the surface of the water, soil, sewage, and plants. They are also known to colonize on the mucosal surface of mammals and in plants(4,5). As a saprophyte, K. pneumoniae is present in the nasopharynx region and also in the gut(4). The primary target of the Klebsiella species is geriatric and pediatric individuals, with low immunity and those who are suffering from various diseases like diabetes, pulmonary obstruction, and cardiac diseases. Klebsiella infections may be Community-acquired or nosocomial with a high mortality rate if it’s not treated correctly(4,6).
pneumoniae is a pulmonary pathogen, with sudden inception having a high fever, blood coughing (7), infections were seen in lungs, abdominal cavity, urinary tract, surgery sites, and also subsequent bacteria in blood are common clinical conditions of K. pneumoniae(1) in humans. Most of the endemic and epidemic (Nosocomial) Infections caused by K. pneumonia are hospital acquired (8). Epidemic infections spread through neonatal intensive care units infecting the bloodstream. In endemic infections, the urinary tract is the main site and is through medicine and surgery amenities(8). The common sources in hospital settings like water, tables, floors, bedpans, and stethoscopes are responsible for the transmission of K. pneumoniae outbreaks apart from person to person transmission as in hospital-associated urinary tract infections. Along with this contaminated: a) aerosol used for respiratory therapy in pulmonary wards, b) breast milk, and lipid emulsions are given to newborns in neonatal ICUs, c) sampling probes used in blood culture analyzers also causes pneumonia(8). The spread of K. pneumoniae infections can be prevented through good hygienic practices in hospital settings.
About 3.6 million episodes of severe pneumonia are found to dread the country every year in children younger than 5 years of age. About 4% of the Indian population is affected by pneumonia each year, making it one of the major causes of community-acquired diseases and also contributes most to the fatality rate with a high number of deaths in the country.
Disease types
Pneumonia is a condition in which infection and inflammation are seen in the lower respiratory tract due to bacterial invasions. Pneumonia that occurs outside the hospital is called community-acquired pneumonia (CAP). Contaminated hospital settings being a reason for pneumonia occurrence in a patient, when admitted to the hospital are called Nosocomial or Hospital Acquired Pneumonia (HAP) (9). If pneumonia arises in a patient receiving endotracheal intubation for more than 48-72 hours then it is called Ventilator-associated pneumonia (VAP)(10).
Pneumonia diagnosis
For the detection of pneumonia routine laboratory evaluation methods like microscopy, respiratory tract specimen’s cultures, blood cultures, antigens detection in urine and respiratory specimens, and also detection of antibodys in blood serum has been employed. Detection of Nucleic acid using polymerase chain reaction (PCR) have been established for the detection of biomarkers in the sample(11).
According to the Clinical and Laboratory Standards Institute guidelines clinically relevant bacterial isolates were subjected to antimicrobial susceptibility testing. Culturing of blood, sputum, pleural fluid, lung aspirates, and postmortem tissues for bacterial culture were done using standard microbiological methods. Urine, pleural fluid, Nasopharyngeal, and Oropharyngeal specimens are used for antigen detection. Antibodies are detected by serum samples (11,12).
Commonly, pneumonia pathogens will colonize in the upper respiratory tracts of healthy individuals. Only their presence in the patients doesn’t mean that is the cause of pneumonia. So, distinguishing infection from colonization from is the major challenge even with recent advanced technologies and various diagnostic methods. The interpretation of diagnosed test results for pneumonia detection remains the expert’s act (11).
The depiction of Klebsiella
The genus Klebsiella belonging to the family Enterobacteriaceae is a gram-negative, non-motile, rod-shaped bacteria with the dimensions of 0.3-1 µm in diameter and 0.6-6 µm in length, surrounded by a capsule(4). Genus Klebsiella is classified into five species, namely K. pneumoniae, K. oxytoca, K. terrigena, K. planticola. The species K. pneumoniae comprises three subspecies, K. pneumoniae subsp. pneumoniae, K.pneumoniae subsp. Ozaenae and K .pneumoniae subsp. rhinoscleromatis(13,14). Klebsiella grows readily on nutrient agar, tryptic casein soy agar, blood agar, and bromocresol purple lactose agar(3).
