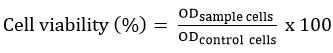Manuscript accepted on :13-05-2022
Published online on: 09-06-2022
Plagiarism Check: Yes
Reviewed by: Dr. Deepak Pharmacy
Second Review by: Dr. Aditya Ganeshpurkar
Final Approval by: Dr. H Fai Poon
Saraswati Ramadhani Priyono , Sutriyo*
, Sutriyo* and Ratika Rahmasari
and Ratika Rahmasari
Faculty of Pharmacy, Universitas Indonesia, Depok, Indonesia, 16424.
Corresponding Author E-mail: sutriyo@farmasi.ui.ac.id
DOI : https://dx.doi.org/10.13005/bpj/2410
Abstract
Covid-19 was mainly treated by a broad-spectrum antiviral called Remdesivir. A truncated cone molecular structure of Hydroxypropyl-β-cyclodextrin can enhance the solubility and cellular uptake of the poorly soluble drug's through biological membranes. This study aimed to synthesize, characterize, observe cellular uptake and evaluate the cytotoxicity of remdesivir-hydroxypropyl-β-cyclodextrin (RDV-HPβCD) inclusion complex. The RDV-HPβCD inclusion complex was synthesized by the solvent evaporation method. Furthermore, the inclusion complex characteristic was evaluated by ultraviolet-visible (UV-Vis) spectrophotometry; particle size analyzer (PSA); Fourier infrared spectrophotometry (FTIR); X-ray diffraction (XRD); and differential scanning calorimetry (DSC). Further, fluorescence microscopy was used to evaluate the cellular uptake and 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used in the cytotoxicity study. In the UV-Vis spectrum, both the inclusion complex and pure remdesivir showed a maximum peak at 246 nm. The inclusion complex has a particle size of 1697 ± 738.02 nm with -22.4 ± 1.58 mV of zeta potential. Shifted FTIR spectrum, broad XRD peak, and broad DSC thermogram peak at 72.93 °C indicated the successful formation of the RDV-HPβCD inclusion complex. Furthermore, cellular uptake observation of RDV-HPβCD inclusion complex conjugated to FITC showed better intensity inside the Vero cell than pure remdesivir conjugated to FITC. Further, Inclusion complex showed higher cell viability than pure remdesivir at a certain concentration.
Keywords
Cellular Uptake; Cytotoxicity; Hydroxypropyl-β-Cyclodextrin; Inclusion Complex; MTT assay; Remdesivir
Download this article as:| Copy the following to cite this article: Priyono S. R, Sutriyo S, Rahmasari R. Preparation, Cellular Uptake, and Cytotoxic Evaluation of Remdesivir-Hydroxypropyl-β-Cyclodextrin Inclusion Complex. Biomed Pharmacol J 2022;15(2). |
| Copy the following to cite this URL: Priyono S. R, Sutriyo S, Rahmasari R. Preparation, Cellular Uptake, and Cytotoxic Evaluation of Remdesivir-Hydroxypropyl-β-Cyclodextrin Inclusion Complex. Biomed Pharmacol J 2022;15(2). Available from: https://bit.ly/3zrL7id |
Introduction
Broad-spectrum antiviral called remdesivir was mainly used to treated Ebola virus infection1. An active nucleoside triphosphate derivative was processed from monophosphate derivatives that previously were a prodrug called remdesivir which metabolized in the cell to become an alanine metabolites2. SARS, MERS, and SARS-CoV-2 was RdRp coronaviruses whose viral RNA synthesis can be inhibited by a specific mechanism, which is in delaying chain termination by triphosphate form of remdesivir3. Hospitalized COVID-19 patients, both adult and pediatric are treated with remdesivir, which has been granted permission for use by the Food and Drug Administration (FDA)4.
From commercial preparation, remdesivir was known to be insoluble in water, in the form of a lyophilized powder injection dosage form at a dose of 100 mg per vial, it is necessary to add sulfobutylether-β-cyclodextrin (SBECD) as a solubility enhancer with complex formation5. The presence of SBECD in remdesivir preparations may result in the accumulation of SBECD sodium salts so it is not recommended for patients with a glomerular filtration rate (GFR) of less than 30 mL/min5,6. When administered intravenously, remdesivir will distribute to tissues and blood cells through passive diffusion7. Remdesivir is a nucleotide analog that has poor cell permeability, when it is inside the cell it must undergo several phosphorylation processes to become the active form of the nucleoside triphosphate.