pneumonia and K. oxytoca colonies are dome-shaped, mucoid and having stickiness with 3-4mm diameter with an overnight incubation at 300 C / 370 C. K. terrigena and K. planticola are also have dome-shaped, less mucoid colonies with 1.5-2.5mm diameter. K. pneumoniae subsp. pneumoniae, K. Ozaenae, and K. Rhinoscleromatis will show voluminous, mucoid, and also rounded, translucent, and confluent colonies at 300 C / 370 C when it is kept for 48h (3).
pneumoniae is facultative anaerobe with both respiratory and fermentative type of metabolism. They grow on meat extract medium, producing more or less dome-shaped and glistening colonies with stickiness. They show a positive test for lactose, catalase, and Voges-Proskauer (VP) and negative test for methyl red and oxidase tests. As a sole carbon source, K. pneumoniae strains can utilize citrate and glucose but cannot utilize L-Sorbose. For most of the strains when glucose is supplied to a media, fermentation is done with the production of acid, gas, and 2, 3 butanediols as an end product (3,14). K. pneumoniae is alsinvolved in nitrogen fixation (15). Strains can be freeze-dried or cultured in broth medium with 10-50% (V/V) glycerol at -800C. At room temperature, they can be stored in semi-solid meat extract medium(3). Usually, the identification and differentiation of Klebsiella species are done based on the results of their biochemical tests.
Individuals with a weakened immune system are always prone to nosocomial infection which is caused by an opportunistic pathogen, Klebsiella pneumoniae. K. pneumoniae possesses around 78 capsular serotypes (K antigen), some of which constitute hyper-virulence due to hyper-secretion of capsule polysaccharide forming hyper-mucoviscous phenotype which is the most virulent form of K. pneumoniae known. Hyper-mucoviscous strains can invade multiple organs post-infection leading to multiple organ failures. Apart from these hyper-virulent forms of K. pneumoniae causing pneumonia in healthy individuals, they also cause life-threatening community-acquired infections, such as pyogenic liver abscess, meningitis, necrotizing fasciitis, endophthalmitis and severe pneumonia(16).
Virulence Factors of Klebsiella
Klebsiella pneumoniae has to overcome the innate and humoral immunity (17) of host defense systems to enter and colonize inside the host. The following are the virulence factors possed by the Klebsiella pneumoniae to spread pathogenesis.
Adherence
Klebsiella pneumoniae has two types of fimbriae type 1 and type3. Type 1 fimbriae, a virulence protein responsible for urinary tract infections in K. pneumonia. Type 1 mediate the adhesion to the mannose containing structures on host cell and in the extracellular matrix of the host. Type 3 fimbriae play a major role in biofilm formation. (18)
Biofilm Formation
pneumoniae possess a complex, un-deciphered signaling mechanism which, when present in large bio-films, the entire mass of bacteria present in the biofilm acts as one entity and has a comprehensive and virulent mechanism to infect the host and shield itself from various antibacterial agents which is a predominant phenomenon seen in drug resistance conditions.
Biofilm formation on medical devices is one of the sources of nosocomial infections that are difficult to treat due to acquired bacterial resistance. K. pneumoniae clinical isolates express type 3 fimbriae, they are thin, non-channeled belongs to chaperone- usher class of fimbriae (19). Type 3 fimbriae, a virulence factor in K. pneumoniae mediates the biofilm formation by adhering to the host structures and also plays an important role in infections associated with biofilm formation. Type 3 fimbriae are encoded by Mrk gene clusters, which contain genes like MrkA (Major fimbrial subunit), MrkB (Chaperone), MrkC (Usher), MrkD (Adhesin), MrkF (Minor fimbrial subunit) (19). Kpc is a type of fimbriae that possess the biofilm forming activity .It is encoded by kpcISABCDEFG operon like type 1 and type 3 fimbriae.
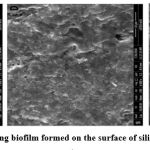 |
Figure 1: SEM images of strong biofilm formed on the surface of silicon coated latex catheter (20). |
Efflux pump
Efflux pumps are the channels developed by the bacterial system to eliminate antibiotics, dyes, detergents, and help to gain bacterial resistance. Certain strains of K. pneumoniae are composed of the AcrAB multidrug-resistant efflux system coded by acrRAB operon (21).
Immune evasion
The capsular polysaccharide is an acidic sugar polymers layer that encapsulates K. pneumoniae and plays an important role in virulence by giving protection against macrophage phagocytosis and complement-mediated killing (22,23). Capsules produced by the Klebsiella pneumoniae strains is of the serotypes K1 to K78. Capsules made up of K1 and K2 serotypes exhibit higher pathogenicity(17).
Iron Acquisition
Iron is the requirement during infection for Klebsiella pneumoniae, which is not readily available from the host. In the host, iron is found in the bound form to the transporter protein called transferrin. To get iron from the host, bacteria have to synthesis the siderophores, a molecule that has more affinity than the host transferrin. K. pneumoniae possesses different siderophores like enterobactin, yersiniabactin, salmochelin, and aerobactin (17).
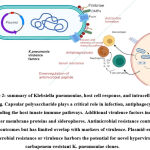 |
Figure 2: summary of Klebsiella pneumoniae, host cell response, and intracellular signaling. |
Table 1: Genes responsible for the Virulence factors of K. pneumoniae.
| Virulence factor | Virulence gene | Reference |
| Hypermucoviscous phenotype and mucoviscosity related genes | rmpA, rmpA2, allS, wabG
|
(24)
|
| Biosynthesis of lipopolysaccharide
|
Uge, wcaG
|
(25) |
| Iron uptake and transport | iutA,icuA,iroN,iroB,ybtA,irp2,kfu, entB | (25) (24) |
| Adhesion | Cf29a,fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, fimK | (26) (25) (24) |
| Efflux pump (AcrAB) | acrA; acrB | (17) |
| Biofilm formation (Type 3 fimbriae) | mrkA, mrkB, mrkC, mrkD, mrkF, mrkH, mrkI, mrkJ | (17) |
| Serum resistance | glf; kfoC; wbbM; wbbN; wbbO; wzm; wzt; uge; wabG | (17) |
Use of Antibiotics
Antibiotics are either chemically synthesized or antimicrobial compounds that are bactericidal or bacteriostatic with a different mode of action and target sites. Commonly used antibiotics against K. pneumoniae are a class of Penicillins, Cephalosporins, Monobactams, and Carbapenems which have the β-lactam ring in common with variable side chains R(27).
Antibiotics are commonly used to treat bacterial respiratory infections, but they can exacerbate inflammation by releasing microbial components that overstimulate the immune system, leading to greater tissue damage(28). cAMP (3′-5′ cyclic adenosine monophosphate) controls a variety of biological activities, including inflammation. Proinflammatory cytokines and chemokines, as well as reactive oxygen species, are reduced by cAMP-elevating drugs such as phosphodiesterase (PDE) 4 inhibitors. Furthermore, inhibiting PDE4 reduces tissue damage , and it has been suggested that this class of medicines be used in conjunction with antibiotics to treat pneumonia(29).Combined treatment of rolipram and antibiotic is said to activate AnX 1 that reduce inflammation.
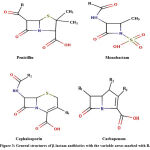 |
Figure 3: General structures of β-lactam antibiotics with the variable areas marked with R. |
Mechanism of actions of Beta-Lactam Antibiotics:
The strength and shape of the bacteria are based on the cell wall structures formed by the polymerization of glycans (30). The β-lactam antibiotics act on penicillin-binding proteins (PBPs) like D-Ala-D-Ala-carboxy peptidase/transpeptidase (DD-peptidase), which are essential enzymes in the bacterial cell wall biosynthesis by cross-linking peptidoglycans. β-lactam antibiotics interception with peptidoglycan cross-linking, will weaken the cell integrity and eventually results in the death of a bacterial cell due to lysis (30).