High polarity and was not diffuse back into the cell membrane or be trapped inside the cell was the characteristic of monophosphate derivative of remdesivir. While nucleoside triphosphate form which has a half-life of 14 to 24 hours was trapped in the cell because it has a negative charge2,7. Antiviral activity of remdesivir may be achieved when nucleoside triphosphate accumulates in cells at high amounts.
Hydroxypropyl-β-cyclodextrin (HPβCD) has a very good solubility of 600 mg/mL, low price, and low toxicity8. Enhancing drug solubility, dissolution rate, bioavailability, and; decreasing side effects, and also contributing to stabilizing the pharmaceutical formulations were the advantages of HPβCD9. HPβCD was a cyclodextrin derivative that has a structure that looks like a truncated cone-like which consists of hydrophilic molecules on the outer surface and lipophilic molecules on the inner cavity. This structure makes it possible to carry drug molecules in the cavity resulting in inclusion complex10. The drug/HPβCD complex formed can enhance drug solubility in water, improve chemical and physical stability, and improve drug delivery through biological membranes11. In addition, the solubility increase was caused by the increased frequency of partitioning into the plasma membrane due to the formation of inclusion complexes12.
This study aimed to obtain the RDV-HPβCD inclusion complex which has better solubility, cellular uptake than pure remdesivir. The RDV-HPβCD inclusion complex formed will be characterized by performing UV-Vis spectroscopic analysis, particle size, PDI, zeta potential, chemical structure, and functional groups using FTIR, XRD, and DSC. Then, the cellular uptake evaluation of the Remdesivir-HPβCD inclusion complex was carried out in Vero cells which were analyzed using fluorescence microscope and determine resulting cytotoxic activity against HeLa cancer cells.
Materials and Methods
Reagents and instruments
Remdesivir (Glentham Life Sciences), Hydroxypropyl-β-cyclodextrin (Shaanxi Ciyuan Biotech Co., Ltd), Dimethyl sulfoxide (Merck, Germany), Vero (ATCC® CCL-81™) normal kidney epithelial cell line from African green monkey, Dulbecco’s modified eagle medium (DMEM) that enriched by 10% fetal bovine serum (FBS), 1% penicillin-streptomycin; HeLa (ATCC® CCL-2™) from human cervical cancer cell line, fluorescein isothiocyanate (FITC), [3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] (MTT) 0,5%, Dulbecco’s phosphate buffered saline (DPBS), phosphate-buffered saline (PBS).
Methods
Synthesis of the inclusion complex
The RDV-HPβCD inclusion complex was prepared with a molar ratio of Remdesivir: HPβCD (1:1) by the solvent evaporation method 11,13. To prepare the remdesivir solution the remdesivir powder was dissolved into the water for injection that contains 10% DMSO. HPβCD solution was prepared by dissolving the HPβCD powder in water for injection. Then each solution was stirred slowly with a spin bar on a magneti stirrer until it dissolved. Remdesivir solution was added to the HPβCD solution little by little and the mixture was stirred by a magnetic stirrer at 250 rpm for 10 minutes. After that, to collect inclusion complex powder the mixture was evaporated. The inclusion complex then was stored at 4°C and kept away from light.
UV-Vis absorption analysis
The UV-Vis absorption spectrum of pure redeliver, HPβCD, and Remdesivir-HPβCD inclusion complex in water for injection solution was acquired using Shimadzu UV-1800 UV-Vis spectrophotometer (Japan) at a wavelength between 200 to 400 nm.
Particle size, PDI, and zeta potential measurement
1 mL of pure remdesivir, HPβCD, and inclusion complex RDV-HPβCD was prepared to see the particle size, PDI, and zeta potential at 25°C using NANO-ZS series from Malvern Zetasizer (UK).
FTIR analysis
The infrared spectrum of pure remdesivir, HPβCD, and inclusion complex RDV-HPβCD was recorded using FTIR spectrophotometer 8400 (Shimadzu, Japan). FT-IR spectrum was obtained at 4000-400 cm-1.
X-ray Diffraction (XRD) analysis
X-ray diffraction patterns of pure remdesivir, HPβCD, and inclusion complex RDV-HPβCD were obtained by an X-ray diffractometer with CuKα radiation. The voltage set of 30 mA, a current 30 kV and 2° min-1 of the scan rate with 2θ or diffraction angle in the range of 5-50°. The data was processed using OriginLab.