Beta-lactams like penicillins, cephalosporins, carbapenems, monobactams inhibits bacterial cell wall synthesis. Tetracyclines, Aminoglycosides, Oxazolidinones (linezolid), Streptogramins (quinupristin-dalfopristin). Ketolides, Macrolides, Lincosamides acts by inhibiting bacterial protein synthesis. Fluoroquinolones inhibit bacterial DNA synthesis, Rifampicin acts by inhibiting RNA synthesis. Polymyxin acts by (Polymyxin-B, Colistin) disorganizing membrane agents. Competitive inhibition of folic acid synthesis is done by Sulfonamides, Trimethoprim (31,32).
The gain of antibiotic resistance
The resistance is the escape mechanism employed by the K. pneumoniae to survive within the host against the complement-mediated killing(5). Resistance shown by the K. pneumoniae against commercially available antibiotics due to overuse and misuse is the main reason behind the increase of pneumonia incidence every year. The increase of resistance to antimicrobial drugs leads to an increase in illness and mortality(33).
Generally, the discovery of antibiotics has modernized the field of treatment and medicine, through which many lives have been saved(34). Antibiotic resistance can be natural or acquired and can be transmitted. The Natural form of antibiotic resistance is caused when bacteria undergo a spontaneous gene mutation in the presence of antibiotics due to a lack of selective pressure. Natural resistance is less common but can play a role in the development of resistance. Development of acquired resistance is seen when antibiotics are used against a heterogeneous group of bacteria’s in a colony, the susceptible bacteria will die and the resistant strain will survive. These surviving bacteria carry a genetic determinant that codifies a gene that expresses a resistance against these antibiotics(31,32).
Resistance can be through numerous biochemical and/or physiological mechanisms. Treatment options for Klebsiella pneumoniae infections are difficult as they possess intrinsic resistance to several classes of antibiotics(5). When the strains of K. pneumoniae having resistant genes against antibiotics undergoes a division, eventually the resistances spread (25). Extended-Spectrum Beta-Lactamases (ESBLs) and carbapenemases producing strains of K. pneumoniae have spread globally and are the reason for the spread of resistance(5).
Table 2: Antimicrobial agents and the resistant genes.
| Antimicrobial Class | Resistant Genes |
| b-Lactamases (bla genes) ESBLs Carbapenemases | CTX-M, SHV, TEM, VEB, OXA, GES, OXA, NDM, CARB Others CphA, AmpC, CMY, DHA, FOX, MIR, VIM, SIM, IMP, SCO, PSE |
| Aminoglycosides | aac, aadAB, aph, armA, ant |
| Folate pathway inhibitors | sul1, sul2, sul3, dfrAB |
| Polymyxins | mcr-1, mcr-3, mcr-8 |
Beta-Lactamases
Klebsiella pneumoniae carbapenemases (KPCs) are β-lactamases which was first identified in 1996 in the USA(35). These β-lactamases are the class of bacterial enzymes, act by hydrolyzing the β-lactam rings present in the penicillin, cephalosporin, and other β-lactam antibiotics (36,37). The structure formed by the hydrolysis of the β-lactam rings will not be suitable for the antibiotics to bind to the active sites of Penicillin Binding Proteins (PBP). This is how the action of β-lactamases makes the drug ineffective and hence bacterial resistance is achieved.
 |
Figure 4: Hydrolysis of the β-lactam ring by the action of β-lactamases(38). |
β-lactamases are differentiated based on a range of antibiotic action and type of amino acids present at their active sites. Based on the range of antibiotics activity, β-lactamases are classified into Narrow-spectrum (Penicillinase), Broadspectrum (Amicillinases), Extended-spectrum β-lactamases (ESBls) and Carbapenemases (38,39). Based on the structural homology of amino acids, the Ambler classification scheme separates β-lactamases into four major classes (A-D). Class A, C, and D have serine at their active site, however class B (also known as Metallo- β-lactamases) has zinc amino acid at their active site (40–43).
Acknowledgment
The authors PH, BS, CD, and CS acknowledge the support and infrastructure provided by JSS AHER. SPK thank Amrita Vishwa Vidyapeetham for support extended towards the collaborations.
Conflict of Interest
The authors declare no conflict of interest.
Funding Sources
There was no funding support utilized to carry out the current research.