Differential Scanning Calorimetry (DSC) analysis
DSC thermogram of pure remdesivir, HPβCD, and inclusion complex RDV-HPβCD was performed using Shimadzu Differential Scanning Calorimetry type 60 (Japan) under N2 stream that heated from 25 °C to 450 °C with a heating rate of 10 °C min-1.
Cellular uptake study
RDV-HPβC-Fluorescein isothiocyanate (RDV-HPβC-FITC) and Remdesivir-Fluorescein isothiocyanate (FITC) were prepared (1:1) by dissolving 1 mL of FITC (10 g/mL) in 1 mL of RDV-HPβC-FITC and pure remdesivir solutions. These mixtures were incubated overnight in the dark at 4°C.14 The Vero cells were cultured in 500 µl of DMEM as a growth medium with 104 density per well in an 8-well slide chamber, then incubated under 5% CO2 stream at 37°C for 24 hours. The sample (RDV-HPβC-FITC, Remdesivir-FITC, and FITC) was prepared according to the desired concentration. The cell medium was discarded, then rinsed with phosphate-buffered saline (PBS) once and sterile PBS three times. After that, 4% formaldehyde was added and incubated at 25°C for an hour. Formaldehyde was taken out and the slide chamber was washout with PBS once and sterile PBS three times. Sample + FITC was added to the cells as much as 500 µl and incubated under 5% CO2 stream at 37°C. The slide chamber was washout with sterile PBS three times. The slide chamber was dried and observed under a fluorescence microscope.
Cytotoxicity study
Using MTT assay the cytotoxicity of RDV-HPβCD inclusion complex against HeLa cells (human cervical cancer cell lines) was obtained. In 96-wells HeLa cells were seeded (5 x 103), then incubated under 5% CO2 stream at 37°C, overnight. The DMEM medium was discarded, and with DPBS the cell were washed. Then, RDV-HPβCD inclusion complex and pure remdesivir were added to cells that has reached 50% confluency, triplicate, after that for additional 24 hours the cells were incubated. After the medium was disposed of, then the cells were washout with PBS once, and to each well 10 μL MTT reagent was added, including the control medium (without cells). Until formazan crystal was formed, the cells were incubated in an incubator for 4 hours under a 5% CO2 stream with the temperature at 37°C. To the formazan crystals, the sodium dodecyl sulfate was added which was a stop solution, then incubated overnight in a dark place at 25°C and covered with aluminum foil. When the wrapping was opened, the plate was inserted into the ELISA reader and at 595 nm the absorbance was read for each sample.15–17 The percentage of cell viability resulting from the cytotoxic test according to the following formula:
where ODsample cells were the optical density of cells treated with pure remdesivir or RDV-HPβCD inclusion complex and ODcontrol cells were the optical density of untreated cells.
Result and Discussion
Synthesis of the inclusion complex
The stability of remdesivir was affected by its poor solubility in water. To overcome this problem, an inclusion complex between remdesivir and HPβCD was formed by solvent evaporation method with 1:1 molar ratio11,18. The 1:1 ratio molar was chosen because of the success of previous studies with the hope that every drug molecule, namely remdesivir, enters the HPβCD11.
The physical stability of the RDV-HPβCD inclusion complex was evaluated for 24 hours at 25°C. The inclusion complex was observed in clear-stated conditions (figure 1C dan 1D), Figure 1(D) explained that the inclusion complex can increase the solubility and maintain the dispersion of remdesivir. The poor solubility of remdesivir in water affects its stability. However, at pure remdesivir solution, remdesivir powder became settled at the bottom of the solution after 24 hours as shown in figure 1B19.
 |
Figure 1: (A) Remdesivir solution 0 hours, (B) Remdesivir solution after 24 hours, (C) Inclusion complex solution 0 hours, and (D) Inclusion complex solution after 24 hours. |
Particle size, Polydispersity Index (PDI), and Zeta potential measurement
The particle size, PDI, and zeta potential of pure remdesivir, pure HPβCD, and the RDV-HPβCD inclusion complex was measured using a Malvern Particle Size Analyzer and the result was shown in table 1, respectively.
It was known that pure remdesivir, pure HPβCD, and the RDV-HPβCD inclusion complex each have the average particle size of 1191 ± 189.95 nm, 650.5 ± 123.81 nm, and 1697 ± 738.02 nm (n = 3). For the polydispersity index (PDI) value of remdesivir, HPβCD and RDV-HPβCD inclusion complex were 0.850 ± 0.28, 0.632 ± 0.09, and 0.028 ± 0.08, respectively. PDI is an index that calculates the heterogeneous particle size of substances in an aqueous solution. The smaller PDI value, the lower possibility of the complex forming aggregates 8,14. Since the obtained PDI value of the RDV-HPβCD inclusion complex was smaller than 0.5, this indicated that the particle size of the inclusion complex was distributed homogeneously but not with pure remdesivir and HPβCD16. Meanwhile, the negative zeta potential values of remdesivir (-20±2.08 mv), HPβCD (-29,1 ± 3,66), and RDV-HPβCD inclusion complexes (-22,4 ± 1,58) were observed. The negative value may result from the presence of hydroxyl (OH¯), phosphate (PO3¯) and cyano (C≡N) groups from remdesivir and hydroxyl (OH¯) group of HPβCD.
Table 1: The particle size, PDI, and zeta potential value of pure remdesivir, HPβCD, and Remdesivir-HPβCD inclusion complexes.
| Particle size ± SD (nm) | PDI* ± SD | Zeta Potential ± SD (mV) | |
| Remdesivir | 1191 ± 189.95 | 0.850 ± 0.28 | -20.0 ± 2.08 |
| HPβCD | 650.5 ± 123.81 | 0.632 ± 0.09 | -29.1 ± 3.66 |
| Remdesivir-HPβCD Inclusion Complex | 1697 ± 738.02 | 0.028 ± 0.08 | -22.4 ± 1.58 |
Each experiment were performed in triplicate. *PDI means polydispersity index
UV-Vis Analysis
Figure 2 presents the UV-Vis absorption spectrum of pure remdesivir, pure HPβCD, and the RDV-HPβCD inclusion complex at 220-290 nm wavelength. Pure HPβCD did not show an absorption peak because HPβCD was a molecule without chromophores20. According to the existing literature, remdesivir shows the highest absorption peak at a wavelength of 246 nm5.
After remdesivir binds to HPβCD, the absorption peak of remdesivir which has formed an inclusion complex is still visible at the same wavelength as pure remdesivir, which is at a wavelength of 246 nm. This corresponds to the formation of hydrogen bonds, where a bond is formed between the two molecules, resulting in a decrease in the electron density of remdesivir and a change in the electron transition level21. Therefore, it is presumed that the RDV-HPβCD inclusion complex has been formed.
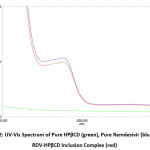 |
Figure 2: UV-Vis Spectrum of Pure HPβCD (green), Pure Remdesivir (blue), and RDV-HPβCD Inclusion Complex (red). |
FTIR Analysis
FTIR was a technique that can be used to ensure that an inclusion complex between the active substance and the cyclodextrin that has been prepared has been formed. The infrared spectrum can be used to predict what kind of bond is formed from the strain vibration band of the pure compound and the cyclodextrin changes in the complexation mechanism. The presence of changes, widening, loss of peak intensity, and displacement can indicate the formation of inclusion complexes. This can occur due to a reduction in the stretching vibration of the active substance molecules that have moved into the cyclodextrin cavity or it can be said that there has been a weak interatomic bond 10. Figure 3 shows the infrared spectrum of pure remdesivir, pure HPβCD, and RDV-HPβCD inclusion complex.
The infrared spectrum of pure remdesivir (Figure 3, blue line) showed the presence of nitrile group (C≡N) in the range 2260-2240 cm-1 and the absorption peak was seen at 2260.65 cm-1. In the range of 2000-1650 cm-1 there was C-H of the aromatic group which shows a peak at 1859.44 cm-1. In the range 1690-1640 cm-1 there was C=N vibrations which shows a peak at 1662.69 cm-1. In the range 1680-1600 cm-1 there was C=C vibrations which shows a peak at 1604.83 cm-1. In the range of 1350-1000 cm-1 there were C-N vibrations, which showed a peak at 1010.73 cm-1 22.