References
- Shon AS, Bajwa RPS, Russo TA. Hypervirulent ( hypermucoviscous ) Klebsiella pneumoniae A new and dangerous breed. 2013;107–18.
CrossRef - Sikarwar AS, Batra HV. Prevalence of Antimicrobial Drug Resistance of Klebsiella pneumoniae in India. 2011;1(3):211–5.
CrossRef - Brisse S, Grimont F, Grimont PAD. The Genus Klebsiella. The Prokaryotes. 2006;159–96.
CrossRef - Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998 Oct;11(4):589–603.
CrossRef - Doorduijn DJ, Rooijakkers SHM, van Schaik W, Bardoel BW. Complement resistance mechanisms of Klebsiella pneumoniae. Immunobiology. 2016 Oct;221(10):1102–9.
CrossRef - Damian M, Usein C, Palade A, Ceciu S, Cosman M. Molecular Epidemiology and Virulence Characteristics of Klebsiella pneumoniae Strains Isolated from Hospital-Associated Infections. Open Epidemiol J. 2009;2:69–78.
CrossRef - Ko W, Paterson DL, Sagnimeni AJ, Hansen DS, Gottberg A Von, Mohapatra S, et al. Community-Acquired Klebsiella pneumoniae Bacteremia : Global Differences in Clinical Patterns. 2002;8(2):160–6.
CrossRef - Jarvis WR, Munn VP, Highsmith AK, Culver DH, Hughes JM. The Epidemiology of Nosocomial Infections Caused by Klebsiella pneumoniae. Infect Control. 1985;6(2):68–74.
CrossRef - J.Bruyere H. Bacterial pneumonia. In: 100 Case Studies in Pathophysiology. 1st ed. Lippincott Williams & Wilkins; 2008. p. 1–11.
- American Thoracic Society IDS of A. Guidelines for the Management of Adults with Hospital-acquired, Ventilator-associated, and Healthcare-associated Pneumonia. Am J Respir Crit Care Med. 2005;171:388–416.
CrossRef - Driscoll AJ, Karron RA, Morpeth SC, Bhat N, Levine OS, Baggett HC, et al. Standardization of laboratory methods for the PERCH study. Clin Infect Dis. 2017;64(Suppl 3):S245–52.
CrossRef - Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing ; Twenty-First Informational Supplement. Vol. 31. 2011.
- Drancourt M, Bollet C, Carta A, Rousselier P. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int J Syst Evol Microbiol. 2001;51(3):925–32.
CrossRef - Carter JS, Bowden FJ, Bastian I, Myers GM, Sriprakash KS, Kemp DJ. Phylogenetic evidence for reclassification of Calymmatobacterium granulomatis as Klebsiella granulomatis comb. nov. Int J Syst Bacteriol. 1999;49:1695–700.
CrossRef - Ladha JK, Barraquio WL, Watanabe I. Isolation and identification of nitrogen-fixing Enterobacter cloacae and Klebsiella planticola associated with rice plants. Can J Microbiol. 1983 Oct;29(10):1301–8.
CrossRef - Li B, Zhao Y, Liu C, Chen Z, Zhou D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014;9(9):1071–81.
CrossRef - Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev. 2016;80(3):629–61.
CrossRef - Struve C, Bojer M, Krogfelt KA. Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect Immun. 2008;76(9):4055–65.
CrossRef - Struve C, Bojer M, Krogfelt KA. Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect Immun. 2009;77(11):5016–24.
CrossRef - Desai S, Sanghrajka K, Gajjar D. High adhesion and increased cell death contribute to strong biofilm formation in Klebsiella pneumoniae. Pathogens. 2019;8(4):277.
CrossRef - Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother. 2010;54(1):177–83.
CrossRef - Lawlor MS, Handley SA, Miller VL. Comparison of the host responses to wild-type and cpsB mutant Klebsiella pneumoniae infections. Infect Immun. 2006;74(9):5402–7.
CrossRef - Lawlor MS, Hsu J, Rick PD, Miller VL. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol. 2005;58(4):1054–73.