The pure HPβCD infrared spectrum (Figure 3, black line) shows a wide band in the range of 3600-3200 cm-1 which was the O-H of the hydroxyl group and shows an absorption peak at 3381.33 cm-1. There were C-H and CH2 vibrations in the range of 3100-2800 cm-1 which shows an absorption peak at 2928.04 cm-1. There was H-O-H vibrations in the range 1651-1646 cm-1, and the peak shows at 1647.26 cm-1 23. There was a C-O-C bond in the range 1200-1030 cm-1 which shows an absorption peak at 1201.69 cm-1 24. There was C-O vibration of the ether group in the range of 1150-1085 cm-1 and the absorption peak is at 1155.4 cm-1 23,25. There was a vibration at ± 855 which was an α-glycosidase bond that represents the glucose unit of HPβCD and shows an absorption peak at 856.42 cm-1 21,25,26.
Meanwhile, in the RDV-HPβCD inclusion complex infrared spectrum (figure 3, red line), absorption peaks are seen as similar to those of the pure HPβCD spectrum. There was a wide peak in the range of 3600-3200 cm-1 centered on 3381.33 cm-1 which is O-H. There were C-H and CH2 vibrations in the range of 3100-2800 cm-1 which shows a peak at 2926.11 cm-1. There was H-O-H in the range 1651-1646 cm-1 which shows a peak at 1645.33 cm-1. There was C-O-C in the range of 1200-1030 cm-1 which shows a peak at 1209.41 cm-1. C-O from the ether group shows a peak at 1153.47 cm-1. There was an α-glycosidase bond which is the unit of HPβCD at ± 855 cm-1 and shows a peak at 852.56 cm-1.
When the electron-rich cavity of the HPβCD is penetrated by the hydrophobic remdesivir, the increase in electron density was due to the increase in frequency. The stable inclusion complex was caused by hydrogen bonds and non-covalent forces associated with the shifted absorbance frequency between the inclusion complex and pure remdesivir. The success of remdesivir entering the HPβCD cavity to form an inclusion complex can be proved by FTIR analysis 19,21.
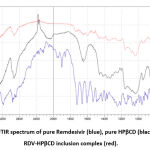 |
Figure 3: FTIR spectrum of pure Remdesivir (blue), pure HPβCD (black), and RDV-HPβCD inclusion complex (red). |
XRD Analysis
The inclusion complex formation can be confirmed by the XRD method. When the guest molecule has formed an inclusion complex by entering the cyclodextrin cavity, it is no longer in crystal form because the guest molecule is separated from each other by the cyclodextrin27.
From figure 4 the HPβCD XRD pattern did not show a clear peak or can be said to be flat, this is a characteristic of amorphous compounds because of the absence of crystallinity properties. Meanwhile, in the XRD pattern of remdesivir, 6 peaks were quite sharp, namely at (2Ø) = 5.37°, 8.45°, 12.61°, 14.45°, 16.05°, and 22.20°. sharp peaks indicate that remdesivir is pure crystalline13.
RDV-HPβCD inclusion complex and remdesivir XRD pattern were different because there was a lack of crystallinity of remdesivir. i.e. no sharp peak was seen so it could be said that the inclusion complex had no crystallinity. The lack of crystallinity can be used as a reference to presume that the inclusion complex has been formed, where the crystalline remdesivir molecule turns into an amorphous form or it can be said that remdesivir molecules are dispersed in the HPβCD cavity in an amorphous state13.
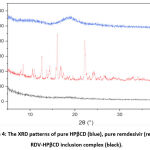 |
Figure 4: The XRD patterns of pure HPβCD (blue), pure remdesivir (red), and RDV-HPβCD inclusion complex (black). |
DSC Analysis
DSC was a useful method for determining the thermodynamic properties of inclusion complexes during the complexation process, such as crystallization characteristics, thermal stability, melting point, and others28–30. Figure 5 shows the DSC thermogram of hydroxypropyl-β-Cyclodextrin, remdesivir, and the RDV-HPβCD inclusion complex.
The DSC thermogram of HPβCD (figure 5.A) showed 2 peaks, the first endothermic peak (upwards) was at 274.29 °C and the second was the exothermic peak (downwards) at 389.56 °C. The DSC thermogram of the Remdesivir (figure 5.B) showed 2 exothermic peaks (downward) at 138.98 °C and 239.05 °C. The sharpness of the visible peak indicates that remdesivir is crystalline. The DSC thermogram of the RDV-HPβCD inclusion complex (figure 5.C) only showed one broad peak, at 72.93 °C, and no sharp peak was seen from remdesivir From that, we can conclude that remdesivir which was in the HPβCD cavity will no longer be seen or a shift of peak occurs and it can also be said that remdesivir which was in the inclusion complex was already in an amorphous form27.
 |
Figure 5: DSC thermogram of HPβCD (A), Remdesivir (B), and RDV-HPβCD inclusion complex (C). |
Cellular Uptake study
A cellular uptake study was done to decide the capability of the RDV-HPβCD inclusion complexes, pure remdesivir, and control to penetrate into Vero cells. Vero cells were typical cells from African green monkey kidney 31. The green color seen under the fluorescein microscopy was from the fluorescein isothiocyanate (FITC) stain conjugated to each sample. The Observation was carried out qualitatively which only compared the visible intensity.
Figure 6 showed the intensity of Vero cells that were conjugated to FITC. Control cells contained Vero cells that were treated only with FITC (Figure 6A). cellular uptake of RDV-HPβCD inclusion complex conjugated to FITC in Vero cells showed better intensity than pure remdesivir conjugated to FITC. Hence, the formation of the RDV-HPβCD inclusion complex can enhance the cellular uptake of remdesivir12.
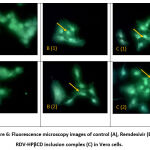 |
Figure 6: Fluorescence microscopy images of control (A), Remdesivir (B), and RDV-HPβCD inclusion complex (C) in Vero cells. |
Cytotoxicity analysis
The cell viability that was investigated by MTT assay was to show the influence of complexation with HPβCD in the cytotoxicity activity of remdesivir. Figure 7 shows the viability of HeLa the human cervical cancer cell line that was influenced by pure remdesivir and RDV-HPβCD inclusion complex. The cells were incubated with pure remdesivir and inclusion complex at a certain concentration (ranging between 0 and 14 μM) for 24 h. Reduction in cell viability of both samples (pure remdesivir and RDV-HPβCD inclusion complex) was observed in a dose-dependent manner. Pure remdesivir showed a reduction of cell viability starting from the lowest dose. Meanwhile, the RDV-HPβCD inclusion complex showed maximum cell viability at a concentration of 1.75 μM at 179.52% ± 6,31 but at higher concentrations, there was a reduction in cell viability. The significant improvement of HeLa cell viability was due to the solubilizing effect of the HPβCD inclusion complex, which validated its superiority as a carrier. When the concentration at 14 μM it showed the lowest cell viability (52,82% ± 2,69) of inclusion complex. But inclusion complex had a higher viability cell than pure remdesivir. Therefore, both pure remdesivir and inclusion complex were not toxic at HeLa cells32.
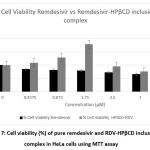 |
Figure 7: Cell viability (%) of pure remdesivir and RDV-HPβCD inclusion complex in HeLa cells using MTT assay. |
Conclusion
The RDV-HPβCD inclusion complex with molar ratio (1:1) had been successfully synthesized using the solvent evaporation method as it was confirmed by physical characterization using UV-Vis, XRD, DSC, and FTIR. Further, the complexation of RDV- HPβCD was proved to enhance remdesivir solubility and chemical stability as observed by a clear-state solution of inclusion complex and the presence of remdesivir peak at 246 nm in inclusion complex UV-Vis spectrum after 24 hours incubation, transformation of crystalline structure into an amorphous form that can be seen from XRD pattern, a broad DSC thermogram of inclusion complex at 72.93 °C and the intermolecular hydrogen bonds occurred between HPβCD and remdesivir from FTIR. In addition, the RDV-HPBCD inclusion complex improved the cellular uptake at the Vero cell and better Hela cell viability.
Author’s Contributions
The authors have carried out their obligations and responsibilities.
Conflict of Interests
There is no conflict of interest stated by the author.
Funding Sources
There is no funding sources.