CrossRef - Liu Z, Gu Y, Li X, Liu Y, Ye Y, Guan S, et al. Identification and Characterization of NDM-1-producing Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae in China. Ann Lab Med [Internet]. 2019 Mar;39(2):167–75. Available from: https://pubmed.ncbi.nlm.nih.gov/30430779
CrossRef - Kim D, Park BY, Choi MH, Yoon E-J, Lee H, Lee KJ, et al. Antimicrobial resistance and virulence factors of Klebsiella pneumoniae affecting 30 day mortality in patients with bloodstream infection. J Antimicrob Chemother. 2019 Jan;74(1):190–9.
CrossRef - Huang Y-H, Chou S-H, Liang S-W, Ni C-E, Lin Y-T, Huang Y-W, et al. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother. 2018 Aug;73(8):2039–46.
CrossRef - Holten KB, Onusko EM, College C. Appropriate Prescribing of Oral Beta Lactam Antibiotics. 2000;
- Steel HC, Cockeran R, Anderson R, Feldman C. Overview of community-acquired pneumonia and the role of inflammatory mechanisms in the immunopathogenesis of severe pneumococcal disease. Mediators Inflamm. 2013;2013.
CrossRef - Garcia CC, Russo RC, Guabiraba R, Fagundes CT, Polidoro RB, Tavares LP, et al. Platelet-activating factor receptor plays a role in lung injury and death caused by Influenza A in mice. PLoS Pathog. 2010;6(11):e1001171.
CrossRef - Mcdonough M a, Lee W, Silvaggi NR, Kotra LP, Takeda Y, Kelly J a. Structure of a Penicillin Binding Protein Complexed with a Cephalosporin-Peptidoglycan Mimic. NSLS Act Rep. 2001;36–8.
- Alanis AJ. Resistance to antibiotics: Are we in the post-antibiotic era? Arch Med Res. 2005;36(6):697–705.
CrossRef - Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. NatMed. 2004;10:S122–9.
CrossRef - Sanchez G V., Master RN, Clark RB, Fyyaz M, Duvvuri P, Ekta G, et al. Klebsiella pneumoniae Antimicrobial Drug Resistance ,. Emerg Infect Dis. 2013;19(1):133–6.
CrossRef - Davies J, Davies D. Origins and Evolution of Antibiotic Resistance. Microbiol Mol Biol Rev. 2010;74(3):417–33.
CrossRef - Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, et al. Clinical epidemiology of the global expansion of kpn carbapenemases. Lancet Infect Dis. 2013;13(9):785–96.
CrossRef - Medeiros AA. Beta- Lactamases. Br Med Bull. 1984;40(1):18–27.
CrossRef - Lomaestro BM, Tobin EH, Shang W, Gootz T. The Spread of Klebsiella pneumoniae Carbapenemase–Producing K. pneumoniae to Upstate New York . Clin Infect Dis. 2006;43(3):e26–8.
CrossRef - Alibi S, Ferjani A, Boukadida J. Molecular characterization of extended spectrum beta-lactamases produced by Klebsiella pneumoniae clinical strains from a Tunisian hospital. Med Mal Infect. 2015;45(4):139–43.
CrossRef - Pessoa-Silva CL, Meurer Moreira B, Câmara Almeida V, Flannery B, Almeida Lins MC, Mello Sampaio JL, et al. Extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a neonatal intensive care unit: Risk factors for infection and colonization. J Hosp Infect. 2003;53(3):198–206.
CrossRef - Hirsch EB, Tam VH. Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): An emerging cause of multidrug-resistant infection. J Antimicrob Chemother. 2010;65(6):1119–25.
CrossRef - Paterson DL, Bonomo RA. Extended-Spectrum Beta Lactamases : a Clinical Update. Clin Microbiol Rev. 2005;18(4):657–86.
CrossRef - Queenan AM, Bush K. Carbapenemases: The versatile β-lactamases. Clin Microbiol Rev. 2007;20(3):440–58.
CrossRef - Paterson DL. Resistance in Gram-Negative Bacteria: Enterobacteriaceae. Am J Med. 2006;119(6 SUPPL. 1):20–8.
CrossRef