References
- Liang C, Tian L, Liu Y, et al. A promising antiviral candidate drug for the COVID-19 pandemic: A mini-review of remdesivir. Eur J Med Chem. 2020;201:112527. doi:10.1016/j.ejmech.2020.112527
CrossRef - Eastman RT, Roth JS, Brimacombe KR, et al. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent Sci. 2020;6(5):672-683. doi:10.1021/acscentsci.0c00489
CrossRef - Saha A, Sharma AR, Bhattacharya M, Sharma G, Lee SS, Chakraborty C. Probable Molecular Mechanism of Remdesivir for the Treatment of COVID-19: Need to Know More. Arch Med Res. 2020;51(6):585-586. doi:10.1016/j.arcmed.2020.05.001
CrossRef - COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. Disponible en: https://covid19treatmentguidelines.nih.gov/. Natl Inst Heal. 2020;2019:130. https://www.covid19treatmentguidelines.nih.gov/.
- Sahakijpijarn S, Moon C, Koleng JJ, Christensen DJ, Williams RO. Development of remdesivir as a dry powder for inhalation by thin film freezing. Pharmaceutics. 2020;12(11):1-27. doi:10.3390/pharmaceutics12111002
CrossRef - Malin JJ, Suárez I, Priesner V, Fätkenheuer G. Remdesivir against COVID-19 and Other Viral Diseases. Am Soc Microbiol. 2021;34(1):1-21. doi:https://doi.org/10 .1128/CMR.00162-20
CrossRef - Sun D. Remdesivir for Treatment of COVID-19: Combination of Pulmonary and IV Administration May Offer Aditional Benefit. AAPS J. 2020;22(4). doi:10.1208/s12248-020-00459-8
CrossRef - Melo C de O, Rodrigues MS da S, da Silva MVS, et al. Preparation and characterization of spiro-acridine derivative and 2-hydroxypropyl-β-cyclodextrin inclusion complex. J Mol Struct. 2020;1222:128945. doi:10.1016/j.molstruc.2020.128945
CrossRef - Creteanu A, Pamfil D, Vasile C, et al. Study on the Role of the Inclusion Complexes with 2-Hydroxypropyl- β -cyclodextrin for Oral Administration of Amiodarone. Int J Polym Sci. 2019;2019. doi:10.1155/2019/1695189
CrossRef - YURTDAŞ KIRIMLIOĞLU G. Host-guest inclusion complex of desloratadine with 2-(Hydroxy)propyl-β-cyclodextrin (hp-β-cd): Preparation, binding behaviors and dissolution properties. J Res Pharm. 2020;24(5):693-707. doi:10.35333/jrp.2020.224
CrossRef - Saokham P, Muankaew C, Jansook P, Loftsson T. Solubility of cyclodextrins and drug/cyclodextrin complexes. Molecules. 2018;23(5):1-15. doi:10.3390/molecules23051161
CrossRef - Terauchi M, Tamura A, Yamaguchi S, Yui N. Enhanced cellular uptake and osteogenic differentiation efficiency of melatonin by inclusion complexation with 2-hydroxypropyl β-cyclodextrin. Int J Pharm. 2018;547(1-2):53-60. doi:10.1016/j.ijpharm.2018.05.063
CrossRef - Ghosh A, Biswas S, Ghosh T. Preparation and evaluation of silymarin β-cyclodextrin molecular inclusion complexes. J Young Pharm. 2011;3(3):205-210. doi:10.4103/0975-1483.83759
CrossRef - Chen J, Yao J, Ma Z, et al. Delivery of fluorescent-labeled cyclodextrin by liposomes: role of transferrin modification and phosphatidylcholine composition. J Liposome Res. 2017;27(1):21-31. doi:10.3109/08982104.2016.1140184
CrossRef - Ye W, Yao M, Dong Y, et al. Remdesivir (GS-5734) Impedes Enterovirus Replication Through Viral RNA Synthesis Inhibition. Front Microbiol. 2020;11(June):1-9. doi:10.3389/fmicb.2020.01105
CrossRef - Ren Z, Xu Y, Lu Z, et al. Construction of a water-soluble and photostable rubropunctatin/β-cyclodextrin drug carrier. RSC Adv. 2019;9(20):11396-11405. doi:10.1039/c9ra00379g
CrossRef - Rassu G, Fancello S, Roldo M, et al. Investigation of cytotoxicity and cell uptake of cationic beta-cyclodextrins as valid tools in nasal delivery. Pharmaceutics. 2020;12(7):1-13. doi:10.3390/pharmaceutics12070658
CrossRef - Patil JS, Kadam D V., Marapur SC, Kamalapur M V. Inclusion complex system; a novel technique to improve the solubility and bioavailability of poorly soluble drugs: A review. Int J Pharm Sci Rev Res. 2010;2(2):29-34.
- Chouker MA, Abdallah H, Zeiz A, El-Dakdouki MH. Host-quest inclusion complex of quinoxaline-1,4-dioxide derivative with 2-hydroxypropyl-β-cyclodextrin: Preparation, characterization, and antibacterial activity. J Mol Struct. 2021;1235:130273. doi:10.1016/j.molstruc.2021.130273
CrossRef - Vukic MD, Vukovic NL, Popovic SL, et al. Effect of β-cyclodextrin encapsulation on cytotoxic activity of acetylshikonin against HCT-116 and MDA-MB-231 cancer cell lines. Saudi Pharm J. 2020;28(1):136-146. doi:10.1016/j.jsps.2019.11.015
CrossRef - Qiao X, Yang L, Hu X, et al. Characterization and evaluation of inclusion complexes between astaxanthin esters with different molecular structures and hydroxypropyl-β-cyclodextrin. Food Hydrocoll. 2021;110(5):106208. doi:10.1016/j.foodhyd.2020.106208
CrossRef - Qudsiani K, Sutriyo, Rahmasari R. Polyamidoamine-Remdesivir Conjugate: Physical Stability and Cellular Uptake Enhancement. Biomed Pharmacol J. 2021;14(4):2073-2083. doi:10.13005/bpj/2304
CrossRef - Tambe A, Pandita N, Kharkar P, Sahu N. Encapsulation of boswellic acid with β- and hydroxypropyl-β-cyclodextrin: Synthesis, characterization, in vitro drug release and molecular modelling studies. J Mol Struct. 2018;1154:504-510. doi:10.1016/j.molstruc.2017.10.061
CrossRef - Sadaquat H, Akhtar M. Comparative effects of β-cyclodextrin, HP-β-cyclodextrin and SBE7-β-cyclodextrin on the solubility and dissolution of docetaxel via inclusion complexation. J Incl Phenom Macrocycl Chem. 2020;96(3-4):333-351. doi:10.1007/s10847-020-00977-0
CrossRef - Menezes PP, Serafini MR, Quintans-Júnior LJ, et al. Inclusion complex of (-)-linalool and β-cyclodextrin. J Therm Anal Calorim. 2014;115(3):2429-2437. doi:10.1007/s10973-013-3367-x
CrossRef - Yuan C, Liu B, Liu H. Characterization of hydroxypropyl-β-cyclodextrins with different substitution patterns via FTIR, GC-MS, and TG-DTA. Carbohydr Polym. 2015;118:36-40. doi:10.1016/j.carbpol.2014.10.070
CrossRef - Celebioglu A, Uyar T. Metronidazole/Hydroxypropyl-β-Cyclodextrin inclusion complex nanofibrous webs as fast-dissolving oral drug delivery system. Int J Pharm. 2019;572(August):118828. doi:10.1016/j.ijpharm.2019.118828
CrossRef - Qiao X, Yang L, Hu X, et al. Food Hydrocolloids Characterization and evaluation of inclusion complexes between astaxanthin esters with different molecular structures and hydroxypropyl- β -cyclodextrin. Food Hydrocoll. 2021;110(5):106208. doi:10.1016/j.foodhyd.2020.106208
CrossRef - Kasapoglu-Calik M, Ozdemir M. Synthesis and controlled release of curcumin-β-cyclodextrin inclusion complex from nanocomposite poly(N-isopropylacrylamide/sodium alginate) hydrogels. J Appl Polym Sci. 2019;136(21):1-11. doi:10.1002/app.47554
CrossRef - Ghosh R, Roy N, Saha S, et al. Synthesis and characterization of an industrially significant ionic liquid and its inclusion complex with β -cyclodextrin and its soluble derivative for their advanced applications. Chem Phys Lett. 2021;769(February):138401. doi:10.1016/j.cplett.2021.138401
CrossRef - Chu H, Chan JF-W, Yuen TT-T, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. The Lancet Microbe. 2020;1(1):e14-e23. doi:10.1016/s2666-5247(20)30004-5
CrossRef - Kulkarni AD, Belgamwar VS. Inclusion complex of chrysin with sulfobutyl ether-β-cyclodextrin (Captisol®): Preparation, characterization, molecular modelling and in vitro anticancer activity. J Mol Struct. 2017;1128:563-571. doi:10.1016/j.molstruc.2016.09.025
CrossRef